Abstract
The olive tree is a venerable Mediterranean plant and often used in traditional medicine. The main aim of the present study was to evaluate the effect of Olea europaea L. cv. Arbosana leaf extract (OLE) and its encapsulation within a spanlastic dosage form on the improvement of its pro-oxidant and antiproliferative activity against HepG-2, MCF-7, and Caco-2 human cancer cell lines. The LC-HRESIMS-assisted metabolomic profile of OLE putatively annotated 20 major metabolites and showed considerable in vitro antiproliferative activity against HepG-2, MCF-7, and Caco-2 cell lines with IC50 values of 9.2 ± 0.8, 7.1 ± 0.9, and 6.5 ± 0.7 µg/mL, respectively. The encapsulation of OLE within a (spanlastic) nanocarrier system, using a spraying method and Span 40 and Tween 80 (4:1 molar ratio), was successfully carried out (size 41 ± 2.4 nm, zeta potential 13.6 ± 2.5, and EE 61.43 ± 2.03%). OLE showed enhanced thermal stability, and an improved in vitro antiproliferative effect against HepG-2, MCF-7, and Caco-2 (IC50 3.6 ± 0.2, 2.3 ± 0.1, and 1.8 ± 0.1 µg/mL, respectively) in comparison to the unprocessed extract. Both preparations were found to exhibit pro-oxidant potential inside the cancer cells, through the potential inhibitory activity of OLE against glutathione reductase and superoxide dismutase (IC50 1.18 ± 0.12 and 2.33 ± 0.19 µg/mL, respectively). These inhibitory activities were proposed via a comprehensive in silico study to be linked to the presence of certain compounds in OLE. Consequently, we assume that formulating such a herbal extract within a suitable nanocarrier would be a promising improvement of its therapeutic potential.
1. Introduction
Olea europaea is a venerable cultural plant habitat in the Mediterranean [1]. The cultivation of this tree extends back many thousands of years and perhaps took place in the Eastern Mediterranean [1]. More than 300 original olive varieties have been developed worldwide to produce olive products [1]. These outputs represent a significant branch of industry in the Mediterranean regions, especially Spain, Italy, and Greece [1]. O. europaea is a critical tree, which requires limited demands on the soil and atmosphere and is several hundred years old [1].
Olive fruits and leaves are well-known for their therapeutic potential in many diseases [2]. The therapeutic utilities of olive trees, particularly their leaves, have been pointed out in folk medicine [3]. Studies have shown that it can reduce blood glucose level, serum cholesterol, and uric acid [3]. Additionally, olive fruits treat diabetes, elevated blood pressure, inflammatory disorders, diarrhea, respiratory tract disorders, urinary tract infections, intestinal diseases, and hemorrhoids [1,2,3,4]. Many phenolic olive leaf-derived compounds, principally iridoids and secoiridoids [4], and their pharmacological potentiality have been the focus of concern for investigation in the last decade [5,6]. Olive fruits are edible [7], with many biologically phenolic compounds [8]. Volatile compounds from O. europaea fruits and leaves and their functions in aroma improvement have also been an intense field of modern research [9].
Despite the vast range of therapeutic effects of such components, their optimum use has been hindered by low stability under different pH and temperature conditions, their hydrophobicity, and compliance due to the bitter taste [10]. Therefore, there was an essential need to design advanced delivery systems to cope with these aspects of these components. Many nanocarriers were developed to promote the bioavailability and therapeutic potential of naturally active products and their components [11,12,13]. The liposome of O. europaea extract was recently formulated by González-Ortega et al., 2020 [14], who reported enhanced stability for the extract components when encapsulated within a lipid-based formulation.
Spanlastics are nanovesicular systems that are formulated using Spans with an edge activator. Many active agents can be enclosed in the bilayer formed by the nonionic surfactants [15]. Spanlastics show comparable stability compared to other forms such as liposomes. Unlike other nanocarriers that contain cationic surfactants, they are nonirritant and provide improved delivery due to their deformable elastic nature [16].
During the last ten years, many reports have outlined the positive correlation between the pro-oxidant capacity and the antiproliferative effects of several chemical substances, particularly herbal extracts and polyphenolic compounds [17,18,19,20,21].
Across the world, cancer has a major impact on society. Cancer reports clearly show the aggressiveness of cancer and the difficulty of investigation in the initial stages. Prevention through new strategies is a challenge. Currently, the recognition of modern remedial strategies concentrating on inhibiting tumor cell proliferation has grown into a significant demand. In recent decades, the research of the anticancer properties of food ingredients has received further attention. Recently, researchers have been focusing on engaging in the benefits of the Mediterranean diet, longevity, and quality of life [21].
Herein, we aimed to evaluate the effect of OLE and its encapsulation within the spanlastic nanoformulation on the improvement of its pro-oxidant and, in turn, its antiproliferative activity toward HepG-2, MCF-7, and Caco-2 cancer cell lines. Additionally, we integrated LC-HRMS metabolic profiling along with several in silico studies (e.g., inverse docking, molecular dynamic simulation, binding free energy calculations) to highlight the main bioactive components and suggest a probable mode of action.
