The Role of the Gut–Biliary–Liver Axis in Primary Hepatobiliary Liver Cancers: From Molecular Insights to Clinical Applications
Abstract
:1. Background
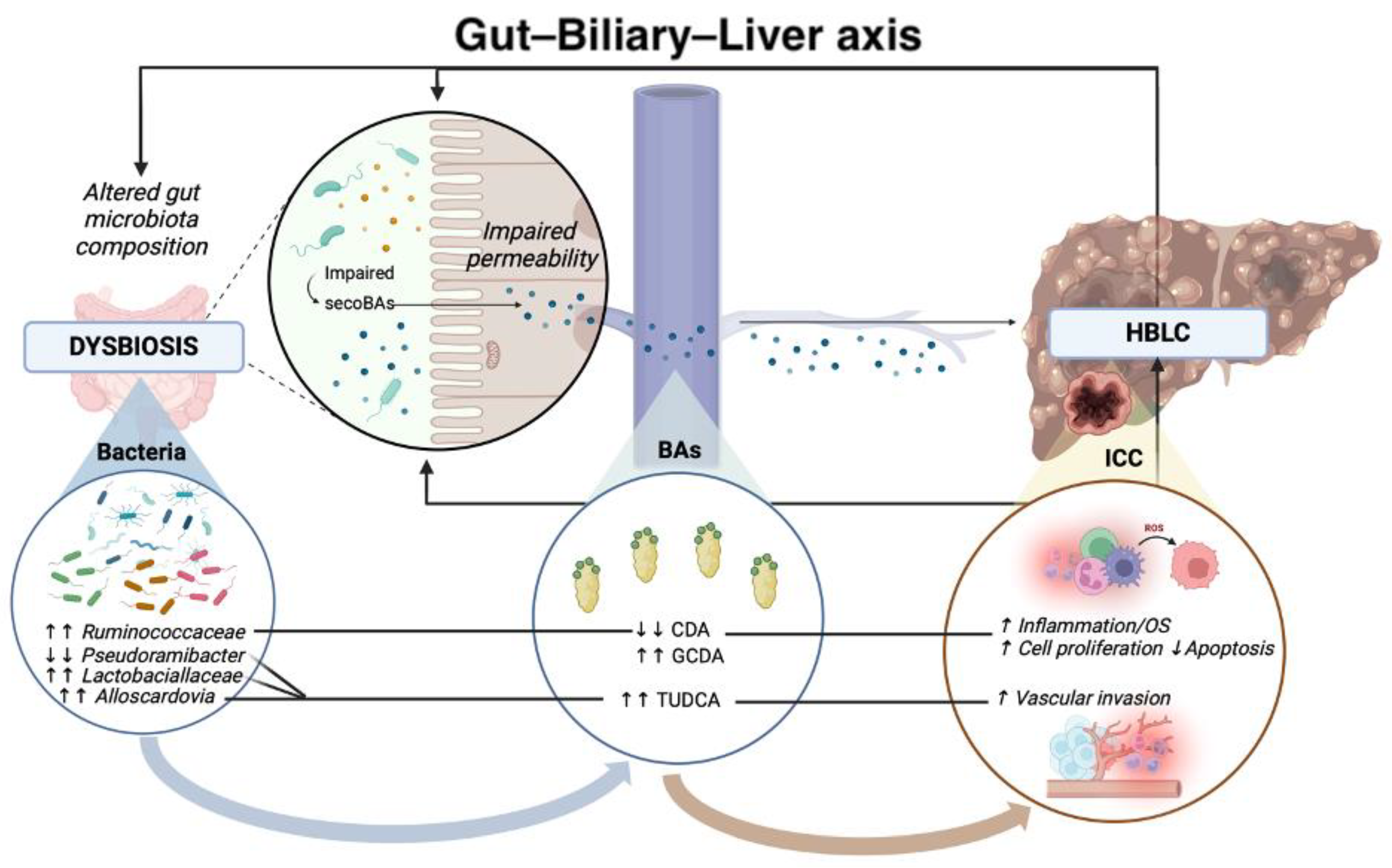
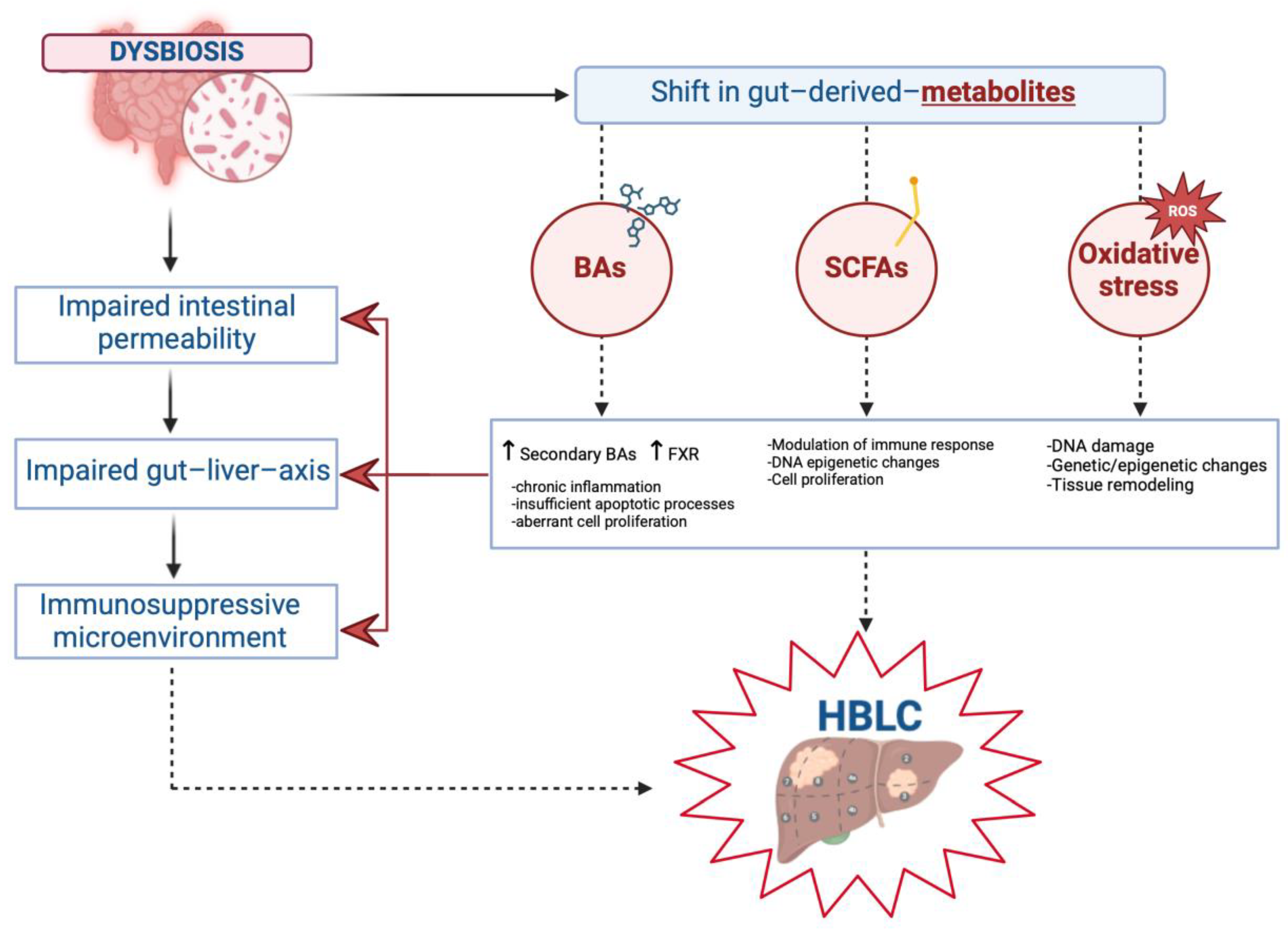
2. Gut–Biliary–Liver Axis in the Pathogenesis of HBLCs
2.1. Impaired Intestinal and Biliary Permeability in the Pathogenesis of Hepatobiliary Liver Cancers
2.2. Principal Alterations in Gut Microbiota Composition in Hepatobiliary Liver Cancers
2.2.1. Altered Gut Microbiota Composition and Hepatocellular Carcinoma
2.2.2. Altered Gut Microbiota Composition and Cholangiocarcinoma
2.3. Gut Microbial Metabolites in the Pathogenesis of Primary Hepatobiliary Liver Cancers
2.3.1. Bile Acids (BAs) in Hepatobiliary Carcinogenesis
2.3.2. Short-Chain Fatty Acids (SCFAs) in Hepatobiliary Carcinogenesis
3. Gut–Biliary–Liver Axis-Related Applications in Managing HBLC
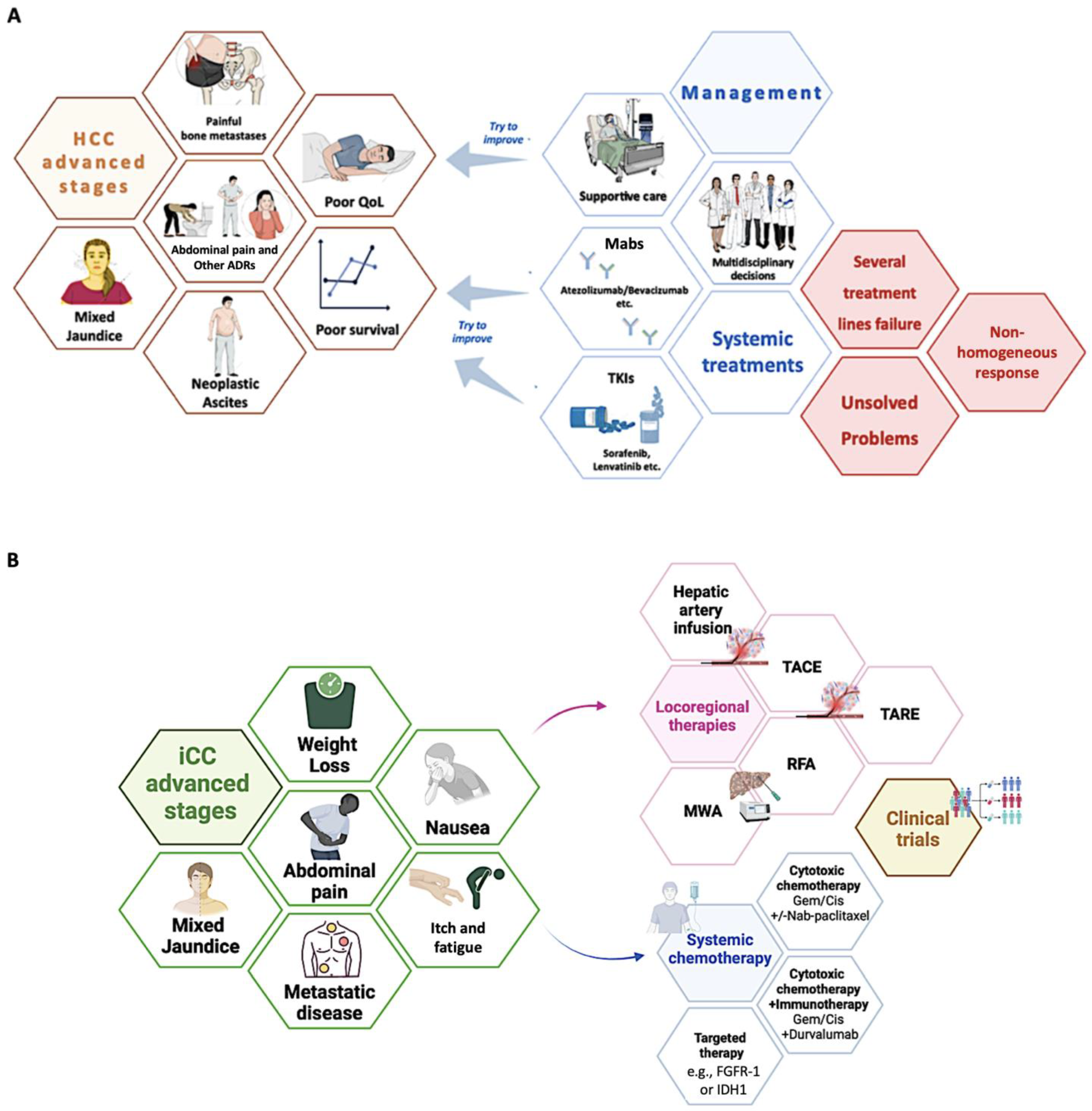
3.1. Current Treatment Strategies: An Overview of Therapeutic Chances for Advanced HBLC Stages
3.1.1. Hepatocellular Carcinoma
3.1.2. Cholangiocarcinoma
3.2. Microbiome–Gut–Biliary–Liver Axis-Related Potential Clinical Applications
3.2.1. Gut Microbiota in Optimizing Early Diagnostic Processes in HBLC
3.2.2. Gut Microbiota as a Novel Tool in Predicting Treatment Response in Advanced HBLC
3.2.3. Future Perspectives: Modulating Gut Microbiota as a Novel Therapeutic Strategy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACLD | advanced chronic liver disease |
| AF | alpha-fetoprotein |
| ASV | amplicon sequence variants |
| BAs | bile acids |
| CC | cholangiocarcinoma |
| CLD | chronic liver disease |
| CMRFs | cardiometabolic risk factors |
| CRC | colorectal cancer |
| DAMPS | damage-associated molecular patterns |
| dCC | distal CC |
| EGF | epidermal growth factor |
| FGFR | fibroblast growth factor receptor |
| FMT | fecal microbiota transplant |
| FXR | farnesoid X receptor |
| GSH | glutathione |
| GVB | gut–vascular barrier |
| HBLC | hepatobiliary liver cancer |
| HCC | hepatocellular carcinoma |
| HDAC | disrupting histone deacetylase |
| HMGB1 | high-mobility group box 1 |
| HNE | 4-hydroxy-trans-2-nonenal |
| ICC | intrahepatic cholangiocarcinoma |
| IDH1 or 2 | isocitrate dehydrogenase 1 or 2 |
| ILC 3 | group 3 innate lymphoid cells |
| IR | insulin resistance |
| JAMs | junctional adhesion molecules |
| LC | liver cirrhosis |
| LPS | lipopolysaccharide |
| Mab | monoclonal antibodies |
| ML | machine learning |
| mMDSC | monocytic myeloid-derived suppressor cells |
| MASLD | Metabolic Dysfunction-Associated Steatotic Liver Disease |
| NAFLD | non-alcoholic fatty liver disease |
| NKT | natural killer T |
| NSCLC | non-small cell lung cancer |
| PAMPs | pathogen-associated molecular patterns |
| PC | pancreatic cancer |
| pCC | perihilar cholangiocarcinoma |
| PDL-1 | programmed cell death ligand 1 |
| PFS | progression-free survival |
| PKC | protein kinase C |
| PKA | protein kinase A |
| PMN-MDSCs | polymorphonuclear myeloid-derived suppressor cells |
| PSC | primary sclerosing cholangitis |
| QoL | quality of life |
| ROM | reactive oxygen metabolite |
| ROS | reactive oxygen species |
| rRNA | ribosomal RNA |
| SCFAs | short-chain fatty acids |
| SOD | superoxide dismutase |
| STING | stimulator of interferon genes |
| TKI | tyrosine kinase inhibitors |
| TJs | tight junctions |
| TLR | toll-like receptor |
| TUDCA | tauroursodeoxycholic acid |
| VEGF-R | vascular endothelial growth factor receptor |
| VI | vascular invasion |
| ZO | zonula occludens |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rich, N.E. Changing Epidemiology of Hepatocellular Carcinoma Within the United States and Worldwide. Surg. Oncol. Clin. N. Am. 2024, 33, 1–12. [Google Scholar] [CrossRef]
- Pascale, A.; Rosmorduc, O.; Duclos-Vallée, J.-C. New Epidemiologic Trends in Cholangiocarcinoma. Clin. Res. Hepatol. Gastroenterol. 2023, 47, 102223. [Google Scholar] [CrossRef] [PubMed]
- Dallio, M.; Sangineto, M.; Romeo, M.; Cipullo, M.; Coppola, A.; Mammone, S.; Di Gioia, G.; Masarone, M.; Persico, M.; Serviddio, G.; et al. The Influence of Acute Lifestyle Changes on NAFLD Evolution in a Multicentre Cohort: A Matter of Body Composition. Nutr. Diabetes 2024, 14, 33. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL-ILCA Clinical Practice Guidelines on the Management of Intrahepatic Cholangiocarcinoma. J. Hepatol. 2023, 79, 181–208. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Dallio, M.; Masarone, M.; Romeo, M.; Tuccillo, C.; Morisco, F.; Persico, M.; Loguercio, C.; Federico, A. PNPLA3, TM6SF2, and MBOAT7 Influence on Nutraceutical Therapy Response for Non-Alcoholic Fatty Liver Disease: A Randomized Controlled Trial. Front. Med. 2021, 8, 734847. [Google Scholar] [CrossRef]
- Dallio, M.; Ventriglia, L.; Romeo, M.; Scognamiglio, F.; Diano, N.; Moggio, M.; Cipullo, M.; Coppola, A.; Ziogas, A.; Netea, M.G.; et al. Environmental Bisphenol A Exposure Triggers Trained Immunity-Related Pathways in Monocytes. Front. Immunol. 2023, 14, 1270391. [Google Scholar] [CrossRef]
- Romeo, M.; Dallio, M.; Scognamiglio, F.; Ventriglia, L.; Cipullo, M.; Coppola, A.; Tammaro, C.; Scafuro, G.; Iodice, P.; Federico, A. Role of Non-Coding RNAs in Hepatocellular Carcinoma Progression: From Classic to Novel Clinicopathogenetic Implications. Cancers 2023, 15, 5178. [Google Scholar] [CrossRef]
