Purification, Identification and Characterization of Antioxidant Peptides from Corn Silk Tryptic Hydrolysate: An Integrated In Vitro-In Silico Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of CS Protein Isolate and Hydrolysate
2.3. Purification of Antioxidant Peptides from T1H
2.4. Identification of Purified Peptides
2.5. Determination of Antioxidant Activities
2.6. In Silico Analysis
2.6.1. Modelling of Peptide Structures
2.6.2. Prediction of Antioxidant Peptides and Docking to ABTS•+
2.6.3. Docking-Based Screening of Potential Inhibitors of Keap1, MPO, and XO
2.6.4. Prediction of Physicochemical Properties, Toxicity, Allergenicity, and Cell-Penetrating Potential
2.7. Statistical Analysis
3. Results and Discussion
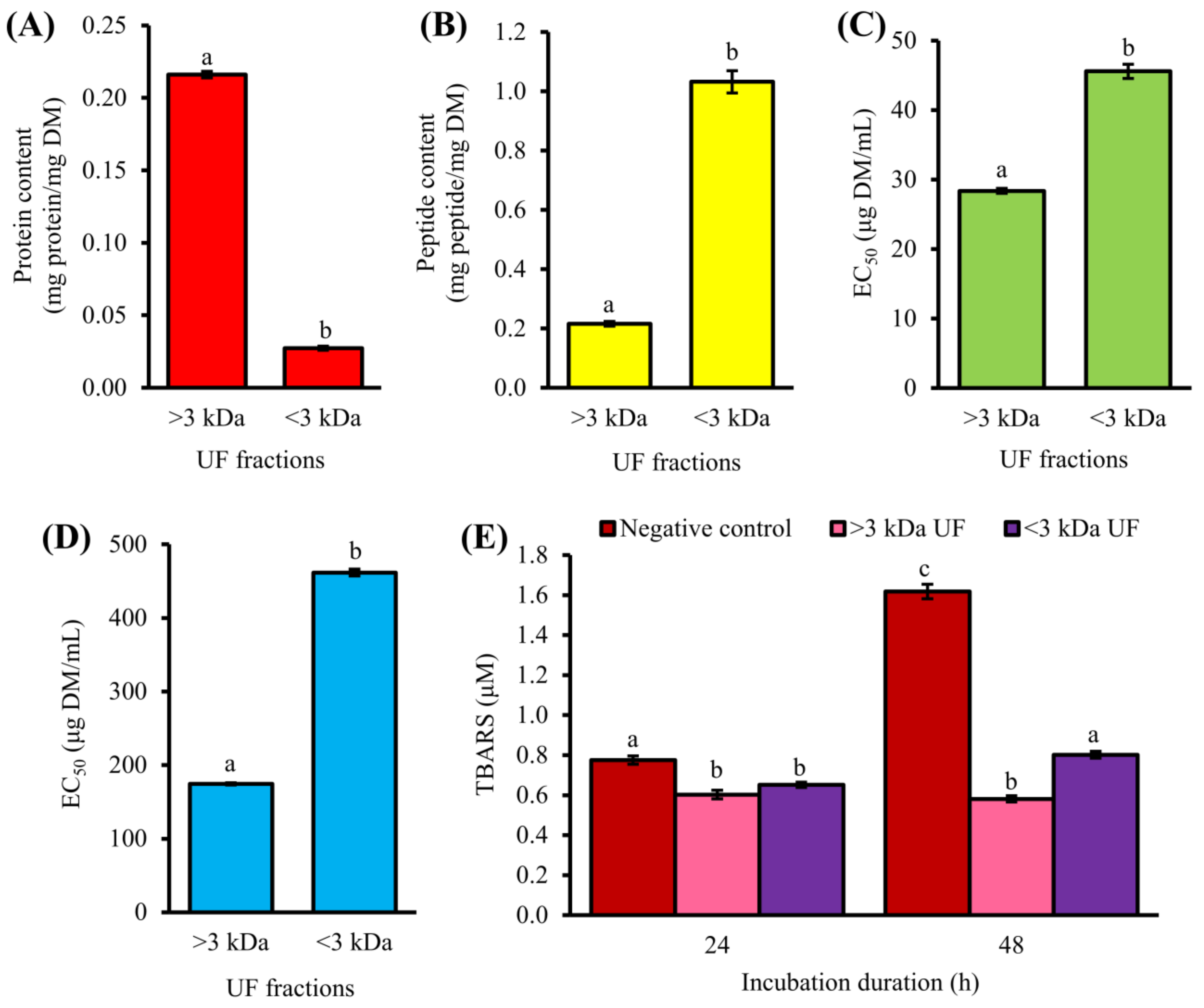
3.1. Purification of T1H by UF
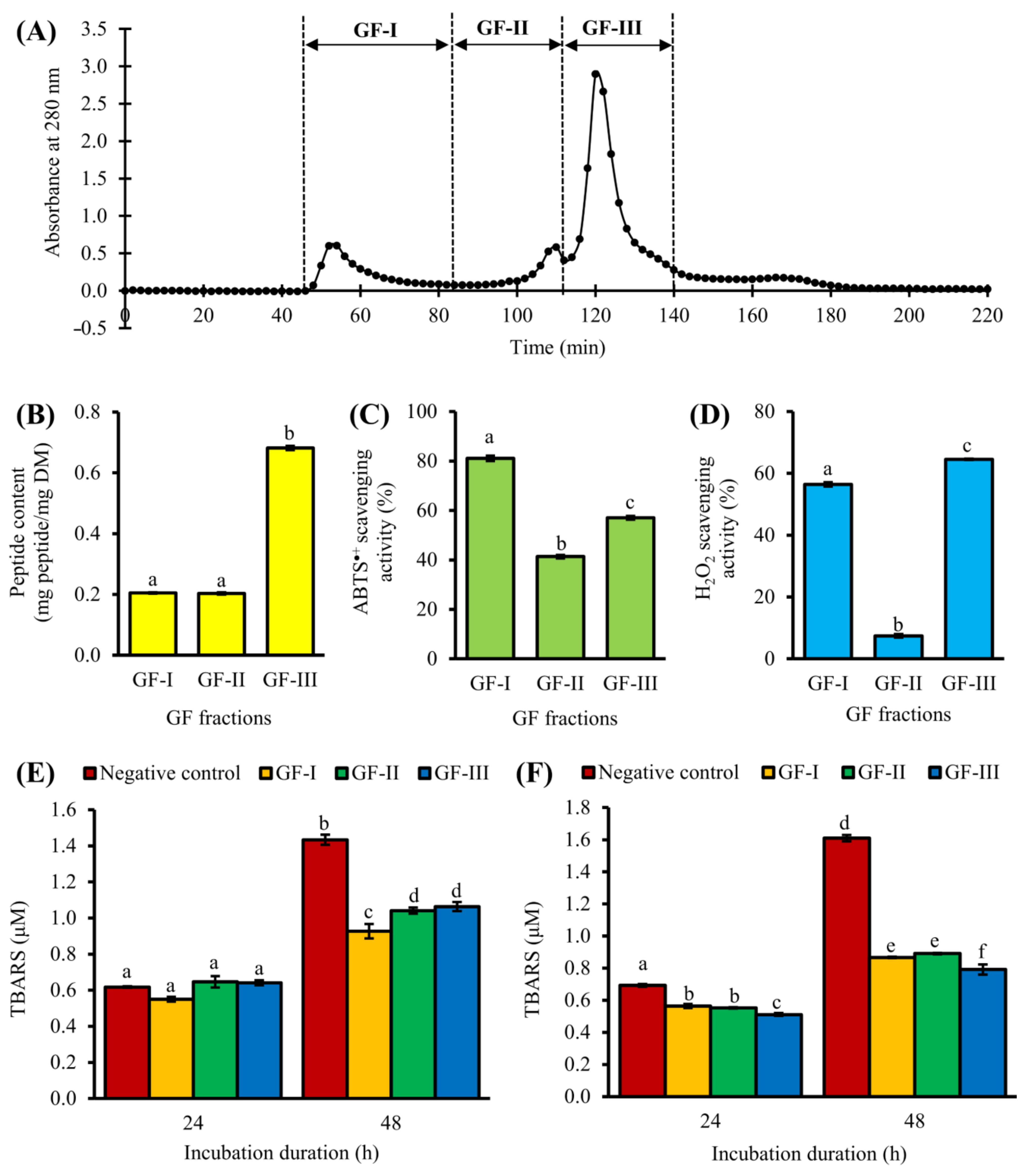
3.2. Purification of <3 kDa Fraction by GFC
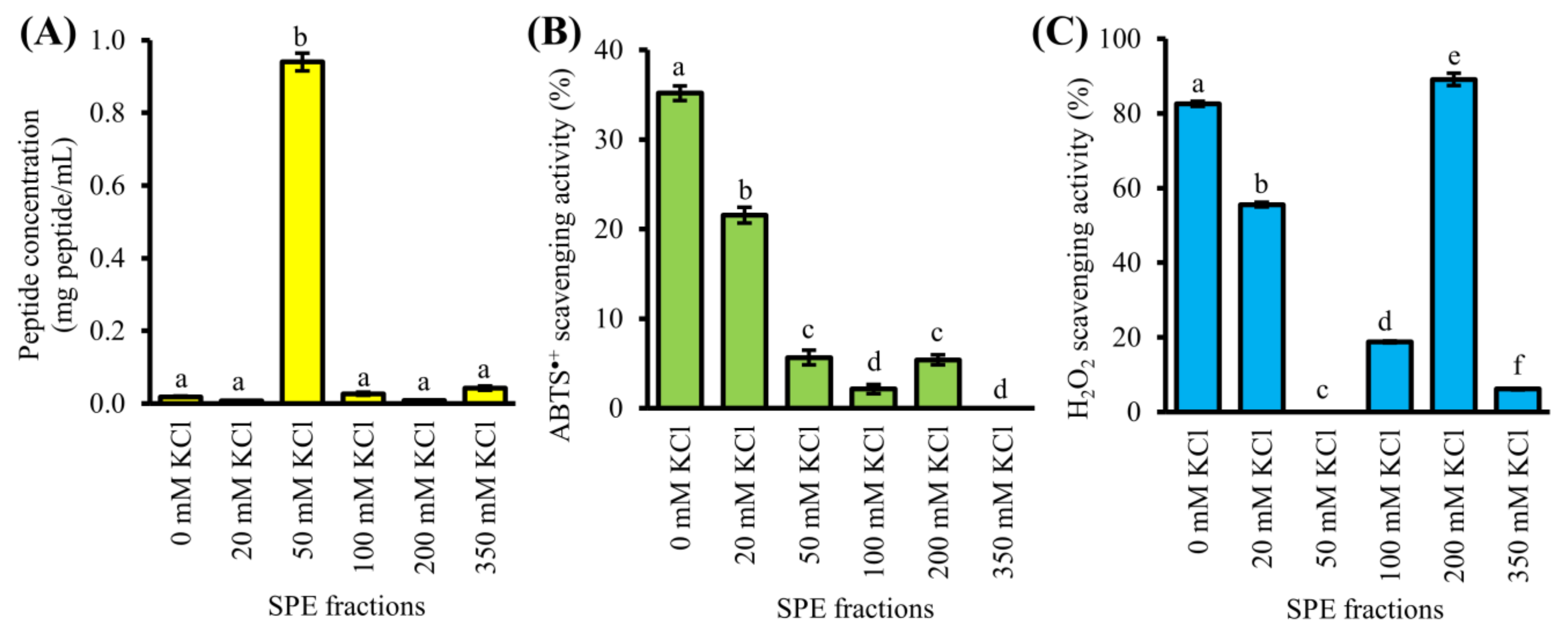
3.3. Purification of GF-III by SCX-SPE
3.4. Identification and Characterization of Antioxidant Peptides
3.5. Molecular Docking between CS Peptides and ABTS•+
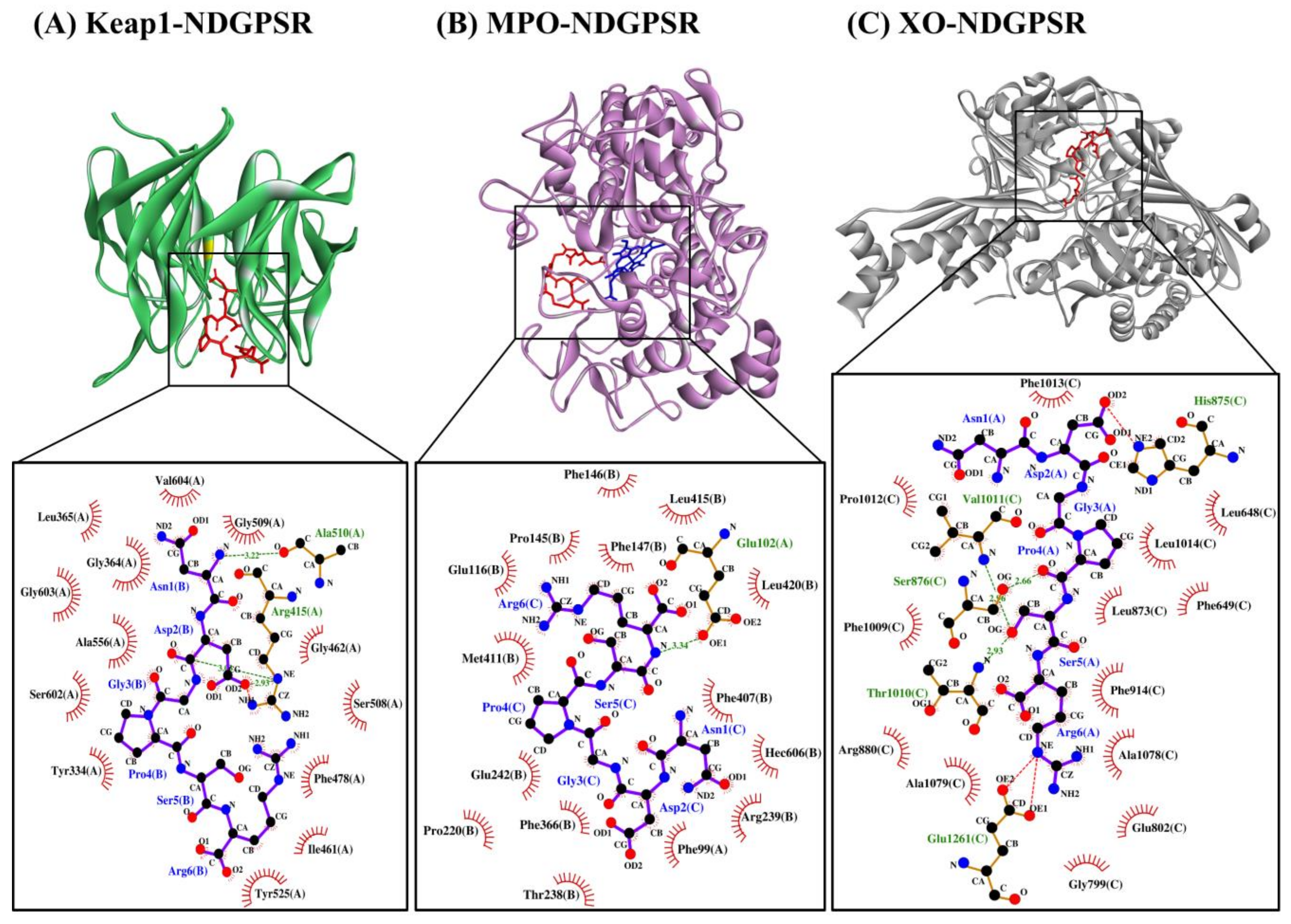
3.6. Molecular Docking of Peptides on Keap1
3.7. Molecular Docking of Peptides on MPO
3.8. Molecular Docking of Peptides on XO
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasanudin, K.; Hashim, P.; Mustafa, S. Corn silk (Stigma maydis) in healthcare: A phytochemical and pharmacological review. Molecules 2012, 17, 9697–9715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.-C.; Lee, Y.-C.; Lo, H.-Y.; Huang, Y.-W.; Hsiang, C.-Y.; Ho, T.-Y. Antihypertensive effects of corn silk extract and its novel bioactive constituent in spontaneously hypertensive rats: The involvement of angiotensin-converting enzyme inhibition. Molecules 2019, 24, 1886. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.-Y.; Li, C.-C.; Lo, H.-Y.; Chen, F.-Y.; Hsiang, C.-Y. Corn silk extract and its bioactive peptide ameliorated lipopolysaccharide-induced inflammation in mice via the nuclear factor-κB signaling pathway. J. Agric. Food Chem. 2017, 65, 759–768. [Google Scholar] [CrossRef]
- Chai, T.-T.; Ang, S.-Y.; Goh, K.; Lee, Y.-H.; Ngoo, J.-M.; Teh, L.-K.; Wong, F.-C. Trypsin-hydrolyzed corn silk proteins: Antioxidant activities, in vitro gastrointestinal and thermal stability, and hematoprotective effects. eFood 2020, 1, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Wong, F.-C.; Xiao, J.; Wang, S.; Ee, K.-Y.; Chai, T.-T. Advances on the antioxidant peptides from edible plant sources. Trends Food Sci. Technol. 2020, 99, 44–57. [Google Scholar] [CrossRef]
