Sex-Specific Expression of Non-Coding RNA Fragments in Frontal Cortex, Hippocampus and Cerebellum of Rats
Abstract
:1. Introduction
2. Results
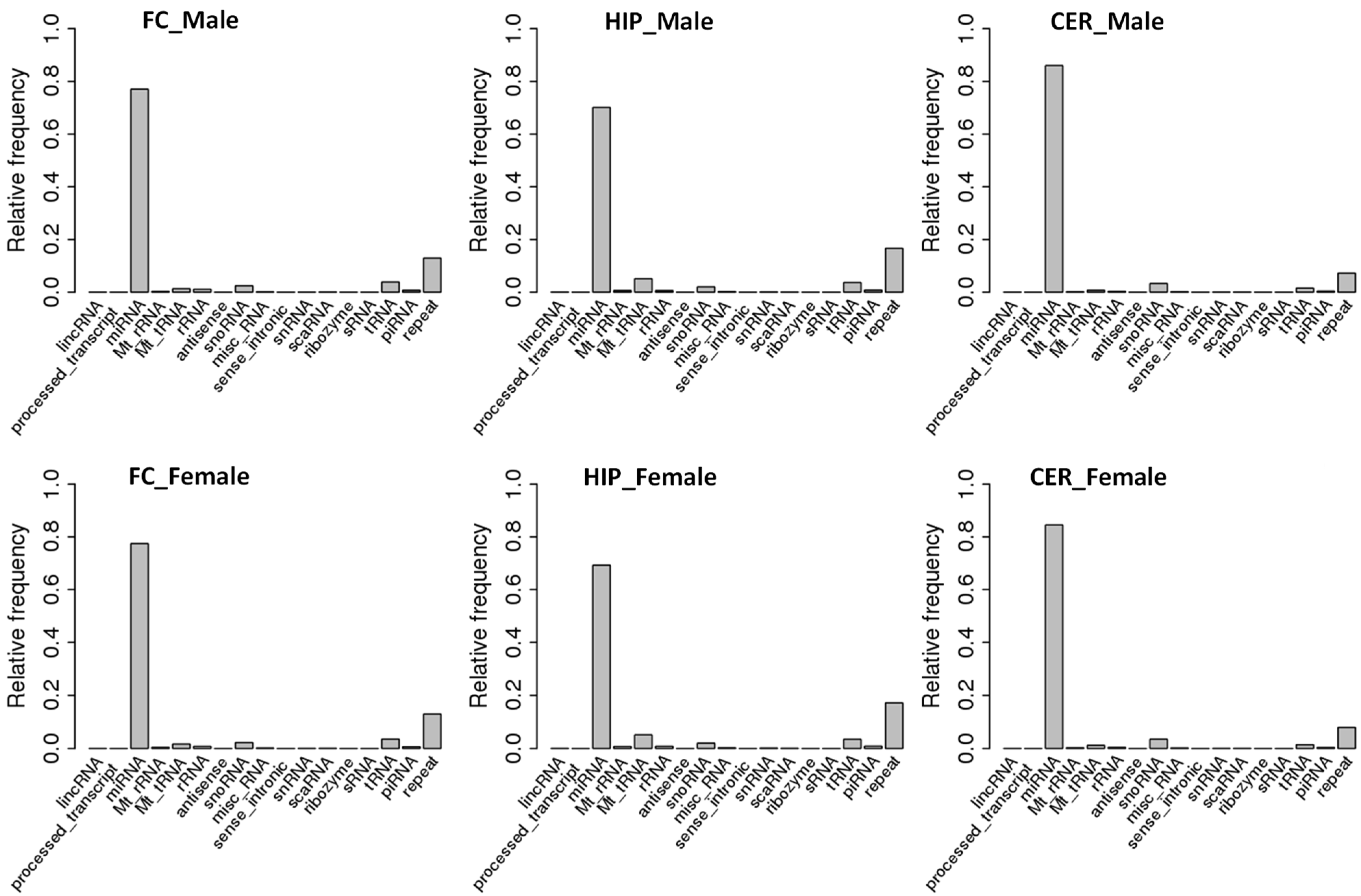
2.1. Comparison of ncRNA Abundance between Different Brain Regions in Male and Female Rats
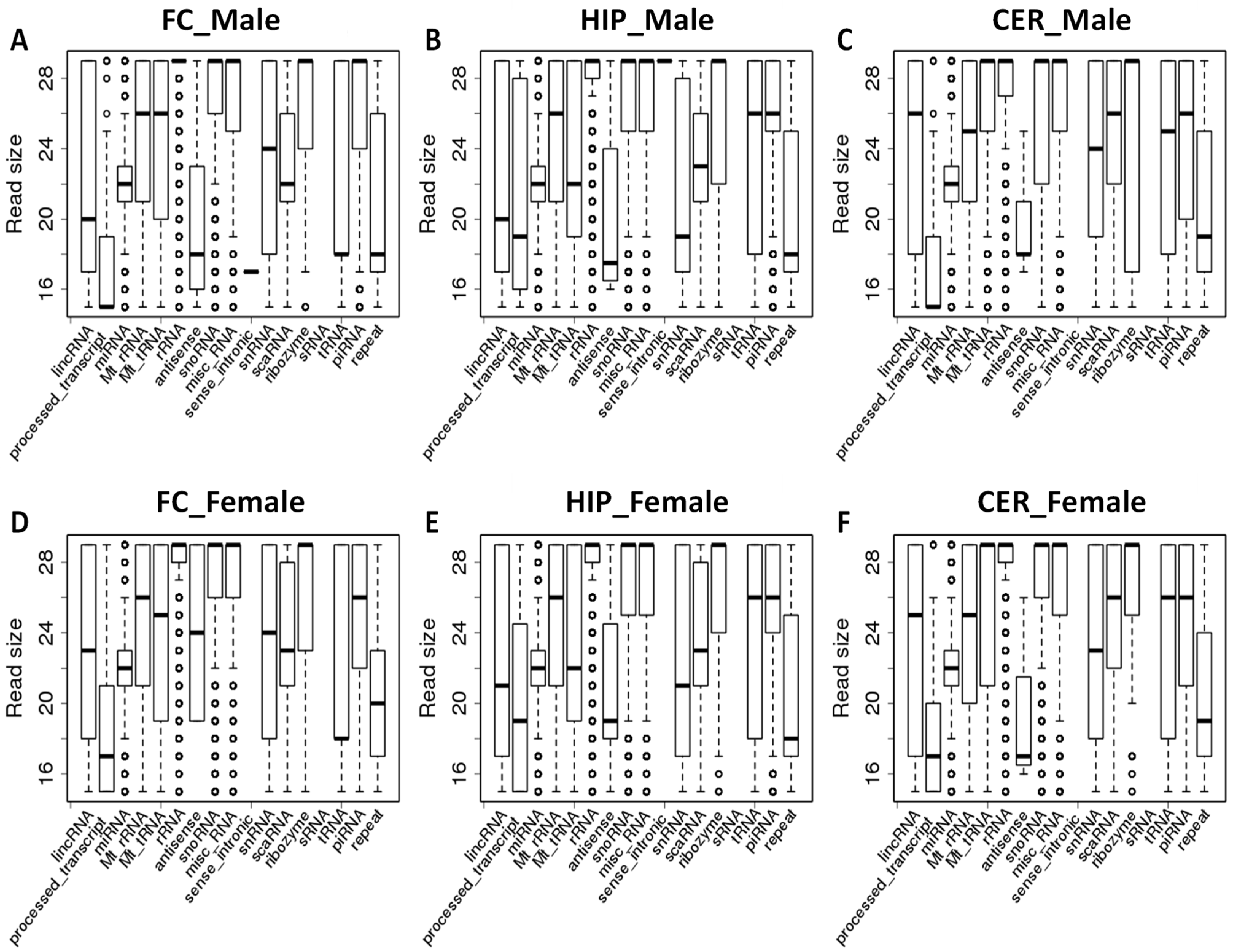
2.2. Comparison of Average Read Size for Various ncRNA Classes between Brain Regions of Male and Female Rats
2.3. Analysis of GC Content in ncRNA Reads
2.4. Analysis of the Distribution of Reads across the Entire Length of ncRNAs
2.5. ncRF Analysis
2.6. Analysis of ncRF Fractions and ncRF Types
2.7. Correlation between ncRNA Reads and ncRF Reads
2.8. Prediction of Targets of ncRFs in Various Brain Regions
2.9. Analysis of Overlapping and Unique Pathways Using DAVID
3. Discussion
3.1. Sex-Specific Differences in ncRNA Expression
3.2. Brain-Specific Differences in ncRNAs and ncRFs
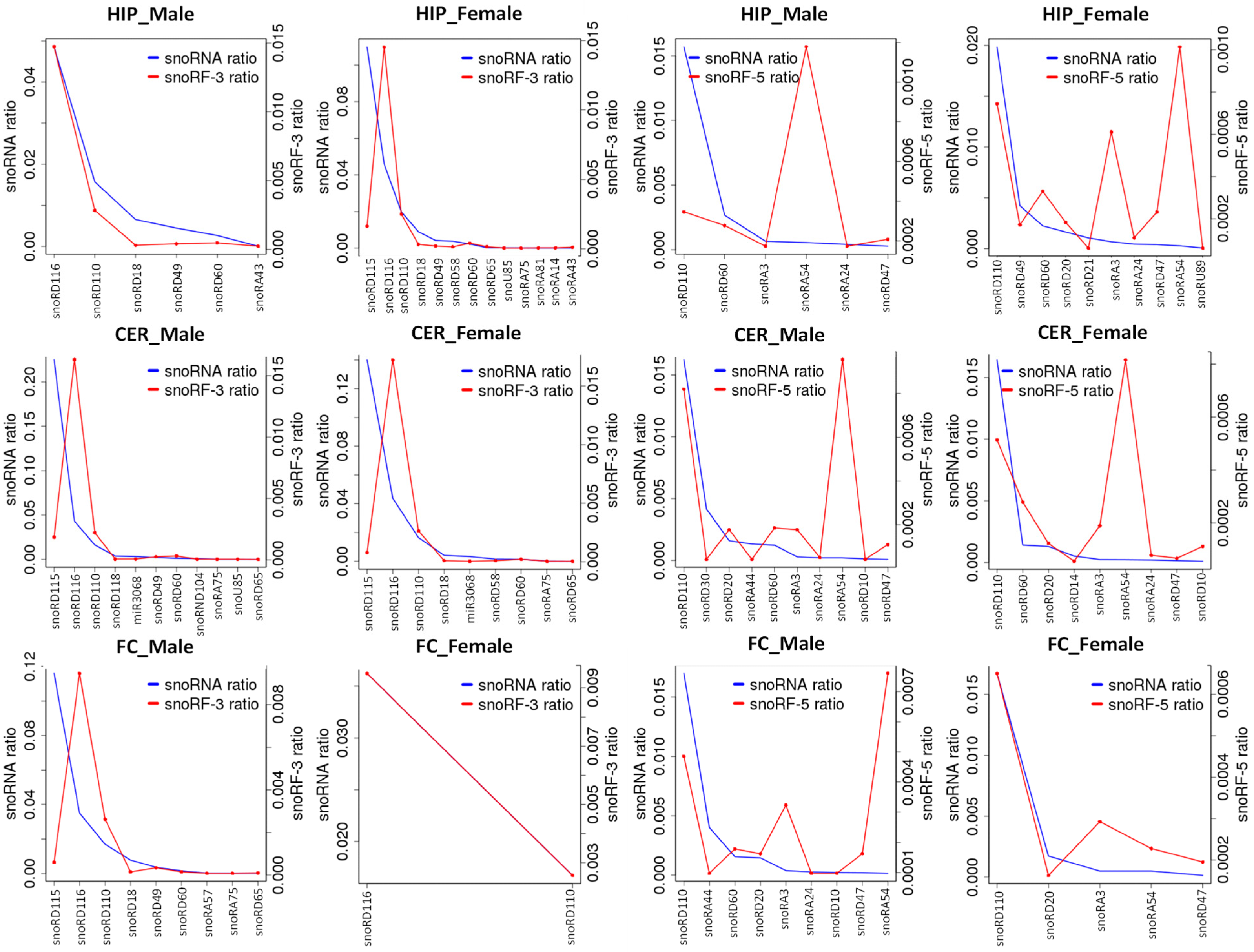
3.3. Difference in Processing of ncRFs
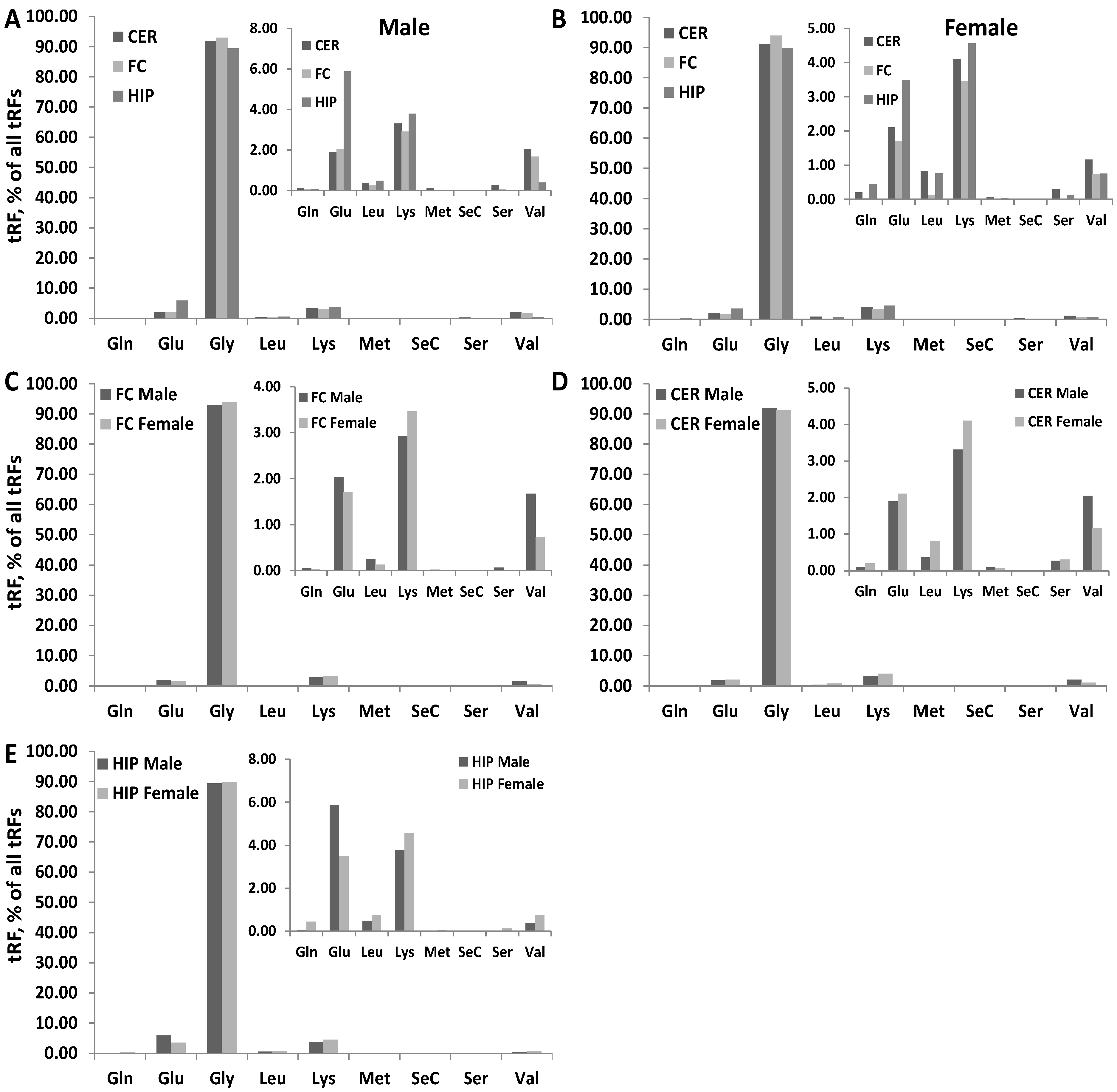
3.4. Differences in the Type of tRFs
3.5. Predicted Targets
4. Materials and Methods
4.1. Animals Used in the Experiment
4.2. Sequencing
4.3. The Identification and Description of ncRFs
4.4. The Analysis of the Distribution of ncRF Reads across the Entire Length of Precursor ncRNAs
4.5. The Analysis of the Enrichment of ncRFs Relative to the Number of ncRNA Precursors
4.6. ncRFs Target Prediction Using miRDB
4.7. ncRFs Target Pathway Prediction Using DAVID
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Gomez-Verjan, J.C.; Vazquez-Martinez, E.R.; Rivero-Segura, N.A.; Medina-Campos, R.H. The RNA world of human ageing. Qual. Life Res. 2018, 137, 865–879. [Google Scholar] [CrossRef]
