The MACROD2 rs6110695 A>G Polymorphism and the Metabolites Indoleacrylic Acid and Butyrylcarnitine Potentially Have Clinical Relevance to WBC Count Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Blood Sample Collection
2.3. Anthropometric and Clinical/Biochemical Parameters
2.4. MACROD2 rs6110695 A>G Polymorphism
2.5. Metabolic Profiling
2.5.1. UHPLC–MS/MS Analysis
2.5.2. Identification of Metabolites
2.6. Statistical Analysis
3. Results
3.1. Anthropometric and Clinical/Biochemical Characteristics
3.2. Total Blood Cell Count and Inflammatory Markers
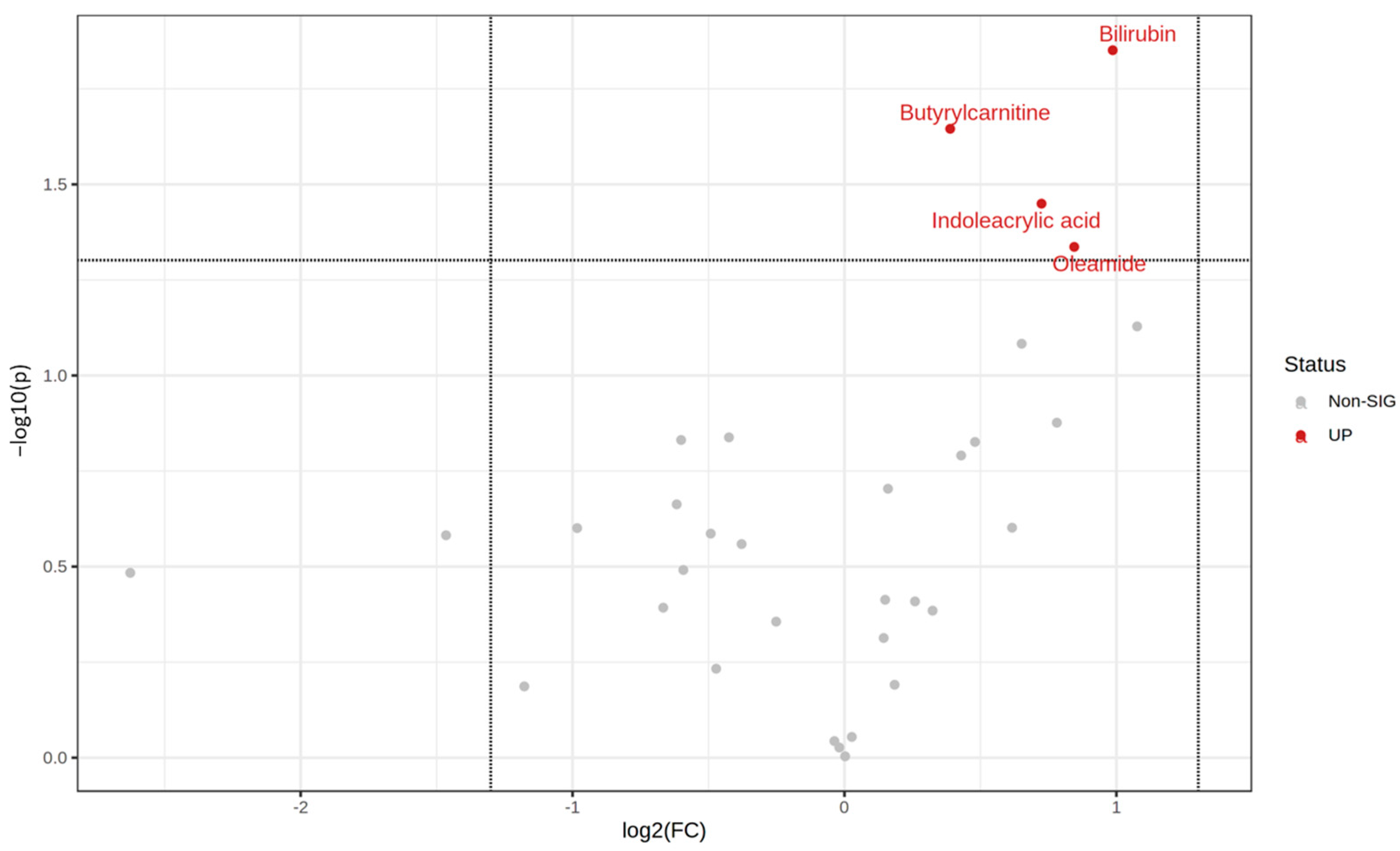
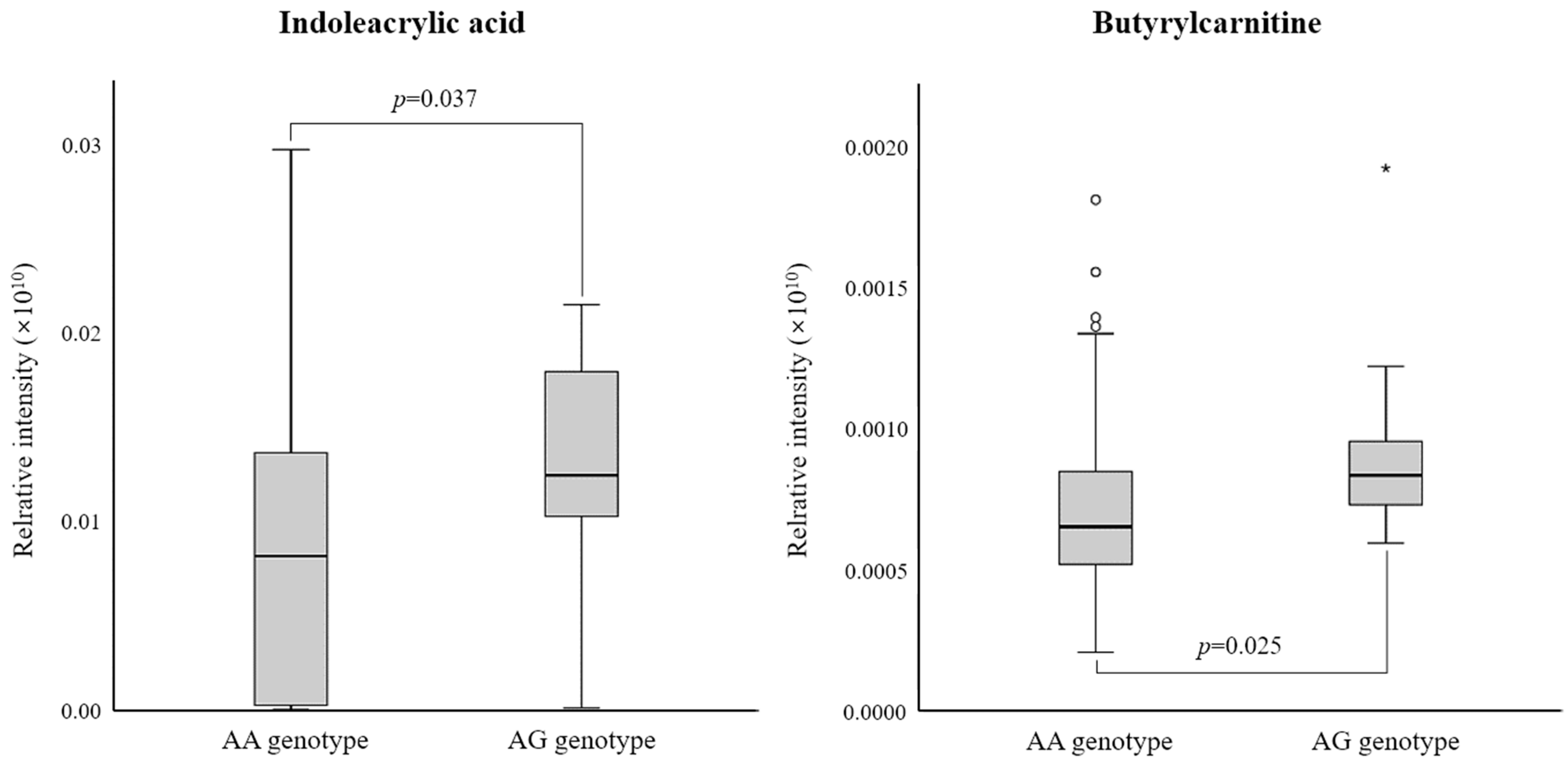
3.3. Differential Metabolites between the Two rs6110695 Genotype Groups
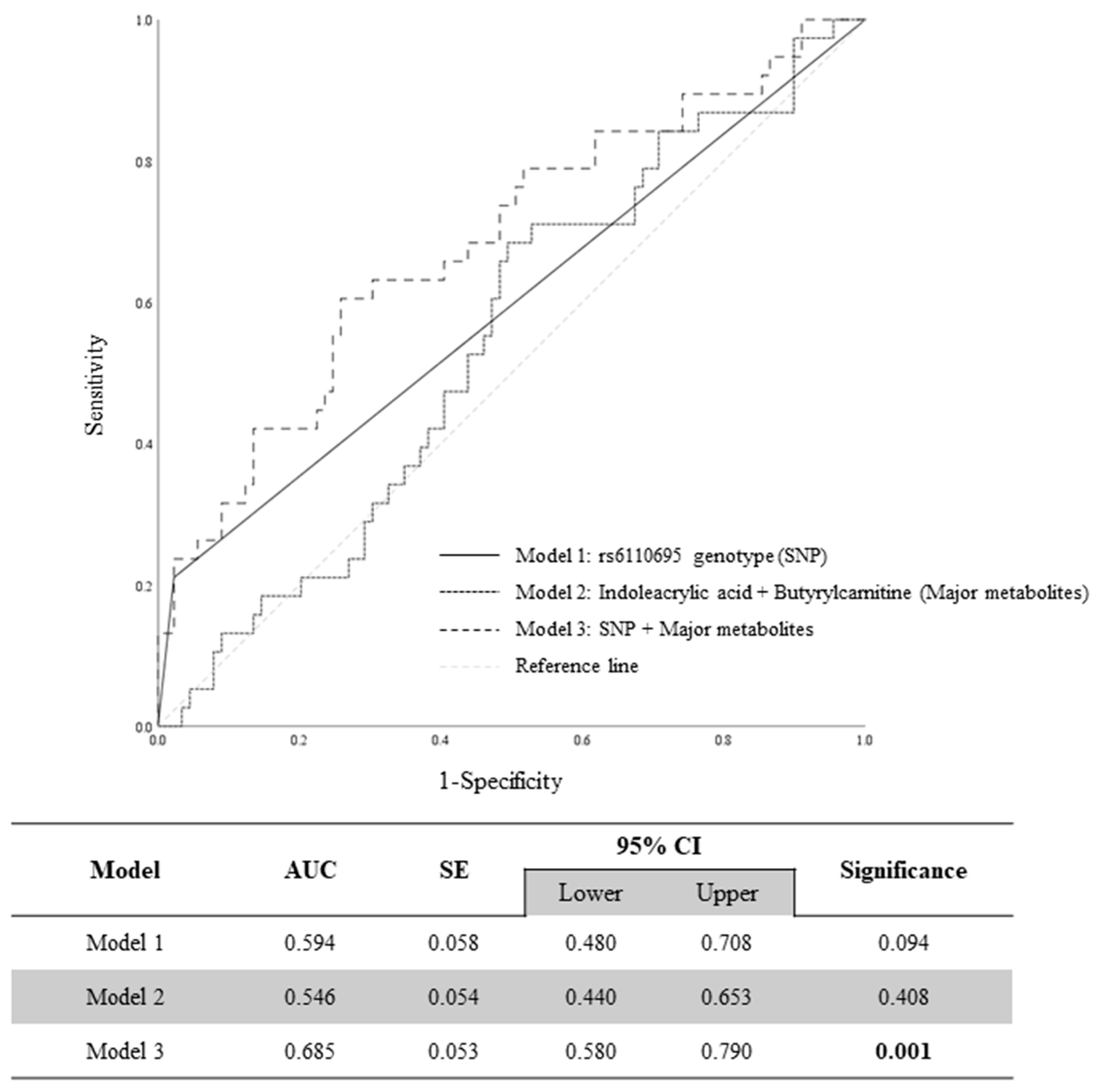
3.4. Evaluation of Prediction Models of WBC Count via ROC Curve Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kawabe, A.; Yasu, T.; Morimoto, T.; Tokushige, A.; Momomura, S.; Sakakura, K.; Node, K.; Inoue, T.; Ueda, S.; CHD Collaborative Investigators. WBC count predicts heart failure in diabetes and coronary artery disease patients: A retrospective cohort study. ESC Heart Fail. 2021, 8, 3748–3759. [Google Scholar] [CrossRef]