- Tonolo, F.; Moretto, L.; Grinzato, A.; Fiorese, F.; Folda, A.; Scalcon, V.; Ferro, S.; Arrigoni, G.; Bellamio, M.; Feller, E.; et al. Fermented soy-derived bioactive peptides selected by a molecular docking approach show antioxidant properties involving the Keap1/Nrf2 pathway. Antioxidants 2020, 9, 1306. [Google Scholar] [CrossRef]
- Winkel, A.F.; Engel, C.K.; Margerie, D.; Kannt, A.; Szillat, H.; Glombik, H.; Kallus, C.; Ruf, S.; Güssregen, S.; Riedel, J.; et al. Characterization of RA839, a noncovalent small molecule binder to Keap1 and selective activator of Nrf2 signaling. J. Biol. Chem. 2015, 290, 28446–28455. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Cui, C.; Wang, Y.; Ni, J.; Zheng, L.; Wei, H.-K.; Peng, J. FSGHF3 and peptides, prepared from fish skin gelatin, exert a protective effect on DSS-induced colitis via the Nrf2 pathway. Food Funct. 2020, 11, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Thaha, A.; Wang, B.-S.; Chang, Y.-W.; Hsia, S.-M.; Huang, T.-C.; Shiau, C.-Y.; Hwang, D.-F.; Chen, T.-Y. Food-derived bioactive peptides with antioxidative capacity, xanthine oxidase and tyrosinase inhibitory activity. Processes 2021, 9, 747. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Ong, M.G.-L.; Pang, M.-J.; Wong, S.-J.; Teh, L.-K.; Chai, T.-T. Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chem. 2019, 271, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chai, T.-T.; Kwek, M.-T.; Ong, H.-C.; Wong, F.-C. Water fraction of edible medicinal fern Stenochlaena palustris is a potent α-glucosidase inhibitor with concurrent antioxidant activity. Food Chem. 2015, 186, 26–31. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster denovo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [Green Version]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.H.; Yesiltas, B.; Marin, F.I.; Pertseva, M.; García-Moreno, P.J.; Gregersen, S.; Overgaard, M.T.; Jacobsen, C.; Lund, O.; Hansen, E.B.; et al. AnOxPePred: Using deep learning for the prediction of antioxidative properties of peptides. Sci. Rep. 2020, 10, 21471. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Schüttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Kochnev, Y.; Hellemann, E.; Cassidy, K.C.; Durrant, J.D. Webina: An open-source library and web app that runs AutoDock Vina entirely in the web browser. Bioinformatics 2020, 36, 4513–4515. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Shaw, S.A.; Vokits, B.P.; Dilger, A.K.; Viet, A.; Clark, C.G.; Abell, L.M.; Locke, G.A.; Duke, G.; Kopcho, L.M.; Dongre, A.; et al. Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg. Med. Chem. 2020, 28, 115723. [Google Scholar] [CrossRef]