- Morgan, C.P.; Bale, T.L. Sex differences in microRNA-mRNA networks: Examination of novel epigenetic programming mechanisms in the sexually dimorphic neonatal hypothalamus. Biol. Sex Differ. 2017, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Pak, T.R.; Rao, Y.S.; Prins, S.A.; Mott, N.N. An emerging role for microRNAs in sexually dimorphic neurobiological systems. Pflug. Arch. 2013, 465, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isakova, A.; Fehlmann, T.; Keller, A.; Quake, S.R. A mouse tissue atlas of small noncoding RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 25634–25645. [Google Scholar] [CrossRef] [PubMed]
- Issler, O.; van der Zee, Y.Y.; Ramakrishnan, A.; Wang, J.; Tan, C.; Loh, Y.-H.E.; Purushothaman, I.; Walker, D.M.; Lorsch, Z.S.; Hamilton, P.J.; et al. Sex-Specific Role for the Long Non-coding RNA LINC00473 in Depression. Neuron 2020, 106, 912–926.e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Z.; Chen, D.; Zhang, B.; Tian, R.-R.; Wu, J.; Zhang, Y.; Xu, K.; Yang, L.-M.; Cheng, C.; et al. Annotation and cluster analysis of spatiotemporal- and sex-related lncRNA expression in rhesus macaque brain. Genome Res. 2017, 27, 1608–1620. [Google Scholar] [CrossRef] [Green Version]
- Pinkard, O.; McFarland, S.; Sweet, T.; Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef] [Green Version]
- Bratkovič, T.; Rogelj, B. Biology and applications of small nucleolar RNAs. Cell. Mol. Life Sci. 2011, 68, 3843–3851. [Google Scholar] [CrossRef]
- Garritano, S.; Gigoni, A.; Costa, D.; Malatesta, P.; Florio, T.; Cancedda, R.; Pagano, A. A novel collection of snRNA-like promoters with tissue-specific transcription properties. Int. J. Mol. Sci. 2012, 13, 11323–11332. [Google Scholar] [CrossRef] [Green Version]