- Pallozzi, M.; De Gaetano, V.; Di Tommaso, N.; Cerrito, L.; Santopaolo, F.; Stella, L.; Gasbarrini, A.; Ponziani, F.R. Role of Gut Microbial Metabolites in the Pathogenesis of Primary Liver Cancers. Nutrients 2024, 16, 2372. [Google Scholar] [CrossRef]
- Brown, Z.J.; Tsilimigras, D.I.; Ruff, S.M.; Mohseni, A.; Kamel, I.R.; Cloyd, J.M.; Pawlik, T.M. Management of Hepatocellular Carcinoma: A Review. JAMA Surg. 2023, 158, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 4317–4345. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Tang, R.; Li, B.; Ma, X.; Schnabl, B.; Tilg, H. Gut Microbiome, Liver Immunology, and Liver Diseases. Cell Mol. Immunol. 2021, 18, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Allam-Ndoul, B.; Castonguay-Paradis, S.; Veilleux, A. Gut Microbiota and Intestinal Trans-Epithelial Permeability. Int. J. Mol. Sci. 2020, 21, 6402. [Google Scholar] [CrossRef]
- Horowitz, A.; Chanez-Paredes, S.D.; Haest, X.; Turner, J.R. Paracellular Permeability and Tight Junction Regulation in Gut Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 417–432. [Google Scholar] [CrossRef]
- Camilleri, M. Leaky Gut: Mechanisms, Measurement and Clinical Implications in Humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Wiest, R.; Garcia-Tsao, G. Bacterial Translocation (BT) in Cirrhosis. Hepatology 2005, 41, 422–433. [Google Scholar] [CrossRef]
- Miura, K.; Ishioka, M.; Minami, S.; Horie, Y.; Ohshima, S.; Goto, T.; Ohnishi, H. Toll-like Receptor 4 on Macrophage Promotes the Development of Steatohepatitis-Related Hepatocellular Carcinoma in Mice. J. Biol. Chem. 2016, 291, 11504–11517. [Google Scholar] [CrossRef]
- Mohamed, F.E.-Z.A.; Hammad, S.; Luong, T.V.; Dewidar, B.; Al-Jehani, R.; Davies, N.; Dooley, S.; Jalan, R. Expression of TLR-2 in Hepatocellular Carcinoma Is Associated with Tumour Proliferation, Angiogenesis and Caspase-3 Expression. Pathol. Res. Pract. 2020, 216, 152980. [Google Scholar] [CrossRef]
- Cowden, J.M.; Yu, F.; Challapalli, M.; Huang, J.-F.; Kim, S.; Fung-Leung, W.-P.; Ma, J.Y.; Riley, J.P.; Zhang, M.; Dunford, P.J.; et al. Antagonism of the Histamine H4 Receptor Reduces LPS-Induced TNF Production In Vivo. Inflamm. Res. 2013, 62, 599–607. [Google Scholar] [CrossRef]
- Zhe, Y.; Li, Y.; Liu, D.; Su, D.-M.; Liu, J.-G.; Li, H.-Y. Extracellular HSP70-Peptide Complexes Promote the Proliferation of Hepatocellular Carcinoma Cells via TLR2/4/JNK1/2MAPK Pathway. Tumour Biol. 2016, 37, 13951–13959. [Google Scholar] [CrossRef]
- Wang, X.; Fang, Y.; Liang, W.; Cai, Y.; Wong, C.C.; Wang, J.; Wang, N.; Lau, H.C.-H.; Jiao, Y.; Zhou, X.; et al. Gut-Liver Translocation of Pathogen Klebsiella Pneumoniae Promotes Hepatocellular Carcinoma in Mice. Nat. Microbiol. 2025, 10, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Baquer, A.; Labiano, I.; Sharif, O.; Agirre-Lizaso, A.; Oakley, F.; Rodrigues, P.M.; Zhuravleva, E.; O’Rourke, C.J.; Hijona, E.; Jimenez-Agüero, R.; et al. TREM-2 Defends the Liver against Hepatocellular Carcinoma through Multifactorial Protective Mechanisms. Gut 2021, 70, 1345–1361. [Google Scholar] [CrossRef]
- Merlen, G.; Tordjmann, T. Tight Junction Proteins and Biliary Diseases. Curr. Opin. Gastroenterol. 2024, 40, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Sheth, P.; Elias, B.C.; Rao, R. Protein Phosphatases 2A and 1 Interact with Occludin and Negatively Regulate the Assembly of Tight Junctions in the CACO-2 Cell Monolayer. J. Biol. Chem. 2007, 282, 11487–11498. [Google Scholar] [CrossRef]
- Guntaka, S.R.; Samak, G.; Seth, A.; LaRusso, N.F.; Rao, R. Epidermal Growth Factor Protects the Apical Junctional Complexes from Hydrogen Peroxide in Bile Duct Epithelium. Lab. Investig. 2011, 91, 1396–1409. [Google Scholar] [CrossRef]
- Rao, R.K.; Samak, G. Bile Duct Epithelial Tight Junctions and Barrier Function. Tissue Barriers 2013, 1, e25718. [Google Scholar] [CrossRef] [PubMed]
- Patonai, A.; Erdélyi-Belle, B.; Korompay, A.; Somorácz, A.; Straub, B.K.; Schirmacher, P.; Kovalszky, I.; Lotz, G.; Kiss, A.; Schaff, Z. Claudins and Tricellulin in Fibrolamellar Hepatocellular Carcinoma. Virchows Arch. 2011, 458, 679–688. [Google Scholar] [CrossRef]
- Bunthot, S.; Obchoei, S.; Kraiklang, R.; Pirojkul, C.; Wongkham, S.; Wongkham, C. Overexpression of Claudin-4 in Cholangiocarcinoma Tissues and Its Possible Role in Tumor Metastasis. Asian Pac. J. Cancer Prev. 2012, 13, 71–76. [Google Scholar]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia Muciniphila-Derived Extracellular Vesicles Influence Gut Permeability through the Regulation of Tight Junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Yu, J.; Chen, X.; Yang, X.; Zhang, B. Understanding Gut Dysbiosis for Hepatocellular Carcinoma Diagnosis and Treatment. Trends Endocrinol. Metab. 2024, 35, 1006–1020. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ding, C.; Gu, Y.; He, Y.; Chen, B.; Zheng, S.; Li, Q. Association between Gut Microbiota and Hepatocellular Carcinoma from 2011 to 2022: Bibliometric Analysis and Global Trends. Front. Oncol. 2023, 13, 1120515. [Google Scholar] [CrossRef]
- Rajapakse, J.; Khatiwada, S.; Akon, A.C.; Yu, K.L.; Shen, S.; Zekry, A. Unveiling the Complex Relationship between Gut Microbiota and Liver Cancer: Opportunities for Novel Therapeutic Interventions. Gut Microbes 2023, 15, 2240031. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ren, Z.; Li, A.; Zhang, H.; Jiang, J.; Xu, S.; Luo, Q.; Zhou, K.; Sun, X.; Zheng, S.; et al. Deep Sequencing Reveals Microbiota Dysbiosis of Tongue Coat in Patients with Liver Carcinoma. Sci. Rep. 2016, 6, 33142. [Google Scholar] [CrossRef]