- Cao, H.; Pauff, J.M.; Hille, R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J. Nat. Prod. 2014, 77, 1693–1699. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.-C.; Ong, J.-H.; Chai, T.-T. Identification of putative cell-entry-inhibitory peptides against SARS-CoV-2 from edible insects: An in silico study. eFood 2020, 1, 357–368. [Google Scholar] [CrossRef]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2--a server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- de Oliveira, E.C.L.; Santana, K.; Josino, L.; Lima, E.; Lima, A.H.; de Souza Sales Júnior, C. Predicting cell-penetrating peptides using machine learning algorithms and navigating in their chemical space. Sci. Rep. 2021, 11, 7628. [Google Scholar] [CrossRef] [PubMed]
- Sharmila, M.D.; Chai, T.-T.; Wong, F.-C. Antioxidant and protein protection potentials of fennel seed-derived protein hydrolysates and peptides. Mod. Food Sci. Technol. 2019, 35, 22–29. [Google Scholar]
- Hu, R.; Dunmire, K.M.; Truelock, C.N.; Paulk, C.B.; Aldrich, G.; Li, Y. Antioxidant performances of corn gluten meal and DDGS protein hydrolysates in food, pet food, and feed systems. J. Sci. Food Agric. 2020, 2, 100030. [Google Scholar] [CrossRef]
- Chai, T.-T.; Law, Y.-C.; Wong, F.-C.; Kim, S.-K. Enzyme-assisted discovery of antioxidant peptides from edible marine invertebrates: A review. Mar. Drugs 2017, 15, 42. [Google Scholar] [CrossRef]
- Feng, Y.-X.; Ruan, G.-R.; Jin, F.; Xu, J.; Wang, F.-J. Purification, identification, and synthesis of five novel antioxidant peptides from Chinese chestnut (Castanea mollissima Blume) protein hydrolysates. LWT 2018, 92, 40–46. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, X.; Kopparapu, N.; Cong, W.; Deng, Y.; Sun, X.; Liu, X. Purification and evaluation of a novel antioxidant peptide from corn protein hydrolysate. Process Biochem. 2014, 49, 1562–1569. [Google Scholar] [CrossRef]
- Chai, T.-T.; Xiao, J.; Mohana Dass, S.; Teoh, J.-Y.; Ee, K.-Y.; Ng, W.-J.; Wong, F.-C. Identification of antioxidant peptides derived from tropical jackfruit seed and investigation of the stability profiles. Food Chem. 2021, 340, 127876. [Google Scholar] [CrossRef]