- Juhila, J.; Sipilä, T.; Icay, K.; Nicorici, D.; Ellonen, P.; Kallio, A.; Korpelainen, E.; Greco, D.; Hovatta, I. MicroRNA Expression Profiling Reveals MiRNA Families Regulating Specific Biological Pathways in Mouse Frontal Cortex and Hippocampus. PLoS ONE 2011, 6, e21495. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keam, S.P.; Sobala, A.; Ten Have, S.; Hutvagner, G. tRNA-Derived RNA Fragments Associate with Human Multisynthetase Complex (MSC) and Modulate Ribosomal Protein Translation. J. Proteome Res. 2017, 16, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Levitz, R.; Chapman, D.; Amitsur, M.; Green, R.; Snyder, L.; Kaufmann, G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990, 9, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Cura das Neves, R.F.; Souto-Padrón, T.; De Souza, W.; Cayota, A. Gene expression changes induced by Trypanosoma cruzi shed microvesicles in mammalian host cells: Relevance of tRNA-derived halves. BioMed Res. Int. 2014, 2014, 305239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [Green Version]
- Martinez, G.; Choudury, S.G.; Slotkin, R.K. Choudury, and R.K. Slotkin, tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017, 45, 5142–5152. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef] [Green Version]
- Yeung, M.L.; Bennasser, Y.; Watashi, K.; Le, S.-Y.; Houzet, L.; Jeang, K.-T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: Evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009, 37, 6575–6586. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Yang, H.; Cheng, X.; Wang, D.; Fu, S.; Shen, W.; Zhang, Q.; Zhang, L.; Xue, Z.; Li, Y.; et al. tRF/miR-1280 Suppresses Stem Cell–like Cells and Metastasis in Colorectal Cancer. Cancer Res. 2017, 77, 3194–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.-H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [Green Version]