- Schneider, K.M.; Mohs, A.; Gui, W.; Galvez, E.J.C.; Candels, L.S.; Hoenicke, L.; Muthukumarasamy, U.; Holland, C.H.; Elfers, C.; Kilic, K.; et al. Imbalanced Gut Microbiota Fuels Hepatocellular Carcinoma Development by Shaping the Hepatic Inflammatory Microenvironment. Nat. Commun. 2022, 13, 3964. [Google Scholar] [CrossRef]
- Zhang, H.-L.; Yu, L.-X.; Yang, W.; Tang, L.; Lin, Y.; Wu, H.; Zhai, B.; Tan, Y.-X.; Shan, L.; Liu, Q.; et al. Profound Impact of Gut Homeostasis on Chemically-Induced pro-Tumorigenic Inflammation and Hepatocarcinogenesis in Rats. J. Hepatol. 2012, 57, 803–812. [Google Scholar] [CrossRef]
- Grąt, M.; Wronka, K.M.; Krasnodębski, M.; Masior, Ł.; Lewandowski, Z.; Kosińska, I.; Grąt, K.; Stypułkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A Multisociety Delphi Consensus Statement on New Fatty Liver Disease Nomenclature. J. Hepatol. 2023, 79, 1542–1556. [Google Scholar] [CrossRef]
- Behary, J.; Amorim, N.; Jiang, X.-T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut Microbiota Impact on the Peripheral Immune Response in Non-Alcoholic Fatty Liver Disease Related Hepatocellular Carcinoma. Nat. Commun. 2021, 12, 187. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global Epidemiology of NAFLD-Related HCC: Trends, Predictions, Risk Factors and Prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.G.; Kim, S.M.; Caussy, C.; Fu, T.; Guo, J.; Bassirian, S.; Singh, S.; Madamba, E.V.; Bettencourt, R.; Richards, L.; et al. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020, 32, 878–888.e6. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-Invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2019, 30, 607. [Google Scholar] [CrossRef]
- Caussy, C.; Hsu, C.; Lo, M.-T.; Liu, A.; Bettencourt, R.; Ajmera, V.H.; Bassirian, S.; Hooker, J.; Sy, E.; Richards, L.; et al. Link between Gut-Microbiome Derived Metabolite and Shared Gene-Effects with Hepatic Steatosis and Fibrosis in NAFLD. Hepatology 2018, 68, 918–932. [Google Scholar] [CrossRef]
- Pettinelli, P.; Arendt, B.M.; Schwenger, K.J.P.; Sivaraj, S.; Bhat, M.; Comelli, E.M.; Lou, W.; Allard, J.P. Relationship Between Hepatic Gene Expression, Intestinal Microbiota, and Inferred Functional Metagenomic Analysis in NAFLD. Clin. Transl. Gastroenterol. 2022, 13, e00466. [Google Scholar] [CrossRef]
- Huang, J.-H.; Wang, J.; Chai, X.-Q.; Li, Z.-C.; Jiang, Y.-H.; Li, J.; Liu, X.; Fan, J.; Cai, J.-B.; Liu, F. The Intratumoral Bacterial Metataxonomic Signature of Hepatocellular Carcinoma. Microbiol. Spectr. 2022, 10, e0098322. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver Cirrhosis Contributes to the Disorder of Gut Microbiota in Patients with Hepatocellular Carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, S.; Yamada, T.; Takemura, N.; Kokudo, N.; Hase, K.; Kawamura, Y.I. Profiling of Tumour-Associated Microbiota in Human Hepatocellular Carcinoma. Sci. Rep. 2021, 11, 10589. [Google Scholar] [CrossRef]
- Ma, J.; Li, J.; Jin, C.; Yang, J.; Zheng, C.; Chen, K.; Xie, Y.; Yang, Y.; Bo, Z.; Wang, J.; et al. Association of Gut Microbiome and Primary Liver Cancer: A Two-Sample Mendelian Randomization and Case-Control Study. Liver Int. 2023, 43, 221–233. [Google Scholar] [CrossRef]
- Park, S.-Y.; Hwang, B.-O.; Lim, M.; Ok, S.-H.; Lee, S.-K.; Chun, K.-S.; Park, K.-K.; Hu, Y.; Chung, W.-Y.; Song, N.-Y. Oral–Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers 2021, 13, 2124. [Google Scholar] [CrossRef]
- Elghannam, M.T.; Hassanien, M.H.; Ameen, Y.A.; Turky, E.A.; Elattar, G.M.; ElRay, A.A.; Eltalkawy, M.D. Oral Microbiota and Liver Diseases. Clin. Nutr. ESPEN 2023, 54, 68–72. [Google Scholar] [CrossRef]
- Clements, O.; Eliahoo, J.; Kim, J.U.; Taylor-Robinson, S.D.; Khan, S.A. Risk Factors for Intrahepatic and Extrahepatic Cholangiocarcinoma: A Systematic Review and Meta-Analysis. J. Hepatol. 2020, 72, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Elvevi, A.; Laffusa, A.; Gallo, C.; Invernizzi, P.; Massironi, S. Any Role for Microbiota in Cholangiocarcinoma? A Comprehensive Review. Cells 2023, 12, 370. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Lu, S.; Zeng, Z.; Liu, Q.; Dong, Z.; Chen, Y.; Zhu, Z.; Hong, Z.; Zhang, T.; Du, G.; et al. Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatology 2020, 71, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, C.; Duan, Y.; Heinrich, B.; Rosato, U.; Diggs, L.P.; Ma, L.; Roy, S.; Fu, Q.; Brown, Z.J.; et al. Gut Microbiome Directs Hepatocytes to Recruit MDSCs and Promote Cholangiocarcinoma. Cancer Discov. 2021, 11, 1248–1267. [Google Scholar] [CrossRef]
- Veglia, F.; Perego, M.; Gabrilovich, D. Myeloid-Derived Suppressor Cells Coming of Age. Nat. Immunol. 2018, 19, 108–119. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Chaisaingmongkol, J.; Budhu, A.; Dang, H.; Rabibhadana, S.; Pupacdi, B.; Kwon, S.M.; Forgues, M.; Pomyen, Y.; Bhudhisawasdi, V.; Lertprasertsuke, N.; et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 2017, 32, 57–70.e3. [Google Scholar] [CrossRef]