- He, R.; Ju, X.; Yuan, J.; Wang, L.; Girgih, A.T.; Aluko, R.E. Antioxidant activities of rapeseed peptides produced by solid state fermentation. Food Res. Int. 2012, 49, 432–438. [Google Scholar] [CrossRef]
- Mishra, M.; Nagarajan, K. Assessment of antioxidant influence of short series peptides using hydrogen peroxide scavenging assay and superoxide radical scavenging activity. World J. Pharm. Pharm. Sci. 2018, 7, 1064–1070. [Google Scholar]
- Chen, T.; Chen, Z.; Wang, H.; Chen, X.; Yang, J.; Han, A.; Lin, D.-H.; Hong, J. Underlying action mechanism of a novel antioxidant peptide derived from Allium tuberosum Rottler protein hydrolysates and its protective effects on hydrogen peroxide induced cell injury. J. Funct. Foods 2018, 40, 606–613. [Google Scholar] [CrossRef]
- Kovacs-Nolan, J.; Zhang, H.; Ibuki, M.; Nakamori, T.; Yoshiura, K.; Turner, P.V.; Matsui, T.; Mine, Y. The PepT1-transportable soy tripeptide VPY reduces intestinal inflammation. Biochim. Biophys. Acta 2012, 1820, 1753–1763. [Google Scholar] [CrossRef]
- He, S.; Zhang, Y.; Sun, H.; Du, M.; Qiu, J.; Tang, M.; Sun, X.; Zhu, B. Antioxidative peptides from proteolytic hydrolysates of false abalone (Volutharpa ampullacea perryi): Characterization, identification, and molecular docking. Mar. Drugs 2019, 17, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; He, S.; Bonneil, É.; Simpson, B.K. Generation of antioxidative peptides from Atlantic sea cucumber using alcalase versus trypsin: In vitro activity, de novo sequencing, and in silico docking for in vivo function prediction. Food Chem. 2020, 306, 125581. [Google Scholar] [CrossRef]
- Zhong, H.; Abdullah; Zhang, Y.; Deng, L.; Zhao, M.; Tang, J.; Zhang, H.; Feng, F.; Wang, J. Exploring the potential of novel xanthine oxidase inhibitory peptide (ACECD) derived from Skipjack tuna hydrolysates using affinity-ultrafiltration coupled with HPLC-MALDI-TOF/TOF-MS. Food Chem. 2021, 347, 129068. [Google Scholar] [CrossRef] [PubMed]
| SPE Fractions | Peptides | Measured m/z [M + 2H]2 | Molecular Mass (Da) a | Aromatic Residues (%) b | Basic Residues (%) b | Hydrophobic Residues (%) b | Aliphatic Residues (%) b | Aliphatic Index b |
|---|---|---|---|---|---|---|---|---|
| 0 mM KCl | KRYFKR | 449.28 | 896.57 | 33 | 67 | 33 | 0 | 0 |
| PRVRVAGR | 455.79 | 909.58 | 0 | 38 | 63 | 38 | 85 | |
| PVVWAAKR | 463.79 | 925.57 | 13 | 25 | 75 | 50 | 98 | |
| QVASGPLQR | 478.28 | 954.55 | 0 | 11 | 56 | 33 | 87 | |
| MAPRTPRK | 478.78 | 955.57 | 0 | 38 | 50 | 13 | 13 | |