- Sarker, G.; Sun, W.; Rosenkranz, D.; Pelczar, P.; Opitz, L.; Efthymiou, V.; Wolfrum, C.; Peleg-Raibstein, D. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc. Natl. Acad. Sci. USA 2019, 116, 10547–10556. [Google Scholar] [CrossRef] [Green Version]
- Byeon, B.; Bilichak, A.; Kovalchuk, I. Transgenerational Response to Heat Stress in the Form of Differential Expression of Noncoding RNA Fragments in Brassica rapa Plants. Plant Genome 2019, 12, 180022. [Google Scholar] [CrossRef] [Green Version]
- Byeon, B.; Bilichak, A.; Kovalchuk, I. Computational Characterization of ncRNA Fragments in Various Tissues of the Brassica rapa Plant. Non-Coding RNA 2017, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Jehn, J.; Treml, J.; Wulsch, S.; Ottum, B.; Erb, V.; Hewel, C.; Kooijmans, R.N.; Wester, L.; Fast, I.; Rosenkranz, D. 5′ tRNA halves are highly expressed in the primate hippocampus and might sequence-specifically regulate gene expression. RNA 2020, 26, 694–707. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, L.M.; Edelstein, L.C.; Nagalla, S.; Woodley, A.B.; Chen, E.S.; Kong, X.; Ma, L.; Fortina, P.; Kunapuli, S.; Holinstat, M.; et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood 2014, 123, e37–e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mentzel, C.; Anthon, C.; Jacobsen, M.J.; Karlskov-Mortensen, P.; Bruun, C.S.; Jorgensen, C.B.; Gorodkin, J.; Cirera, S.; Fredholm, M. Gender and Obesity Specific MicroRNA Expression in Adipose Tissue from Lean and Obese Pigs. PLoS ONE 2015, 10, e0131650. [Google Scholar] [CrossRef] [PubMed]