- Ishida, S.; Kondo, S.; Funakoshi, S.; Satoh, A.; Maeda, T.; Kawazoe, M.; Yoshimura, C.; Tada, K.; Takahashi, K.; Ito, K.; et al. White blood cell count and incidence of hypertension in the general Japanese population: ISSA-CKD study. PLoS ONE 2021, 16, e0246304. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Lee, H.S.; Park, J.Y.; Jung, D.H.; Lee, J.W. White blood cell count as a predictor of incident type 2 diabetes mellitus among non-obese adults: A longitudinal 10-year analysis of the Korean Genome and Epidemiology Study. J. Inflamm. Res. 2021, 14, 1235–1242. [Google Scholar] [CrossRef]
- Kashima, S.; Inoue, K.; Matsumoto, M.; Akimoto, K. White blood cell count and c-reactive protein independently predicted incident diabetes: Yuport Medical Checkup Center Study. Endocr. Res. 2019, 44, 127–137. [Google Scholar] [CrossRef]
- Korea Disease Control and Prevention Agency. Chronic Disease Management Research & Development. 2021. Available online: https://www.kdca.go.kr/contents.es?mid=a30322000000 (accessed on 7 November 2022).
- Chae, J.; Kim, M.; Lee, J.H.; Yoo, H.J. Body fat composition enhances the predictive ability of changes in white blood cell levels associated with the risk of chronic disease development. J. Immunol. 2021, 207, 389–397. [Google Scholar] [CrossRef]
- Shin, G.; Jang, K.; Kim, M.; Lee, J.H.; Yoo, H.J. Inflammatory markers and plasma fatty acids in predicting WBC level alterations in association with glucose-related markers: A cross-sectional study. Front. Immunol. 2020, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Han, Y.; Lee, J.H.; Yoo, H.J. Association of the MACROD2 rs6110695 A>G polymorphism with an increasing WBC count in a Korean population. Immun. Inflamm. Dis. 2022, 10, e669. [Google Scholar] [CrossRef]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.; Lopata, A.L.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics toward personalized medicine. Mass Spectrom. Rev. 2019, 38, 221–238. [Google Scholar] [CrossRef]
- dbSNP. rs6110695. 2024. Available online: https://www.ncbi.nlm.nih.gov/snp/rs6110695 (accessed on 23 January 2024).
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell. 2015, 58, 935–946. [Google Scholar] [CrossRef]