| NKVVKLMR | 494.31 | 986.62 | 0 | 38 | 50 | 38 | 121 | |
| KVPLAVFSR | 508.82 | 1015.64 | 11 | 22 | 67 | 44 | 119 | |
| LKKGSPLKR | 513.84 | 1025.69 | 0 | 44 | 44 | 22 | 87 | |
| FQLKPVFR | 517.82 | 1033.63 | 25 | 25 | 63 | 25 | 85 | |
| THAVKGVVHK | 538.34 | 1074.67 | 20 | 40 | 50 | 40 | 97 | |
| YTWKFKGR | 543.31 | 1084.61 | 38 | 38 | 50 | 0 | 0 | |
| ARVPQQSYR | 552.80 | 1103.61 | 11 | 22 | 44 | 22 | 43 | |
| VHFNKGKKR | 557.34 | 1112.69 | 22 | 56 | 33 | 11 | 32 | |
| TAPLSSKALKR | 586.37 | 1170.73 | 0 | 27 | 45 | 36 | 89 | |
| FSCPLVMKGPNGLR | 759.91 | 1517.81 | 7 | 14 | 71 | 21 | 76 | |
| 20 mM KCl | RHGSGR | 335.18 | 668.37 | 17 | 50 | 33 | 0 | 0 |
| NMVPGR | 337.17 | 672.34 | 0 | 17 | 67 | 17 | 48 | |
| FMFFVYK | 491.25 | 980.50 | 57 | 14 | 86 | 14 | 41 | |
| MCFHHHFHK | 612.27 | 1222.53 | 67 | 56 | 44 | 0 | 0 | |
| 200 mM KCl | DFPGAK | 317.66 | 633.33 | 17 | 17 | 67 | 17 | 17 |
| NDGPSR | 323.15 | 644.29 | 0 | 17 | 33 | 0 | 0 | |
| AGFPLGK | 345.20 | 688.41 | 14 | 14 | 86 | 29 | 70 | |
| AMQQDK | 360.66 | 719.32 | 0 | 17 | 33 | 17 | 17 | |
| NLEGYR | 376.19 | 750.38 | 17 | 17 | 50 | 17 | 65 | |
| YETLNR | 398.20 | 794.41 | 17 | 17 | 33 | 17 | 65 | |
| MPPKSTR | 408.72 | 815.43 | 0 | 29 | 43 | 0 | 0 | |
| TAGASLVAR | 423.25 | 844.49 | 0 | 11 | 67 | 56 | 109 | |
| SSPATGGSLR | 466.74 | 931.49 | 0 | 10 | 50 | 20 | 49 | |
| NANSLAGPQR | 514.27 | 1026.55 | 0 | 10 | 50 | 30 | 59 |
| Peptides | SPE Fractions | FRS Scores |
|---|---|---|
| MCFHHHFHK | 20 mM KCl | 0.68068 |
| VGPWQK * | - | 0.52254 |
| MYPGLA * | - | 0.49386 |
| NLEGYR | 200 mM KCl | 0.48158 |
| AGFPLGK | 200 mM KCl | 0.44866 |
| FMFFVYK | 20 mM KCl | 0.44397 |
| NMVPGR | 20 mM KCl | 0.44319 |
| PVVWAAKR | 0 mM KCl | 0.43744 |
| DFPGAK | 200 mM KCl | 0.43574 |
| FPLPSF * | - | 0.43352 |
| FSCPLVMKGPNGLR | 0 mM KCl | 0.41864 |
| WAFAPA * | - | 0.41519 |
| RHGSGR | 20 mM KCl | 0.41088 |
| VHFNKGKKR | 0 mM KCl | 0.41055 |
| NANSLAGPQR | 200 mM KCl | 0.40415 |
| QVASGPLQR | 0 mM KCl | 0.40213 |
| MAPRTPRK | 0 mM KCl | 0.39973 |
| NDGPSR | 200 mM KCl | 0.38760 |
| KRYFKR | 0 mM KCl | 0.38352 |
| YETLNR | 200 mM KCl | 0.37938 |
| FQLKPVFR | 0 mM KCl | 0.37599 |
| ARVPQQSYR | 0 mM KCl | 0.37580 |
| YTWKFKGR | 0 mM KCl | 0.36769 |
| AMQQDK | 200 mM KCl | 0.36324 |
| SSPATGGSLR | 200 mM KCl | 0.35382 |
| THAVKGVVHK | 0 mM KCl | 0.35200 |
| MPPKSTR | 200 mM KCl | 0.33529 |
| LKKGSPLKR | 0 mM KCl | 0.32957 |
| PRVRVAGR | 0 mM KCl | 0.32698 |
| KVPLAVFSR | 0 mM KCl | 0.32525 |