- Freitak, D.; Knorr, E.; Vogel, H.; Vilcinskas, A. Gender- and stressor-specific microRNA expression in Tribolium castaneum. Biol. Lett. 2012, 8, 860–863. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Zhang, Q.; Ma, X.; Wang, J.; Liang, T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep. 2017, 7, 39812. [Google Scholar] [CrossRef] [Green Version]
- Dressman, D.; Ahearn, M.E.; Yariz, K.O.; Basterrecha, H.; Martínez, F.; Palau, F.; Barmada, M.M.; Clark, R.D.; Meindl, A.; Wirth, B.; et al. X-linked infantile spinal muscular atrophy: Clinical definition and molecular mapping. Genet. Med. 2007, 9, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Rowland, L.P.; Shneider, N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2009, 344, 1688–1700. [Google Scholar] [CrossRef]
- Kim, D.W.; Yao, Z.; Graybuck, L.T.; Kim, T.K.; Nguyen, T.N.; Smith, K.A.; Fong, O.; Yi, L.; Koulena, N.; Pierson, N.; et al. Multimodal analysis of cell types in a hypothalamic node controlling social behavior. Cell 2019, 179, 713. [Google Scholar] [CrossRef]
- Thomas, S.P.; Hoang, T.T.; Ressler, V.T.; Raines, R.T. Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor. RNA 2018, 24, 1018–1027. [Google Scholar] [CrossRef] [Green Version]
- Forman, R.A.; Deschoolmeester, M.L.; Hurst, R.J.M.; Wright, S.H.; Pemberton, A.D.; Else, K.J. The Goblet Cell Is the Cellular Source of the Anti-Microbial Angiogenin 4 in the Large Intestine Post Trichuris muris Infection. PLoS ONE 2012, 7, e42248. [Google Scholar] [CrossRef]
- Haack, F.; Trakooljul, N.; Gley, K.; Murani, E.; Hadlich, F.; Wimmers, K.; Ponsuksili, S. Deep sequencing of small non-coding RNA highlights brain-specific expression patterns and RNA cleavage. RNA Biol. 2019, 16, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, X.; Liu, C.; Liu, J.; Hu, Q.; Pan, T.; Duan, X.; Liu, B.; Zhang, Y.; Chen, J.; et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. J. Immunol. 