- Itthitaetrakool, U.; Pinlaor, P.; Pinlaor, S.; Chomvarin, C.; Dangtakot, R.; Chaidee, A.; Wilailuckana, C.; Sangka, A.; Lulitanond, A.; Yongvanit, P. Chronic Opisthorchis Viverrini Infection Changes the Liver Microbiome and Promotes Helicobacter Growth. PLoS ONE 2016, 11, e0165798. [Google Scholar] [CrossRef]
- Suyapoh, W.; Tangkawattana, S.; Suttiprapa, S.; Punyapornwithaya, V.; Tangkawattana, P.; Sripa, B. Synergistic Effects of cagA+ Helicobacter Pylori Co-Infected with Opisthorchis viverrini on Hepatobiliary Pathology in Hamsters. Acta Trop. 2021, 213, 105740. [Google Scholar] [CrossRef]
- Deenonpoe, R.; Mairiang, E.; Mairiang, P.; Pairojkul, C.; Chamgramol, Y.; Rinaldi, G.; Loukas, A.; Brindley, P.J.; Sripa, B. Elevated Prevalence of Helicobacter Species and Virulence Factors in Opisthorchiasis and Associated Hepatobiliary Disease. Sci. Rep. 2017, 7, 42744. [Google Scholar] [CrossRef]
- Wheatley, R.C.; Kilgour, E.; Jacobs, T.; Lamarca, A.; Hubner, R.A.; Valle, J.W.; McNamara, M.G. Potential Influence of the Microbiome Environment in Patients with Biliary Tract Cancer and Implications for Therapy. Br. J. Cancer 2022, 126, 693–705. [Google Scholar] [CrossRef]
- Boonyanugomol, W.; Chomvarin, C.; Baik, S.-C.; Song, J.-Y.; Hahnvajanawong, C.; Kim, K.-M.; Cho, M.-J.; Lee, W.-K.; Kang, H.-L.; Rhee, K.-H.; et al. Role of cagA-Positive Helicobacter Pylori on Cell Proliferation, Apoptosis, and Inflammation in Biliary Cells. Dig. Dis. Sci. 2011, 56, 1682–1692. [Google Scholar] [CrossRef]
- Manilla, V.; Santopaolo, F.; Gasbarrini, A.; Ponziani, F.R. Type 2 Diabetes Mellitus and Liver Disease: Across the Gut-Liver Axis from Fibrosis to Cancer. Nutrients 2023, 15, 2521. [Google Scholar] [CrossRef] [PubMed]
- Cadamuro, M.; Lasagni, A.; Sarcognato, S.; Guido, M.; Fabris, R.; Strazzabosco, M.; Strain, A.J.; Simioni, P.; Villa, E.; Fabris, L. The Neglected Role of Bile Duct Epithelial Cells in NASH. Semin. Liver Dis. 2022, 42, 34–47. [Google Scholar] [CrossRef]
- Elshaer, A.M.; El-Kharashi, O.A.; Hamam, G.G.; Nabih, E.S.; Magdy, Y.M.; Abd El Samad, A.A. Involvement of TLR4/CXCL9/PREX-2 Pathway in the Development of Hepatocellular Carcinoma (HCC) and the Promising Role of Early Administration of Lactobacillus Plantarum in Wistar Rats. Tissue Cell 2019, 60, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Xu, B.; Wang, X.; Wan, W.-H.; Lu, J.; Kong, D.; Jin, Y.; You, W.; Sun, H.; Mu, X.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Regulate Group 3 Innate Lymphoid Cells in HCC. Hepatology 2023, 77, 48–64. [Google Scholar] [CrossRef]
- Li, N.; Niu, L.; Liu, Y.; Wang, Y.; Su, X.; Xu, C.; Sun, Z.; Guo, H.; Gong, J.; Shen, S. Taking SCFAs Produced by Lactobacillus Reuteri Orally Reshapes Gut Microbiota and Elicits Antitumor Responses. J. Nanobiotechnol. 2024, 22, 241. [Google Scholar] [CrossRef]
- Hadinia, N.; Yavarmanesh, M.; Edalatian Dovom, M.R. Effects of Fermentation Conditions (Salt Concentration, Temperature, and pH) on Lactobacillus Strains for Induction of Interleukin-12 in the Exposed Murine Splenocytes. Heliyon 2024, 10, e39837. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, N.; Su, X.; Gao, Y.; Yang, R. Gut-Microbiota-Derived Metabolites Maintain Gut and Systemic Immune Homeostasis. Cells 2023, 12, 793. [Google Scholar] [CrossRef]
- Dallio, M.; Sangineto, M.; Romeo, M.; Villani, R.; Romano, A.D.; Loguercio, C.; Serviddio, G.; Federico, A. Immunity as Cornerstone of Non-Alcoholic Fatty Liver Disease: The Contribution of Oxidative Stress in the Disease Progression. Int. J. Mol. Sci. 2021, 22, 436. [Google Scholar] [CrossRef] [PubMed]
- Colosimo, S.; Tomlinson, J.W. Bile Acids as Drivers and Biomarkers of Hepatocellular Carcinoma. World J. Hepatol. 2022, 14, 1730–1738. [Google Scholar] [CrossRef] [PubMed]
- Režen, T.; Rozman, D.; Kovács, T.; Kovács, P.; Sipos, A.; Bai, P.; Mikó, E. The Role of Bile Acids in Carcinogenesis. Cell Mol. Life Sci. 2022, 79, 243. [Google Scholar] [CrossRef] [PubMed]
- D’Aldebert, E.; Biyeyeme Bi Mve, M.-J.; Mergey, M.; Wendum, D.; Firrincieli, D.; Coilly, A.; Fouassier, L.; Corpechot, C.; Poupon, R.; Housset, C.; et al. Bile Salts Control the Antimicrobial Peptide Cathelicidin through Nuclear Receptors in the Human Biliary Epithelium. Gastroenterology 2009, 136, 1435–1443. [Google Scholar] [CrossRef]
- Sung, J.Y.; Costerton, J.W.; Shaffer, E.A. Defense System in the Biliary Tract against Bacterial Infection. Dig. Dis. Sci. 1992, 37, 689–696. [Google Scholar] [CrossRef]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting Bile-Acid Signalling for Metabolic Diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef]
- Chen, W.; Ding, M.; Ji, L.; Yao, J.; Guo, Y.; Yan, W.; Yu, S.; Shen, Q.; Huang, M.; Zheng, Y.; et al. Bile Acids Promote the Development of HCC by Activating Inflammasome. Hepatol. Commun. 2023, 7, e0217. [Google Scholar] [CrossRef]
- Song, Y.; Lau, H.C.; Zhang, X.; Yu, J. Bile Acids, Gut Microbiota, and Therapeutic Insights in Hepatocellular Carcinoma. Cancer Biol. Med. 2023, 21, 144–162. [Google Scholar] [CrossRef]
- Guo, X.; Okpara, E.S.; Hu, W.; Yan, C.; Wang, Y.; Liang, Q.; Chiang, J.Y.L.; Han, S. Interactive Relationships between Intestinal Flora and Bile Acids. Int. J. Mol. Sci. 2022, 23, 8343. [Google Scholar] [CrossRef]