| TAGASLVAR | 200 mM KCl | 0.32285 |
| TAPLSSKALKR | 0 mM KCl | 0.29320 |
| NKVVKLMR | 0 mM KCl | 0.27437 |
| Peptides | SPE Fractions | Binding Affinity (kcal/mol) | Peptide Residues Interacting with ABTS•+ a | |
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | |||
| MCFHHHFHK | 20 mM KCl | −4.8 | - | Phe3, His6, Phe7 |
| VHFNKGKKR | 0 mM KCl | −4.7 | Lys7, Arg9 | Val1, His2, Gly6, Lys7, Arg9 |
| PVVWAAKR | 0 mM KCl | −4.7 | Arg8 (2) | Val2, Trp4, Ala5, Ala6, Arg8 |
| FMFFVYK | 20 mM KCl | −4.4 | Lys7 | Phe1, Phe3, Phe4, Lys7 |
| FSCPLVMKGPNGLR | 0 mM KCl | −4.2 | Arg14 (2) | Leu5, Lys8, Gly9, Pro10, Gly12, Arg14 |
| NMVPGR | 20 mM KCl | −4.1 | Asn1, Arg6 (2) | Asn1, Pro4, Gly5, Arg6 |
| NLEGYR | 200 mM KCl | −4.1 | - | Tyr5, Arg6 |
| RHGSGR | 20 mM KCl | −3.9 | Arg1, Arg6 | Arg1, Gly5, Arg6 |
| AGFPLGK | 200 mM KCl | −3.7 | - | Phe3, Pro4, Leu5 |
| DFPGAK | 200 mM KCl | −3.6 | - | Pro3, Gly4, Lys6 |
| FPLPSF * | - | −4.6 | Phe1, Ser5 | Phe1, Pro2, Leu3, Pro4, Ser5 |
| WAFAPA * | - | −4.3 | - | Trp1, Ala4, Pro5 |
| VGPWQK * | - | −3.9 | - | Pro3, Trp4, Lys6 |
| MYPGLA * | - | −3.8 | Pro3 | Pro3, Leu5, Ala6 |
| Peptides a | Basic Residues | Mutant Peptides | Binding Affinity (kcal/mol) |
|---|---|---|---|
| MCFHHHFHK | His6 | MCFHHAFHK | −4.1 |
| VHFNKGKKR | His2 | VAFNKGKKR | −4.8 |
| Lys7 | VHFNKGAKR | −5.0 | |
| Arg9 | VHFNKGKKA | −4.4 | |
| PVVWAAKR | Arg8 | PVVWAAKA | −4.3 |
| FMFFVYK | Lys7 | FMFFVYA | −4.3 |
| FSCPLVMKGPNGLR | Lys8 | FSCPLVMAGPNGLR | −4.2 |
| Arg14 | FSCPLVMKGPNGLA | −4.7 | |
| NMVPGR | Arg6 | NMVPGA | −3.1 |
| NLEGYR | Arg6 | NLEGYA | −3.7 |
| RHGSGR | Arg1 | AHGSGR | −3.8 |
| Arg6 | RHGSGA | −4.2 | |
| DFPGAK | Lys6 | DFPGAA | −3.4 |
| Peptides | Toxicity | Allergenicity | CPP Prediction |
|---|---|---|---|
| NDGPSR | Non-toxin | Probable non-allergen | CPP |
| NLEGYR | Non-toxin | Probable non-allergen | CPP |
| NMVPGR | Non-toxin | Probable non-allergen | CPP |
| SSPATGGSLR | Non-toxin | Probable non-allergen | CPP |
| NANSLAGPQR | Non-toxin | Probable non-allergen | CPP |
| KRYFKR | Non-toxin | Probable non-allergen | CPP |
| RHGSGR | Non-toxin | Probable non-allergen | CPP |
| YETLNR | Non-toxin | Probable non-allergen | Non-CPP |
| AGFPLGK | Non-toxin | Probable non-allergen | Non-CPP |
| KVPLAVFSR | Non-toxin | Probable non-allergen | Non-CPP |
| TAGASLVAR | Non-toxin | Probable allergen | Non-CPP |
| YTWKFKGR | Non-toxin | Probable allergen | CPP |
| AMQQDK | Non-toxin | Probable allergen | CPP |