2016, 196, 1591–1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varrault, A.; Dantec, C.; Le Digarcher, A.; Chotard, L.; Bilanges, B.; Parrinello, H.; Dubois, E.; Rialle, S.; Severac, D.; Bouschet, T.; et al. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 2017, 45, 10466–10480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, B.C.; Dimova, J.G.; Dakoji, S.; Yuan, L.-L.; Gewirtz, J.C.; Tran, P.V. Deletion of novel protein TMEM35 alters stress-related functions and impairs long-term memory in mice. Am. J. Physiol. Integr. Comp. Physiol. 2016, 311, R166–R178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petta, I.; Bougarne, N.; Vandewalle, J.; Dejager, L.; Vandevyver, S.; Ballegeer, M.; Desmet, S.; Thommis, J.; De Cauwer, L.; Lievens, S.; et al. Glucocorticoid Receptor-mediated transactivation is hampered by Striatin-3, a novel interaction partner of the receptor. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Larrieu, D.; Viré, E.; Robson, S.; Breusegem, S.Y.; Kouzarides, T.; Jackson, S.P. Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the Transportin-1 nuclear import pathway. Sci. Signal. 2018, 11, eaar5401. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Mychasiuk, R.; Muhammad, A.; Hossain, S.; Ilnytskyy, S.; Ghose, A.; Kirkby, C.; Ghasroddashti, E.; Kovalchuk, O.; Kolb, B. Liver irradiation causes distal bystander effects in the rat brain and affects animal behaviour. Oncotarget 2015, 7, 4385–4398. [Google Scholar] [CrossRef] [Green Version]
- Kovalchuk, A.; Mychasiuk, R.; Muhammad, A.; Hossain, S.; Ilnytskyy, Y.; Ghose, A.; Kirkby, C.; Ghasroddashti, E.; Kolb, B.; Kovalchuk, O. Profound and Sexually Dimorphic Effects of Clinically-Relevant Low Dose Scatter Irradiation on the Brain and Behavior. Front. Behav. Neurosci. 2016, 10, 84. [Google Scholar] [CrossRef] [Green Version]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2014, 43, D146–D152. [Google Scholar] [CrossRef]