- Lenci, I.; Milana, M.; Signorello, A.; Grassi, G.; Baiocchi, L. Secondary Bile Acids and the Biliary Epithelia: The Good and the Bad. World J. Gastroenterol. 2023, 29, 357–366. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, W.; Huang, W. FXR and Liver Carcinogenesis. Acta Pharmacol. Sin. 2015, 36, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Ma, L.; Tang, W.; Huang, P.; Yang, B.; Wang, L.; Chen, S.; Gao, Q.; Zhang, S.; Xia, J. FXR Acts as a Metastasis Suppressor in Intrahepatic Cholangiocarcinoma by Inhibiting IL-6-Induced Epithelial-Mesenchymal Transition. Cell Physiol. Biochem. 2018, 48, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Fan, M.; Huang, W. Pleiotropic Roles of FXR in Liver and Colorectal Cancers. Mol. Cell Endocrinol. 2022, 543, 111543. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yao, M.; Yan, Y.; Liu, Y.; Wen, X.; Chen, X.; Lu, F. Deoxycholic Acid Upregulates Serum Golgi Protein 73 through Activating NF-κB Pathway and Destroying Golgi Structure in Liver Disease. Biomolecules 2021, 11, 205. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Greten, T.F. Gut Microbiome in HCC—Mechanisms, Diagnosis and Therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef]
- Maresca, M.; Yahi, N.; Younès-Sakr, L.; Boyron, M.; Caporiccio, B.; Fantini, J. Both Direct and Indirect Effects Account for the Pro-Inflammatory Activity of Enteropathogenic Mycotoxins on the Human Intestinal Epithelium: Stimulation of Interleukin-8 Secretion, Potentiation of Interleukin-1beta Effect and Increase in the Transepithelial Passage of Commensal Bacteria. Toxicol. Appl. Pharmacol. 2008, 228, 84–92. [Google Scholar] [CrossRef]
- Wei, S.; Ma, X.; Zhao, Y. Mechanism of Hydrophobic Bile Acid-Induced Hepatocyte Injury and Drug Discovery. Front. Pharmacol. 2020, 11, 1084. [Google Scholar] [CrossRef]
- Meng, X.; Chang, Z.; Che, N.; Wu, J.; Dang, T.; Chai, J. Acid/Bile Exposure Triggers TRAIL-Mediated Apoptosis in Esophageal Cancer Cells by Suppressing the Decoy Receptors and c-FLIPR. Int. J. Biochem. Cell Biol. 2020, 122, 105736. [Google Scholar] [CrossRef]
- Herraez, E.; Romero, M.R.; Macias, R.I.R.; Monte, M.J.; Marin, J.J.G. Clinical Relevance of the Relationship between Changes in Gut Microbiota and Bile Acid Metabolism in Patients with Intrahepatic Cholangiocarcinoma. Hepatobiliary Surg. Nutr. 2020, 9, 211–214. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- McBrearty, N.; Arzumanyan, A.; Bichenkov, E.; Merali, S.; Merali, C.; Feitelson, M. Short Chain Fatty Acids Delay the Development of Hepatocellular Carcinoma in HBx Transgenic Mice. Neoplasia 2021, 23, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Son, M.-Y.; Cho, H.-S. Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers. J. Microbiol. Biotechnol. 2023, 33, 849–856. [Google Scholar] [CrossRef]
- Che, Y.; Chen, G.; Guo, Q.; Duan, Y.; Feng, H.; Xia, Q. Gut Microbial Metabolite Butyrate Improves Anticancer Therapy by Regulating Intracellular Calcium Homeostasis. Hepatology 2023, 78, 88–102. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut Microbiome Analysis as a Tool towards Targeted Non-Invasive Biomarkers for Early Hepatocellular Carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kaur, R.; Kanthaje, S.; Dhiman, R.K.; Chakraborti, A. Bacterial Metabolite Butyrate in Modulating Sorafenib-Targeted microRNAs to Curtail Its Resistance in Hepatocellular Carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 5823–5839. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Shen, X.; Xiao, X.; Li, L.; Huang, Y. Butyrate Modification Promotes Intestinal Absorption and Hepatic Cancer Cells Targeting of Ferroptosis Inducer Loaded Nanoparticle for Enhanced Hepatocellular Carcinoma Therapy. Small 2023, 19, e2301149. [Google Scholar] [CrossRef]
- Lapidot, Y.; Amir, A.; Nosenko, R.; Uzan-Yulzari, A.; Veitsman, E.; Cohen-Ezra, O.; Davidov, Y.; Weiss, P.; Bradichevski, T.; Segev, S.; et al. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems 2020, 5, 10.1128. [Google Scholar] [CrossRef]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D.; Zhang, L.; Wang, W.-B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694.e22. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Shah, M.M.; Meyer, B.I.; Rhee, K.; NeMoyer, R.E.; Lin, Y.; Tzeng, C.-W.D.; Jabbour, S.K.; Kennedy, T.J.; Nosher, J.L.; Kooby, D.A.; et al. Conditional Survival Analysis of Hepatocellular Carcinoma. J. Surg. Oncol. 2020, 122, 684–690. [Google Scholar] [CrossRef]
- Izquierdo-Sanchez, L.; Lamarca, A.; La Casta, A.; Buettner, S.; Utpatel, K.; Klümpen, H.-J.; Adeva, J.; Vogel, A.; Lleo, A.; Fabris, L.; et al. Cholangiocarcinoma Landscape in Europe: Diagnostic, Prognostic and Therapeutic Insights from the ENSCCA Registry. J. Hepatol. 2022, 76, 1109–1121. [Google Scholar] [CrossRef]
- Oh, D.-Y.; He, A.R.; Bouattour, M.; Okusaka, T.; Qin, S.; Chen, L.-T.; Kitano, M.; Lee, C.-K.; Kim, J.W.; Chen, M.-H.; et al. Durvalumab or Placebo plus Gemcitabine and Cisplatin in Participants with Advanced Biliary Tract Cancer (TOPAZ-1): Updated Overall Survival from a Randomised Phase 3 Study. Lancet Gastroenterol. Hepatol. 2024, 9, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Kipp, B.R.; Voss, J.S.; Kerr, S.E.; Barr Fritcher, E.G.; Graham, R.P.; Zhang, L.; Highsmith, W.E.; Zhang, J.; Roberts, L.R.; Gores, G.J.; et al. Isocitrate Dehydrogenase 1 and 2 Mutations in Cholangiocarcinoma. Hum. Pathol. 2012, 43, 1552–1558. [Google Scholar] [CrossRef]