- Žaja, R.; Aydin, G.; Lippok, B.E.; Feederle, R.; Lüscher, B.; Feijs, K. Comparative analysis of MACROD1, MACROD2 and TARG1 expression, localisation and interactome. Sci. Rep. 2020, 10, 8286. [Google Scholar] [CrossRef] [PubMed]
- Re, O.L.; Mazza, T.; Vinciguerra, M. Mono-ADP-Ribosylhydrolase MACROD2 is dispensable for murine responses to metabolic and genotoxic insults. Front. Genet. 2018, 9, 654. [Google Scholar]
- Chang, Y.C.; Hee, S.; Lee, W.; Li, H.; Chang, T.; Lin, M.; Hung, Y.; Lee, I.; Hung, K.; Assimes, T.; et al. Genome-wide scan for circulating vascular adhesion protein-1 levels: MACROD2 as a potential transcriptional regulator of adipogenesis. J. Diabetes Investig. 2018, 9, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.; Jin, H.S. MACROD2 polymorphisms are associated with hypertension in Korean Population. Korean J. Clin. Lab. Sci. 2019, 51, 57–63. [Google Scholar] [CrossRef]
- The Human Protein Atlas. MACROD2-RNA (Immune). 2021. Available online: https://www.proteinatlas.org/ENSG00000172264-MACROD2/blood (accessed on 15 September 2021).