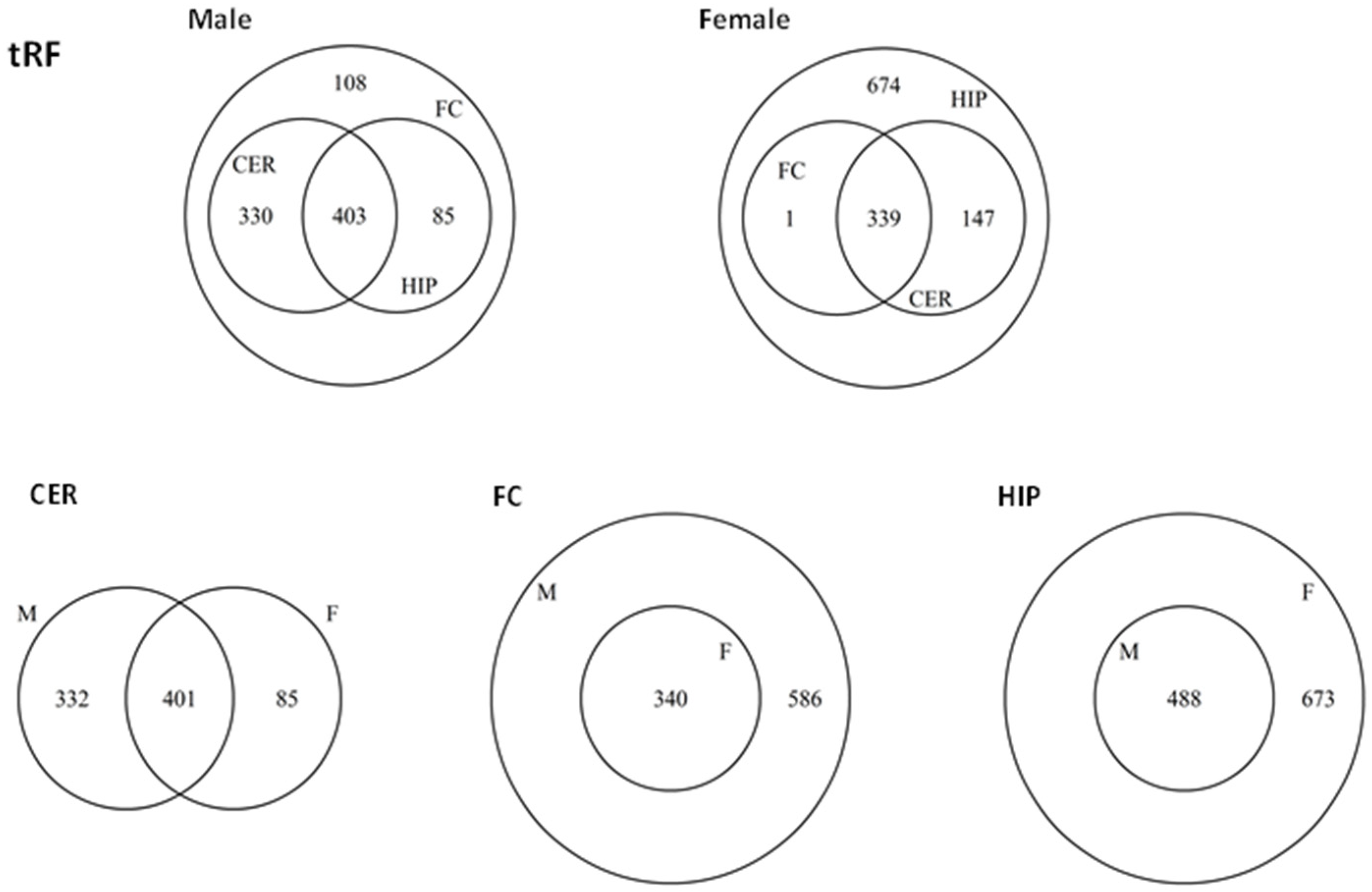
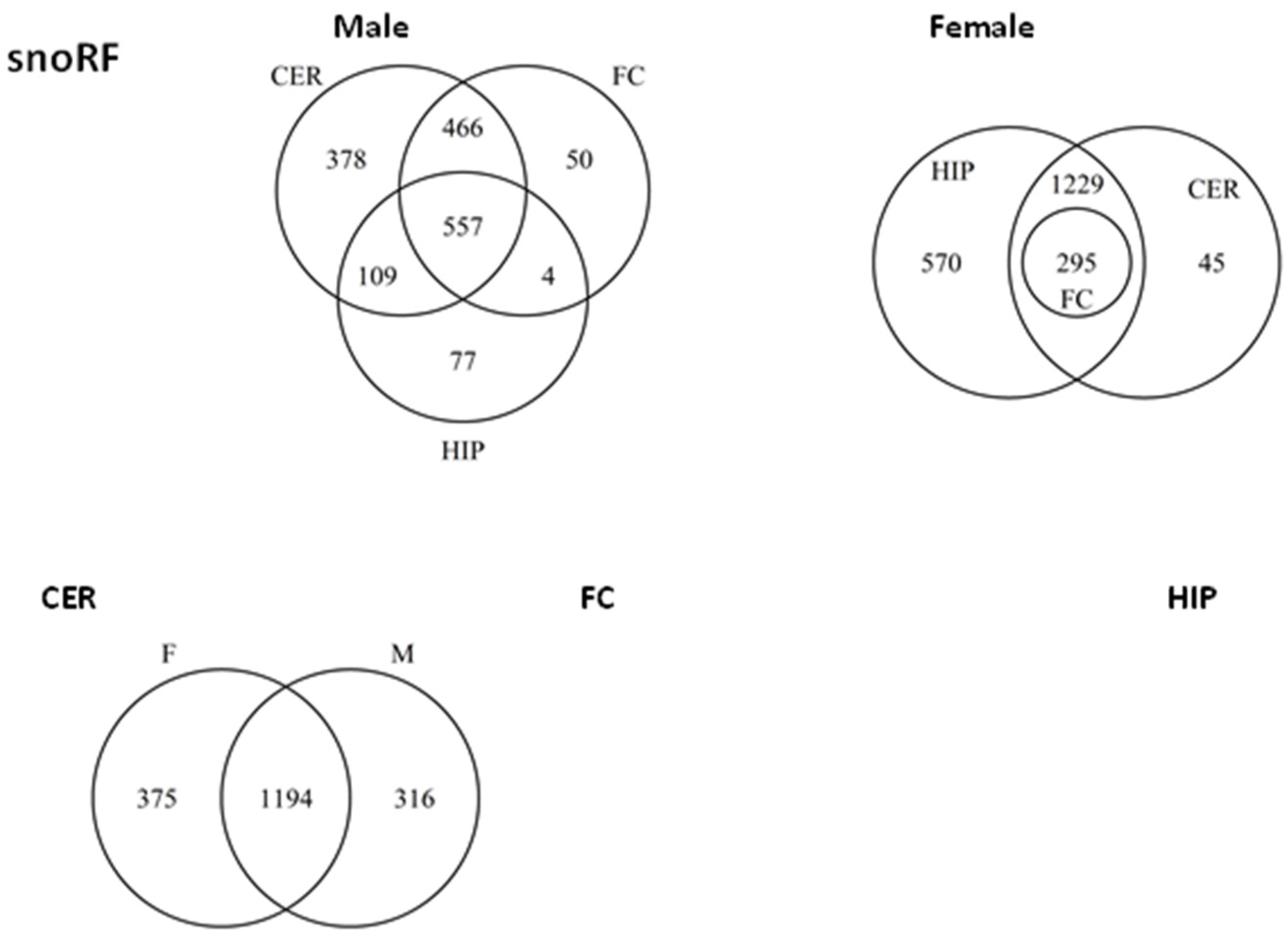
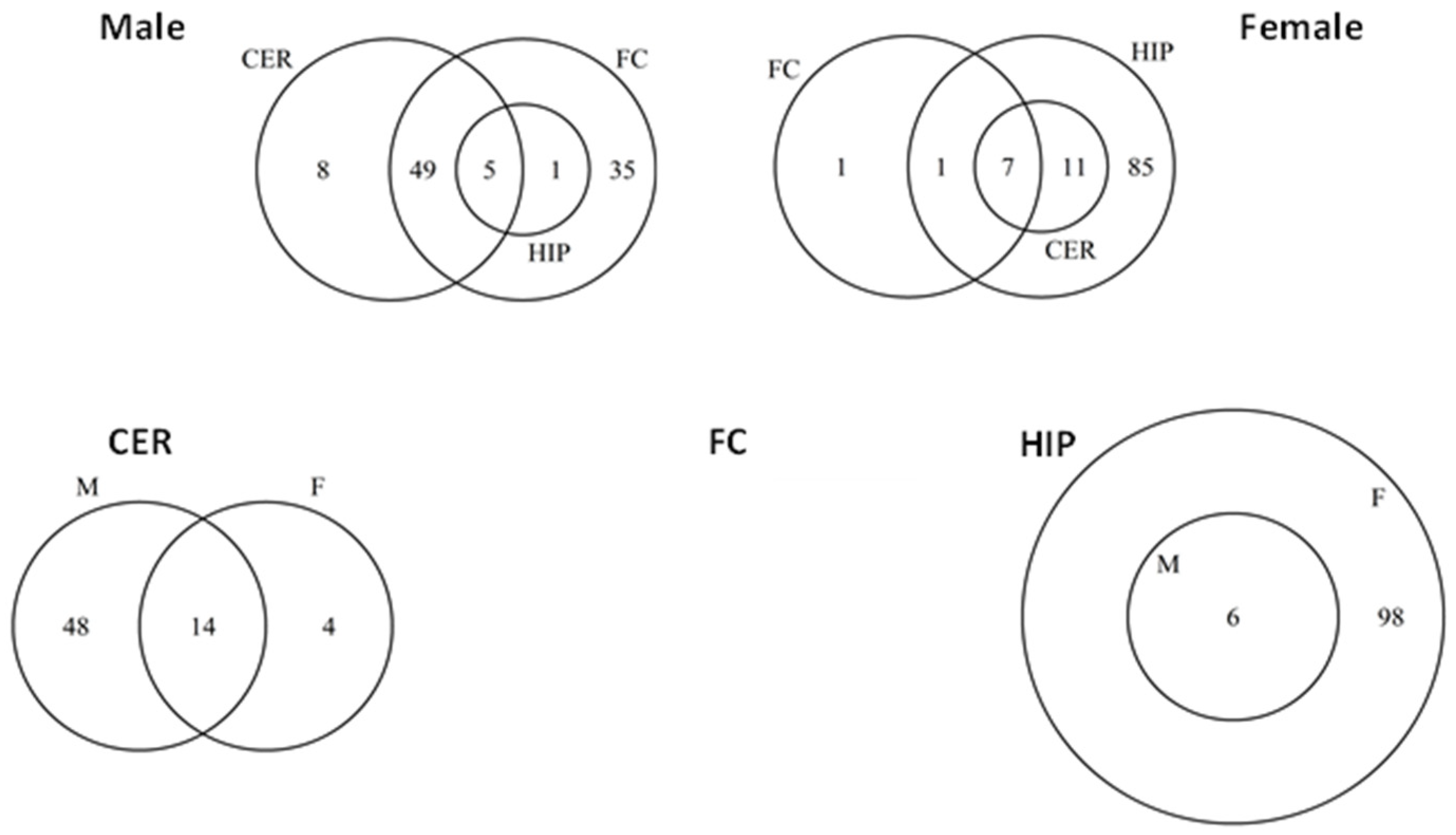
| FC_M | CER_M | HIP_M | FC_F | CER_F | HIP_F | ||
|---|---|---|---|---|---|---|---|
| tRF | Reads | 57,307.00 | 17,374.36 | 37,031.61 | 31,510.58 | 12,117.04 | 28,367.62 |
| % total reads | 98.1 | 88.8 | 94.6 | 97.4 | 83.6 | 93.3 | |
| Ratio/FC | 1.00 | 0.30 * | 0.65 | 1.00 | 0.38 * | 0.90 | |
| Ratio_M/F | 1.82 * | 1.43 | 1.31 | ||||
| rRF | Reads | 42.00 | 23.18 | 17.39 | 10.19 | 0.00 | 81.74 |
| % total reads | 7.2 × 10−4 | 1.2 × 10−3 | 4.4 × 10−4 | 3.1 × 10−4 | 0 | 2.7 × 10−3 | |
| Ratio/FC | 1.00 | 0.55 | 0.41 * | 1.00 | 0.00 | 8.02 * | |
| Ratio_M/F | 4.12 * | 0.21 * | |||||
| snRF | Reads | 87.00 | 117.33 | 893.96 | 24.46 | 196.79 | 746.73 |
| % total reads | 1.5 × 10−3 | 6.0 × 10−3 | 2.3 × 10−2 | 7.6 × 10−4 | 1.4 × 10−2 | 2.5 × 10−2 | |
| Ratio/FC | 1.00 | 1.35 | 10.28 * | 1.00 | 8.05 * | 30.53 * | |
| Ratio_M/F | 3.56 * | 0.60 | 1.20 | ||||
| snoRF | Reads | 989.00 | 2056.82 | 1219.20 | 815.23 | 2178.15 | 1200.29 |
| % total reads | 1.7 × 10−2 | 0.1 | 3.1 × 10−2 | 2.5 × 10−2 | 0.15 | 3.9 × 10−2 | |
| Ratio/FC | 1.00 | 2.08 * | 1.23 | 1.00 | 2.67 * | 1.47 | |
| Ratio_M/F | 1.21 | 0.94 | 1.02 | ||||
| Total | Reads | 58,425.00 | 19,571.69 | 39,162.16 | 32,360.46 | 14,491.98 | 30,396.38 |
| Brain Region | HIP | CER | FC | |||
|---|---|---|---|---|---|---|
| Sex | Male | Female | Male | Female | Male | Female |
| miRNA | = | = | = | = | = | = |
| ncRNA read size, nt | ||||||
| lincRNA | 20 | 21 | 26 | 25 | 20 | 23 |