- Graham, R.P.; Barr Fritcher, E.G.; Pestova, E.; Schulz, J.; Sitailo, L.A.; Vasmatzis, G.; Murphy, S.J.; McWilliams, R.R.; Hart, S.N.; Halling, K.C.; et al. Fibroblast Growth Factor Receptor 2 Translocations in Intrahepatic Cholangiocarcinoma. Hum. Pathol. 2014, 45, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Chmiel, P.; Gęca, K.; Rawicz-Pruszyński, K.; Polkowski, W.P.; Skórzewska, M. FGFR Inhibitors in Cholangiocarcinoma-A Novel Yet Primary Approach: Where Do We Stand Now and Where to Head Next in Targeting This Axis? Cells 2022, 11, 3929. [Google Scholar] [CrossRef]
- Wu, Q.; Ellis, H.; Siravegna, G.; Michel, A.G.; Norden, B.L.; Fece de la Cruz, F.; Balasooriya, E.R.; Zhen, Y.; Silveira, V.S.; Che, J.; et al. Landscape of Clinical Resistance Mechanisms to FGFR Inhibitors in FGFR2-Altered Cholangiocarcinoma. Clin. Cancer Res. 2024, 30, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Chakraborty, E.; Chakraborty, D.; Das, D.; Bai, Y. Discovering Novel Prognostic Biomarkers of Hepatocellular Carcinoma Using eXplainable Artificial Intelligence. Expert Syst. Appl. 2024, 252, 124239. [Google Scholar] [CrossRef]
- Lacalamita, A.; Serino, G.; Pantaleo, E.; Monaco, A.; Amoroso, N.; Bellantuono, L.; Piccinno, E.; Scalavino, V.; Dituri, F.; Tangaro, S.; et al. Artificial Intelligence and Complex Network Approaches Reveal Potential Gene Biomarkers for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 15286. [Google Scholar] [CrossRef]
- Yagin, F.H.; El Shawi, R.; Algarni, A.; Colak, C.; Al-Hashem, F.; Ardigò, L.P. Metabolomics Biomarker Discovery to Optimize Hepatocellular Carcinoma Diagnosis: Methodology Integrating AutoML and Explainable Artificial Intelligence. Diagnostics 2024, 14, 2049. [Google Scholar] [CrossRef]
- Romeo, M.; Dallio, M.; Napolitano, C.; Basile, C.; Di Nardo, F.; Vaia, P.; Iodice, P.; Federico, A. Clinical Applications of Artificial Intelligence (AI) in Human Cancer: Is It Time to Update the Diagnostic and Predictive Models in Managing Hepatocellular Carcinoma (HCC)? Diagnostics 2025, 15, 252. [Google Scholar] [CrossRef]
- Yang, J.; He, Q.; Lu, F.; Chen, K.; Ni, Z.; Wang, H.; Zhou, C.; Zhang, Y.; Chen, B.; Bo, Z.; et al. A Distinct Microbiota Signature Precedes the Clinical Diagnosis of Hepatocellular Carcinoma. Gut Microbes 2023, 15, 2201159. [Google Scholar] [CrossRef]
- Zhang, N.; Zhu, W.; Zhang, S.; Liu, T.; Gong, L.; Wang, Z.; Zhang, W.; Cui, Y.; Wu, Q.; Li, J.; et al. A Novel Bifidobacterium/Klebsiella Ratio in Characterization Analysis of the Gut and Bile Microbiota of CCA Patients. Microb. Ecol. 2023, 87, 5. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, S.; Jin, C.; Lin, Z.; Deng, T.; Xie, X.; Deng, L.; Li, X.; Ma, J.; Ding, X.; et al. A Predictive Model Based on the Gut Microbiota Improves the Diagnostic Effect in Patients With Cholangiocarcinoma. Front. Cell Infect. Microbiol. 2021, 11, 751795. [Google Scholar] [CrossRef]
- Liu, Y.; Baba, Y.; Ishimoto, T.; Gu, X.; Zhang, J.; Nomoto, D.; Okadome, K.; Baba, H.; Qiu, P. Gut Microbiome in Gastrointestinal Cancer: A Friend or Foe? Int. J. Biol. Sci. 2022, 18, 4101–4117. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Li, J.; He, B.; Chen, B.; Liu, F.; Chen, Z.; Zheng, J.; Shi, Z.; Zhang, T.; Deng, L.; et al. Gut Microbiome Alteration as a Diagnostic Tool and Associated with Inflammatory Response Marker in Primary Liver Cancer. Hepatol. Int. 2022, 16, 99–111. [Google Scholar] [CrossRef]
- Llovet, J.M.; Lencioni, R. mRECIST for HCC: Performance and Novel Refinements. J. Hepatol. 2020, 72, 288–306. [Google Scholar] [CrossRef] [PubMed]
- Muscolino, P.; Granata, B.; Omero, F.; De Pasquale, C.; Campana, S.; Calabrò, A.; D’Anna, F.; Drommi, F.; Pezzino, G.; Cavaliere, R.; et al. Potential Predictive Role of Gut Microbiota to Immunotherapy in HCC Patients: A Brief Review. Front. Oncol. 2023, 13, 1247614. [Google Scholar] [CrossRef]
- Lee, P.-C.; Wu, C.-J.; Hung, Y.-W.; Lee, C.J.; Chi, C.-T.; Lee, I.-C.; Yu-Lun, K.; Chou, S.-H.; Luo, J.-C.; Hou, M.-C.; et al. Gut Microbiota and Metabolites Associate with Outcomes of Immune Checkpoint Inhibitor-Treated Unresectable Hepatocellular Carcinoma. J. Immunother. Cancer 2022, 10, e004779. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Chuanbing, Z.; Zhengle, Z.; Ruili, D.; Kongfan, Z.; Jing, T. Genes Modulating Butyrate Metabolism for Assessing Clinical Prognosis and Responses to Systematic Therapies in Hepatocellular Carcinoma. Biomolecules 2022, 13, 52. [Google Scholar] [CrossRef]
- Mao, J.; Wang, D.; Long, J.; Yang, X.; Lin, J.; Song, Y.; Xie, F.; Xun, Z.; Wang, Y.; Wang, Y.; et al. Gut Microbiome Is Associated with the Clinical Response to Anti-PD-1 Based Immunotherapy in Hepatobiliary Cancers. J. Immunother. Cancer 2021, 9, e003334. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhao, R.; Zhou, C.; Zhong, Q.; Shi, J.; Su, C.; Li, Q.; Su, X.; Chi, H.; Lu, X.; et al. Feasibility and Tolerability of Sintilimab plus Anlotinib as the Second-Line Therapy for Patients with Advanced Biliary Tract Cancers: An Open-Label, Single-Arm, Phase II Clinical Trial. Int. J. Cancer 2023, 152, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Ketpueak, T.; Sriwichaiin, S.; Suparan, K.; Kerdphoo, S.; Charoentum, C.; Suksombooncharoen, T.; Chewaskulyong, B.; Chattipakorn, N.; Chattipakorn, S. Alteration of Gut Microbiota Composition in Patients with Cholangiocarcinoma with Non-Responsiveness to First-Line Chemotherapy: A Pilot Study. JCO 2023, 41, 4104. [Google Scholar] [CrossRef]