- Anderson, G.M. The quantitative determination of indolic microbial tryptophan metabolites in human and rodent samples: A systematic review. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1186, 123008. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; D’hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef]
- Modoux, M.; Rolhion, N.; Mani, S.; Sokol, H. Tryptophan metabolism as a pharmacological target. Trends Pharmacol. Sci. 2021, 42, 60–73. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Dong, F.; Hao, F.; Murray, I.A.; Smith, P.B.; Koo, I.; Tindall, A.M.; Kris-Etherton, P.M.; Gowda, K.; Amin, S.G.; Patterson, A.D.; et al. Intestinal microbiota-derived tryptophan metabolites are predictive of Ah receptor activity. Gut Microbes 2020, 12, 1–24. [Google Scholar] [CrossRef]
- Wei, G.Z.; Martin, K.A.; Xing, P.Y.; Agrawal, R.; Whiley, L.; Wood, T.K.; Hejndorf, S.; Ng, Y.Z.; Low, J.Z.Y.; Rossant, J.; et al. Tryptophan-metabolizing gut microbes regulate adult neurogenesis via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2021, 118, e2021091118. [Google Scholar] [CrossRef]
- Dalmas, E.; Venteclef, N.; Caer, C.; Poitou, C.; Cremer, I.; Aron-Wisnewsky, J.; Lacroix-Desmazes, S.; Bayry, J.; Kaveri, S.V.; Clément, K.; et al. T cell-derived IL-22 amplifies IL-1β-driven inflammation in human adipose tissue: Relevance to obesity and type 2 diabetes. Diabetes 2014, 63, 1966–1977. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Zeng, F.; Liu, X.; Wang, Q.J.; Deng, F. Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 2016, 7, e2226. [Google Scholar] [CrossRef] [PubMed]
- Mai, M.; Tönjes, A.; Kovacs, P.; Stumvoll, M.; Fiedler, G.M.; Leichtle, A.B. Serum levels of acylcarnitines are altered in prediabetic conditions. PLoS ONE 2013, 8, e82459. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Zhao, S.; Feng, X.-F.; Huang, T.; Luo, H.-H.; Chen, J.-X.; Zeng, J.; Gu, M.; Li, J.; Sun, X.-Y.; Sun, D.; et al. The association between acylcarnitine metabolites and cardiovascular disease in Chinese patients with type 2 diabetes mellitus. Front. Endocrinol. 2020, 11, 212. [Google Scholar] [CrossRef]
- Locasale, J.W.; Melman, T.; Song, S.; Yang, X.; Swanson, K.D.; Cantley, L.C.; Wong, E.T.; Asara, J.M. Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Mol. Cell. Proteom. 2012, 11, M111.014688. [Google Scholar] [CrossRef]
- Elguindy, M.; Young, J.S.; Mondal, I.; Lu, R.O.; Ho, W.S. Glioma-immune cell crosstalk in tumor progression. Cancers 2024, 16, 308. [Google Scholar] [CrossRef]