| MPPKSTR | Non-toxin | Probable allergen | CPP |
| PVVWAAKR | Non-toxin | Probable allergen | CPP |
| DFPGAK | Non-toxin | Probable allergen | Non-CPP |
| FMFFVYK | Non-toxin | Probable allergen | Non-CPP |
| QVASGPLQR | Non-toxin | Probable allergen | Non-CPP |
| DEQIPSHPPR * | Non-toxin | Probable allergen | Non-CPP |
| DTETGVPT * | Non-toxin | Probable non-allergen | Non-CPP |
| VPY * | Non-toxin | Probable allergen | CPP |
| ACECD * | Non-toxin | Probable allergen | CPP |
| Peptides | Binding Affinity (kcal/mol) | Interaction with Keap1 a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NLEGYR | −8.7 | Arg415, Arg483, Ser508, Gln530, Ser555 | Tyr334, Ser363, Gly364, Leu365, Ala366, Arg415, Ile416, Gly417, Gly462, Phe478, Arg483, Ser508, Gly509, Ala510, Tyr525, Gln530, Ser555, Ala556, Leu557, Tyr572, Phe577, Ser602, Gly603, Val604 | Arg415 |
| NANSLAGPQR | −8.2 | Arg415 (3), Val418, Val465, Arg483 | Ser363, Gly364, Leu365, Arg380, Asn382, Asn414, Arg415, Ile416, Gly417, Ile461, Gly462, Val463, Val465, Phe478, Arg483, Ser508, Gly509, Tyr525, Gln530, Ser555, Ala556, Ile559, Phe577, Gly603 | - |
| NMVPGR | −8.1 | Ser363, Leu365, Asn382, Ser602 | Tyr334, Ser363, Gly364, Leu365, Ala366, Asn382, Arg415, Ile416, Ile461, Gly462, Ser508, Gly509, Ala510, Tyr525, Gln530, Ser555, Ala556, Ser602 | - |
| SSPATGGSLR | −8.1 | Ser363, Arg380, Asn414, Arg415, Ser431, Ser602 | Tyr334, Gly364, Leu365, Arg380, Asn382, Asn414, Arg415, Ile416, Ser431, Gly433, His436, Gly462, Phe478, Arg483, Ser508, Gly509, Ala556, Ser602, Gly603 | - |
| NDGPSR | −8.0 | Arg415 (2), Ala510 | Tyr334, Gly364, Leu365, Arg415, Ile461, Gly462, Phe478, Ser508, Gly509, Tyr525, Ala556, Ser602, Gly603, Val604 | - |
| DEQIPSHPPR * | −8.0 | Tyr334, Asn414, Arg415 (4), Ser431, Arg483 (3), Ser555 | Tyr334, Ser363, Arg380, Asn382, Asn414, Arg415, Ser431, Gly433, His436, Gly462, Phe478, Arg483, Ser508, Gly509, Tyr525, Ser555, Ala556, Tyr572, Phe577, Ser602 | Arg483 (2) |
| Peptides | Binding Affinity (kcal/mol) | Interaction with MPO a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NMVPGR | −6.6 | - | Phe99, Thr100, Glu102, Glu116, Pro145, Phe147, Leu216, Pro220, Arg239, Glu242, Phe366, Phe407, Met411, Arg424, Hec606 | - |
| NLEGYR | −6.5 | His95 | His95, Phe99, Glu102, Glu116, Pro145, Phe146, Phe147, Pro220, Thr238, Arg239, Glu242, Phe407, Val410, Met411, Leu420, Hec606 | - |
| NDGPSR | −6.3 | Glu102 | Phe99, Glu102, Glu116, Pro145, Phe146, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Leu420, Hec606 | - |