- Bednarsch, J.; Czigany, Z.; Heij, L.R.; Luedde, T.; van Dam, R.; Lang, S.A.; Ulmer, T.F.; Hornef, M.W.; Neumann, U.P. Bacterial Bile Duct Colonization in Perihilar Cholangiocarcinoma and Its Clinical Significance. Sci. Rep. 2021, 11, 2926. [Google Scholar] [CrossRef]
- Milosevic, I.; Vujovic, A.; Barac, A.; Djelic, M.; Korac, M.; Radovanovic Spurnic, A.; Gmizic, I.; Stevanovic, O.; Djordjevic, V.; Lekic, N.; et al. Gut-Liver Axis, Gut Microbiota, and Its Modulation in the Management of Liver Diseases: A Review of the Literature. Int. J. Mol. Sci. 2019, 20, 395. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Song, Q.; Zhang, X.; Liu, W.; Wei, H.; Liang, W.; Zhou, Y.; Ding, Y.; Ji, F.; Ho-Kwan Cheung, A.; Wong, N.; et al. Bifidobacterium Pseudolongum-Generated Acetate Suppresses Non-Alcoholic Fatty Liver Disease-Associated Hepatocellular Carcinoma. J. Hepatol. 2023, 79, 1352–1365. [Google Scholar] [CrossRef]
- Nouso, K.; Shiota, S.; Fujita, R.; Wakuta, A.; Kariyama, K.; Hiraoka, A.; Atsukawa, M.; Tani, J.; Tada, T.; Nakamura, S.; et al. Effect of Butyrate-Producing Enterobacteria on Advanced Hepatocellular Carcinoma Treatment with Atezolizumab and Bevacizumab. Cancer Med. 2023, 12, 17849–17855. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Hong, W.; Wang, B.; Chen, Y.; Yang, P.; Zhou, J.; Fan, J.; Zeng, Z.; Du, S. Gut Microbiota Modulate Radiotherapy-Associated Antitumor Immune Responses against Hepatocellular Carcinoma Via STING Signaling. Gut Microbes 2022, 14, 2119055. [Google Scholar] [CrossRef]
- Porcari, S.; Benech, N.; Valles-Colomer, M.; Segata, N.; Gasbarrini, A.; Cammarota, G.; Sokol, H.; Ianiro, G. Key Determinants of Success in Fecal Microbiota Transplantation: From Microbiome to Clinic. Cell Host Microbe 2023, 31, 712–733. [Google Scholar] [CrossRef]
- Kellingray, L.; Gall, G.L.; Defernez, M.; Beales, I.L.P.; Franslem-Elumogo, N.; Narbad, A. Microbial Taxonomic and Metabolic Alterations during Faecal Microbiota Transplantation to Treat Clostridium Difficile Infection. J. Infect. 2018, 77, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal Microbiota Transplantation in Cancer Management: Current Status and Perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Vallianou, N.; Christodoulatos, G.S.; Karampela, I.; Tsilingiris, D.; Magkos, F.; Stratigou, T.; Kounatidis, D.; Dalamaga, M. Understanding the Role of the Gut Microbiome and Microbial Metabolites in Non-Alcoholic Fatty Liver Disease: Current Evidence and Perspectives. Biomolecules 2021, 12, 56. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Badgeley, A.; Anwar, H.; Modi, K.; Murphy, P.; Lakshmikuttyamma, A. Effect of Probiotics and Gut Microbiota on Anti-Cancer Drugs: Mechanistic Perspectives. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188494. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut Microbiota and Intestinal FXR Mediate the Clinical Benefits of Metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Song, W.; Liao, Y.; Wang, X.; Chen, C.; Ming, J.; Cui, J.; Xu, K. Effects of Long-Term Regular Oral Aspirin Combined with Atorvastatin to Prevent Ischemic Stroke on Human Gut Microbiota. Eur. J. Pharmacol. 2023, 951, 175800. [Google Scholar] [CrossRef]
- Li, T.; Ding, N.; Guo, H.; Hua, R.; Lin, Z.; Tian, H.; Yu, Y.; Fan, D.; Yuan, Z.; Gonzalez, F.J.; et al. A Gut Microbiota-Bile Acid Axis Promotes Intestinal Homeostasis upon Aspirin-Mediated Damage. Cell Host Microbe 2024, 32, 191–208.e9. [Google Scholar] [CrossRef]
- Lee, T.-Y.; Hsu, Y.-C.; Ho, H.J.; Lin, J.-T.; Chen, Y.-J.; Wu, C.-Y. Daily Aspirin Associated with a Reduced Risk of Hepatocellular Carcinoma in Patients with Non-Alcoholic Fatty Liver Disease: A Population-Based Cohort Study. EClinicalMedicine 2023, 61, 102065. [Google Scholar] [CrossRef]
- Dallio, M.; Romeo, M.; Di Nardo, F.; Vaia, P.; Napolitano, C.; Ventriglia, L.; Coppola, A.; Silvestrin, A.; Olivieri, S.; Federico, A. FLAME: Training and Validating a Newly Conceived Model Incorporating Alpha-Glutathione-S-Transferase Serum Levels for Predicting Advanced Hepatic Fibrosis and Acute Cardiovascular Events in Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Int. J. Mol. Sci. 2025, 26, 761. [Google Scholar] [CrossRef]
- Gravina, A.G.; Romeo, M.; Pellegrino, R.; Tuccillo, C.; Federico, A.; Loguercio, C. Just Drink a Glass of Water? Effects of Bicarbonate-Sulfate-Calcium-Magnesium Water on the Gut-Liver Axis. Front. Pharmacol. 2022, 13, 869446. [Google Scholar] [CrossRef]
- Yu, L.-X.; Schwabe, R.F. The Gut Microbiome and Liver Cancer: Mechanisms and Clinical Translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Dallio, M.; Romeo, M.; Gravina, A.G.; Masarone, M.; Larussa, T.; Abenavoli, L.; Persico, M.; Loguercio, C.; Federico, A. Nutrigenomics and Nutrigenetics in Metabolic- (Dysfunction) Associated Fatty Liver Disease: Novel Insights and Future Perspectives. Nutrients 2021, 13, 1679. [Google Scholar] [CrossRef] [PubMed]
- Meroni, M.; Longo, M.; Rustichelli, A.; Dongiovanni, P. Nutrition and Genetics in NAFLD: The Perfect Binomium. Int. J. Mol. Sci. 2020, 21, 2986. [Google Scholar] [CrossRef]
| Setting | Lactobacillus spp. Levels | Main Functional Repercussions | References |
|---|---|---|---|