- Wu, H.; Tang, D.; Yun, M.; Liu, H.; Huang, S.; Yun, C.; Hocher, B.; Zhang, X.; Liu, F.; Yin, L.; et al. Metabolic dysfunctions of intestinal fatty acids and tryptophan reveal immuno-inflammatory response activation in IgA nephropathy. Front. Med. 2022, 9, 811526. [Google Scholar] [CrossRef]
- Bian, J.; Sun, J.; Chang, H.; Wei, Y.; Cong, H.; Yao, M.; Xiao, F.; Wang, H.; Zhao, Y.; Liu, J.; et al. Profile and potential role of novel metabolite biomarkers, especially indoleacrylic acid, in pathogenesis of neuromyelitis optica spectrum disorders. Front. Pharmacol. 2023, 14, 1166085. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; Dowdell, A.S.; Henen, M.A.; Lanis, J.M.; Lee, J.S.; Cartwright, I.M.; Schaefer, R.E.M.; Ornelas, A.; Onyiah, J.C.; Vögeli, B.; et al. Microbial-derived indoles inhibit neutrophil myeloperoxidase to diminish bystander tissue damage. FASEB J. 2021, 35, e21552. [Google Scholar] [CrossRef] [PubMed]
- Tomalka, J.; Sharma, A.; Smith, A.G.; Avaliani, T.; Gujabidze, M.; Bakuradze, T.; Sabanadze, S.; Jones, D.P.; Avaliani, Z.; Kipiani, M.; et al. Combined cerebrospinal fluid metabolomic and cytokine profiling in tuberculosis meningitis reveals robust and prolonged changes in immunometabolic networks. Tuberculosis 2024, 144, 102462. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, J.; Wu, T.-T.; Hou, X.-G.; Yang, Y.; Ma, X.; Ma, Y.-T.; Zheng, Y.-Y.; Xie, X. Baseline white blood cell count-to-apolipoprotein A1 ratio as a novel predictor of long-term adverse outcomes in patients who underwent percutaneous coronary intervention: A retrospective cohort study. Lipids Health Dis. 2020, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- van der Vorst, E.P.C. High-density lipoproteins and apolipoprotein A1. Subcell. Biochem. 2020, 94, 399–420. [Google Scholar] [PubMed]
- Kardassis, D.; Mosialou, I.; Kanaki, M.; Tiniakou, I.; Thymiakou, E. Metabolism of HDL and its regulation. Curr. Med. Chem. 2014, 21, 2864–2880. [Google Scholar] [CrossRef] [PubMed]
| Total (n = 139) | p | ||||
|---|---|---|---|---|---|
| AA Genotype (n = 129) | AG Genotype (n = 10) | ||||
| Age (year) § | 56.3 | ±0.59 | 55.4 | ±1.48 | 0.584 |
| Male/Female n, (%) | 16 (12.4)/113 (87.6) | 2 (20.0)/8 (80.0) | 0.618 | ||
| Weight (kg) † | 60.6 | ±0.74 | 63.6 | ±1.43 | 0.051 |
| BMI (kg/m2) | 24.1 | ±0.23 | 24.6 | ±0.61 | 0.515 |
| Waist (cm) | 86.6 | ±0.61 | 87.6 | ±1.75 | 0.662 |
| Waist-to-hip ratio | 0.90 | ±0.00 | 0.90 | ±0.01 | 0.827 |
| Systolic BP (mmHg) † | 121.6 | ±1.17 | 122.8 | ±3.72 | 0.763 |
| Diastolic BP (mmHg) † | 76.5 | ±0.83 | 76.7 | ±2.33 | 0.898 |
| Triglyceride (mg/dL) † | 141.2 | ±7.37 | 159.7 | ±39.6 | 0.723 |
| Total cholesterol (mg/dL) | 213.5 | ±2.82 | 216.8 | ±13.7 | 0.760 |
| HDL-cholesterol (mg/dL) † | 55.3 | ±1.38 | 46.9 | ±5.26 | 0.053 |