| snRNA | 19 | 21 | 24 | 23 | 24 | 24 |
| antisense | 18 | 19 | 18 | 17 | 19 | 24 |
| tRNA | 26 | 26 | 26 | 26 | 18 | 18 |
| tRF-Gly | ~90% | ~90% | ~90% | ~90% | ~90% | ~90% |
| tRF-Glu | 5.8% | 3.5% | 1.8% | 2.0% | 2.0% | 1.7% |
| tRF-Lys | 3.9% | 4.5% | 3.3% | 4.0% | 2.9% | 3.3% |
| tRF-GlyGCC | = | = | +++ | ++ | +++ | +++ |
| tRF-ValCAC | --- | --- | --- | --- | --- | --- |
| tRF-ValACC | --- | --- | --- | --- | --- | --- |
| tRF-LysCTT | --- | --- | --- | --- | --- | --- |
| snoRD116-3′ | = | +++ | +++ | +++ | +++ | +++ |
| snoRA54-5′ | +++ | +++ | +++ | +++ | +++ | + |
| snoRA-3-5′ | +++ | = | = | = | +++ | = |
| 5_8SRNA | -- | = | = | = | = | = |
| U1 | ++ | +++ | = | = | -- | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiselier, A.; Byeon, B.; Ilnytskyy, Y.; Kovalchuk, I.; Kovalchuk, O. Sex-Specific Expression of Non-Coding RNA Fragments in Frontal Cortex, Hippocampus and Cerebellum of Rats. Epigenomes 2022, 6, 11. https://doi.org/10.3390/epigenomes6020011
Fiselier A, Byeon B, Ilnytskyy Y, Kovalchuk I, Kovalchuk O. Sex-Specific Expression of Non-Coding RNA Fragments in Frontal Cortex, Hippocampus and Cerebellum of Rats. Epigenomes. 2022; 6(2):11. https://doi.org/10.3390/epigenomes6020011
Chicago/Turabian StyleFiselier, Anna, Boseon Byeon, Yaroslav Ilnytskyy, Igor Kovalchuk, and Olga Kovalchuk. 2022. "Sex-Specific Expression of Non-Coding RNA Fragments in Frontal Cortex, Hippocampus and Cerebellum of Rats" Epigenomes 6, no. 2: 11. https://doi.org/10.3390/epigenomes6020011
APA StyleFiselier, A., Byeon, B., Ilnytskyy, Y., Kovalchuk, I., & Kovalchuk, O. (2022). Sex-Specific Expression of Non-Coding RNA Fragments in Frontal Cortex, Hippocampus and Cerebellum of Rats. Epigenomes, 6(2), 11. https://doi.org/10.3390/epigenomes6020011