| RHGSGR | −6.2 | Thr100, Thr238 | Phe99, Thr100, Glu102, Pro145, Phe146, Phe147, Leu216, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Hec606 | Glu102 (5) |
| KRYFKR | −5.5 | Thr100, Thr238 | His95, Phe99, Thr100, Glu102, Glu116, Pro145, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Val410, Met411, Leu415, Leu420, Hec606 | Glu102 (2) |
| VPY * | −7.4 | - | His95, Phe99, Thr100, Glu102, Pro220, Thr238, Arg239, Glu242, Phe366, Hec606 | - |
| DTETGVPT * | −5.5 | Thr238 | Phe99, Thr100, Glu102, Phe147, Pro220, Thr238, Arg239, Glu242, Phe366, Phe407, Met411, Leu415, Leu420, Hec606 | - |
| Peptides | Binding Affinity (kcal/mol) | Interaction with XO a | ||
|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | Salt Bridge | ||
| NDGPSR | −5.2 | Ser876, Thr1010, Val1011 | Leu648, Phe649, Gly799, Glu802, Leu873, His875, Ser876, Arg880, Phe914, Phe1009, Thr1010, Val1011, Pro1012, Phe1013, Leu1014, Ala1078, Ala1079, Glu1261 | His875, Glu1261 (2) |
| ACECD * | −5.2 | His875, Ser876 | Leu648, Phe649, Glu802, Leu873, His875, Ser876, Glu879, Phe914, Phe1009, Thr1010, Val1011, Pro1012, Phe1013, Leu1014 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, J.-H.; Koh, J.-A.; Cao, H.; Tan, S.-A.; Abd Manan, F.; Wong, F.-C.; Chai, T.-T. Purification, Identification and Characterization of Antioxidant Peptides from Corn Silk Tryptic Hydrolysate: An Integrated In Vitro-In Silico Approach. Antioxidants 2021, 10, 1822. https://doi.org/10.3390/antiox10111822
Ong J-H, Koh J-A, Cao H, Tan S-A, Abd Manan F, Wong F-C, Chai T-T. Purification, Identification and Characterization of Antioxidant Peptides from Corn Silk Tryptic Hydrolysate: An Integrated In Vitro-In Silico Approach. Antioxidants. 2021; 10(11):1822. https://doi.org/10.3390/antiox10111822
Chicago/Turabian StyleOng, Joe-Hui, Jiun-An Koh, Hui Cao, Sheri-Ann Tan, Fazilah Abd Manan, Fai-Chu Wong, and Tsun-Thai Chai. 2021. "Purification, Identification and Characterization of Antioxidant Peptides from Corn Silk Tryptic Hydrolysate: An Integrated In Vitro-In Silico Approach" Antioxidants 10, no. 11: 1822. https://doi.org/10.3390/antiox10111822
APA StyleOng, J.-H., Koh, J.-A., Cao, H., Tan, S.-A., Abd Manan, F., Wong, F.-C., & Chai, T.-T. (2021). Purification, Identification and Characterization of Antioxidant Peptides from Corn Silk Tryptic Hydrolysate: An Integrated In Vitro-In Silico Approach. Antioxidants, 10(11), 1822. https://doi.org/10.3390/antiox10111822