| LDL-cholesterol (mg/dL) | 130.2 | ±3.08 | 132.6 | ±13.9 | 0.845 |
| Apo A-I (mg/dL) † | 153.3 | ±2.35 | 140.8 | ±4.64 | 0.115 |
| Apo B (mg/dL) † | 109.8 | ±2.41 | 106.3 | ±9.55 | 0.553 |
| WBC-to-apo A-I ratio † | 0.03 | ±0.00 | 0.05 | ±0.01 | <0.001 |
| Glucose (mg/dL) | 92.6 | ±0.91 | 89.3 | ±3.29 | 0.334 |
| Insulin (μIU/mL) § | 8.73 | ±0.48 | 6.94 | ±0.84 | 0.234 |
| HOMA-IR † | 2.01 | ±0.12 | 1.49 | ±0.16 | 0.214 |
| Adiponectin (ng/mL) § | 8.04 | ±0.42 | 6.53 | ±0.90 | 0.344 |
| Total (n = 139) | p | ||||
|---|---|---|---|---|---|
| AA Genotype (n = 129) | AG Genotype (n = 10) | ||||
| Total blood cell count | |||||
| WBC (×103/μL) † | 4.89 | ±0.10 | 6.89 | ±0.50 | <0.001 |
| Lymphocyte count (×103/μL) † | 1.73 | ±0.04 | 2.10 | ±0.16 | 0.018 |
| Monocyte count (×103/μL) § | 0.34 | ±0.02 | 0.59 | ±0.10 | 0.005 |
| Granulocyte count (×103/μL) † | 2.82 | ±0.08 | 4.21 | ±0.41 | <0.001 |
| Lymphocyte (%) | 36.9 | ±0.67 | 31.7 | ±2.16 | 0.039 |
| Monocyte (%) § | 7.99 | ±0.37 | 9.16 | ±1.14 | 0.229 |
| Granulocyte (%) | 55.1 | ±0.88 | 59.1 | ±2.73 | 0.220 |
| Platelet (×103/μL) § | 237.9 | ±5.99 | 242.4 | ±9.98 | 0.427 |
| MLR § | 0.19 | ±0.01 | 0.27 | ±0.03 | 0.028 |
| GLR † | 1.74 | ±0.06 | 2.10 | ±0.27 | 0.116 |
| PLR † | 143.3 | ±3.58 | 120.0 | ±8.43 | 0.068 |
| MPR † | 0.0015 | ±0.0001 | 0.0025 | ±0.0004 | 0.006 |
| Inflammatory markers | |||||
| hs-CRP (mg/L) § | 1.36 | ±0.37 | 0.98 | ±0.26 | 0.764 |
| IL-1β (pg/mL) § | 0.54 | ±0.16 | 0.35 | ±0.12 | 0.593 |
| IL-2 (pg/mL) § | 47.5 | ±2.81 | 63.1 | ±16.9 | 0.286 |
| IL-6 (pg/mL) § | 3.48 | ±0.36 | 3.10 | ±0.81 | 0.696 |
| IL-12 (pg/mL) § | 16.0 | ±4.66 | 6.43 | ±0.65 | 0.529 |
| TNF-α (pg/mL) § | 7.27 | ±2.90 | 4.40 | ±1.77 | 0.332 |
| IFN-γ (pg/mL) § | 31.6 | ±26.0 | 4.08 | ±0.95 | 0.973 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Kim, M.; Yoo, H.J. The MACROD2 rs6110695 A>G Polymorphism and the Metabolites Indoleacrylic Acid and Butyrylcarnitine Potentially Have Clinical Relevance to WBC Count Prediction. J. Pers. Med. 2024, 14, 889. https://doi.org/10.3390/jpm14080889
Han Y, Kim M, Yoo HJ. The MACROD2 rs6110695 A>G Polymorphism and the Metabolites Indoleacrylic Acid and Butyrylcarnitine Potentially Have Clinical Relevance to WBC Count Prediction. Journal of Personalized Medicine. 2024; 14(8):889. https://doi.org/10.3390/jpm14080889
Chicago/Turabian StyleHan, Youngmin, Minjoo Kim, and Hye Jin Yoo. 2024. "The MACROD2 rs6110695 A>G Polymorphism and the Metabolites Indoleacrylic Acid and Butyrylcarnitine Potentially Have Clinical Relevance to WBC Count Prediction" Journal of Personalized Medicine 14, no. 8: 889. https://doi.org/10.3390/jpm14080889