| HCC patients | Reduced (↓↓↓) representation | Promoting inflammation via decreased SCFA production (mouse model) and inducing activation of hepatic TLR4/CXCL9 pathway | [66,67] 3/21/2025 10:11:00 AM. |
| Healthy subjects | Normal (↑) representation | Preserving intestinal integrity and contrasting inflammatory processes via SCFA production | [49,68,69] |
| CC patients | Enhanced (↑↑↑) representation | Impairing specific secondary BA production (correlation between bacterial representation and serum TUDCA levels) | [54] |
| Setting and Research Aims | Specific Microbiota Patterns (“Signature”) | Potential Clinical Applications | References |
|---|---|---|---|
| Discriminating patients with liver cirrhosis and early HCC from individuals exclusively presenting with liver cirrhosis | Gut microbiota: enhanced representation of Actinobacteria; increased levels of specific genera Gemmiger, Parabacteroides, Paraprevotella, Clostridium_XVIII, Erysipelotrichaceae_incertae_sedis, Clostridium_XIVb, Collinsella, Butyricicoccus, Odoribacter Dorea, Acidaminococcus, Holdemania, Eggerthella; decreased butyrate-producing species | Early identification of HCC in patients with liver cirrhosis | [94] 3/21/2025 10:11:00 AM. |
| Discriminating patients with HCC from healthy ones through a ML model | Gut and oral microbiota: enhanced representation of Streptococcus, Shigella, and E. coli | Early HCC detection; improvement of ML model performance after combining AFP serum levels | [111] |
| Discriminating patients with CC from healthy ones | Gut microbiota: enhanced representation of the specific genera Burkholderia-Caballeronia-Paraburkholderia, Faecalibacterium, and Ruminococcus-1, configuring the “B-F-R pattern” | Early CC detection | [113] |
| Discriminating patients with CC from healthy ones | Gut microbiota: reduced representation of phyla Firmicutes and Actinobacteriota simultaneously with increasing levels of Proteobacteria and Bacteroidota; relative abundance of Klebsiella in contrast with the reduction in Bifidobacterium [decreased Bifidobacterium/Klebsiella (B/K) ratio] | Early CC detection | [112] |
| Discriminating patients with HBLC (both HCC and CC) from healthy ones | Gut microbiota: enhanced representation of the specific genera Faecalibacterium, Klebsiella, Ruminococcus Gnavus group, Lactobacillus, Dorea, Veillonella, Burkholderia-Caballeronia-Paraburkholderia, Citrobacter | Early HBLC diagnosis | [115] |
| Setting and Research Aims | Specific Microbiota Patterns (“Signature”) | Potential Clinical Applications | References |
|---|---|---|---|
| Discriminating advanced HCC patients showing response (i.e., radiology-proven disease regression) from non-responders to systemic therapy | Gut microbiota: in responder patients, predominance of Lachnoclostridium, Lachnospiraceae, and Veillonella; in individuals with progressive disease, increased levels of Prevotella 9 [increased Lachnoclostridium/Prevotella 9 (L/P) ratio in long-term OS patients] | Predicting response to systemic therapy (disease progression and OS) in advanced HCC | [118] 3/21/2025 10:11:00 AM. |
| Identifying advanced HCC patients responding to immunotherapy | Gut microbiota: enhanced representation of Akkermansia muciniphila (in turn, promoting the increased relative abundance of SCFA-producer species Lachnospiraceae and Blautia) | Predicting immunotherapy response (disease progression) in advanced HCC | [35,38,119] |
| Identifying advanced ICC patients responding to immunotherapy | Gut microbiota: increased relative abundance of Ruminococcaceae | Predicting response to systemic therapy (disease progression) in advanced ICC | [123] |
| Estimating the risk of postoperative abdominal infections in advanced pCC | Bile duct microbiota: increased levels of Enterococcus faecalis, Enterococcus faecium, Enterobacter cloacae, and Escherichia coli | Predicting the outcomes in the post-surgical period in advanced CC | [124] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, M.; Dallio, M.; Di Nardo, F.; Napolitano, C.; Vaia, P.; Martinelli, G.; Federico, P.; Olivieri, S.; Iodice, P.; Federico, A. The Role of the Gut–Biliary–Liver Axis in Primary Hepatobiliary Liver Cancers: From Molecular Insights to Clinical Applications. J. Pers. Med. 2025, 15, 124. https://doi.org/10.3390/jpm15040124
Romeo M, Dallio M, Di Nardo F, Napolitano C, Vaia P, Martinelli G, Federico P, Olivieri S, Iodice P, Federico A. The Role of the Gut–Biliary–Liver Axis in Primary Hepatobiliary Liver Cancers: From Molecular Insights to Clinical Applications. Journal of Personalized Medicine. 2025; 15(4):124. https://doi.org/10.3390/jpm15040124
Chicago/Turabian StyleRomeo, Mario, Marcello Dallio, Fiammetta Di Nardo, Carmine Napolitano, Paolo Vaia, Giuseppina Martinelli, Pierluigi Federico, Simone Olivieri, Patrizia Iodice, and Alessandro Federico. 2025. "The Role of the Gut–Biliary–Liver Axis in Primary Hepatobiliary Liver Cancers: From Molecular Insights to Clinical Applications" Journal of Personalized Medicine 15, no. 4: 124. https://doi.org/10.3390/jpm15040124
APA StyleRomeo, M., Dallio, M., Di Nardo, F., Napolitano, C., Vaia, P., Martinelli, G., Federico, P., Olivieri, S., Iodice, P., & Federico, A. (2025). The Role of the Gut–Biliary–Liver Axis in Primary Hepatobiliary Liver Cancers: From Molecular Insights to Clinical Applications. Journal of Personalized Medicine, 15(4), 124. https://doi.org/10.3390/jpm15040124