2. Materials and Methods
2.1. Plant Material
O. europaea leaves were collected in April 2020 from Basita farms, Aljouf, KSA. It was identified by Hamdan Ogereef, Camel and Range Research Center, Sakaka, Saudi Arabia. The samples are kept under voucher specimen number (2020-BuPD 75) at Pharmacognosy Department, Faculty of Pharmacy, Beni-Suef University, Egypt. According to the departmental/institutional/local/national guidelines in Egypt, permissions and approvals are not required on plant research.
2.2. Chemicals and Reagents
Chemicals consumed in this study, if not mentioned, were purchased from Sigma-Aldrich (St. Louis, MO, USA). Doxorubicin, the anticancer-positive control, was supplied from Merck Company, Germany. WI-38, HepG-2, MCF-7, and Caco-2 cancer cell lines were from the American Type Culture Collection and continuously subcultured twice per week. WI-32 (normal human fibroblasts) and HepG-2 (human liver cancer) cell lines were cultured in Eagle’s Minimum Essential medium (EMEM) at 37 °C, supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (w/v) L-glutamine, and 0.4% (w/v) antibiotics (50 IU/mL of penicillin, 50 IU/mL of streptomycin).
The MCF-7 (human breast adenocarcinoma) cell line was cultured at 37 °C and 5% (v/v) CO2 in RPMI 1640 medium, and it was supplemented with 5% (v/v) fetal bovine serum (FBS), 1% (w/v) L-glutamine, 1% (v/v) sodium pyruvate, and 0.4% (w/v) antibiotics (50 IU/mL of penicillin, 50 IU/mL of streptomycin).
The Caco-2 (human colon adenocarcinoma) cell line was cultured at 37 °C and 5% (v/v) CO2 in modified Eagle’s medium (MEM) with modified Earle’s salts, and it was supplemented with 20% (v/v) fetal bovine serum (FBS), 1% (w/v) L-glutamine, 1% (v/v) sodium pyruvate, 1% (v/v) nonessential amino acids, and 0.4% (w/v) antibiotics (50 IU/mL of penicillin, 50 IU/mL of streptomycin).
2.3. Extraction of Plant Material
O. europaea leaves (2 kg) were collected, thoroughly washed, and air-dried in shade for 10 days. They were subjected to grinding using an OC-60B/60B herbal grinding machine (60–120 mesh, Henan, Mainland China). The ground plants were macerated for exhaustive extraction with 70% EtOH (5L X3) at room temperature, evaporated, and concentrated in vacuo at 45 °C using a rotary evaporator (Buchi Rotary evaporator R-300, Cole-Parmer, USA) to obtain 80 g of dry extract, which was stored at 4 °C for further biochemical studies. Our investigations of the water-based extracts are underway in order to obtain such a bioactive extract using a greener solvent.
2.4. Metabolic Profiling
O. europaea leaf crude ethanolic extract was developed, and approximately 1 mg/mL of the extract was used for LC-mass investigation. The concentrated ethanolic extract was exposed to metabolic analysis using LC-HRMS, in correspondence with our previous report [22], on an Acquity Ultra Performance Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA). An Acquity ultra-performance liquid chromatography system attached to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA) was employed. Positive and negative ESI ionization modes were employed to obtain the high-resolution mass spectrometry connected with a spray voltage of 4.5 kV, capillary heat of 320 °C, and mass charge ratios of 150–1500 m/z. The MS dataset was processed, and data were obtained using MZmine 2.20 based on the approved parameters. Mass ion peaks were identified and followed by chromatogram builder and chromatogram deconvolution. The local minimum search algorithm was utilized, and isotopes were further determined via the isotopic peaks’ grouper. Missing peaks were illustrated using the gap-filling peak finder. An adduct investigation along with a complex search was performed. The refined dataset was later exposed to molecular formula prediction and peak identification. The positive and negative ionization mode datasets from the respective extract were dereplicated against the DNP (Dictionary of Natural Products) databases.
2.5. Preparation of OLE-Containing Spanlastic
Spanlastics were prepared with the spraying technique reported previously by Refaat et al., 2019 [12]. Briefly, the organic phase was prepared by dissolving Span 60, Tween 80 (4:1 molar ratio), and the alcoholic extract of O. europaea (150 mg) in 2 mL of absolute ethanol. Sucrose was dissolved in 3 mL of deionized water to yield a 9% w/v sucrose solution (aqueous solution). The organic phase was sprayed using a modified long nozzle sprayer on the surface of the aqueous phase (100 µL for 5 s). During spraying, the aqueous phase temperature was kept constant at 60 °C with stirring at 1500 rpm. Stirring at 60 °C was maintained for 20 min until spanlastics were spontaneously formed and ethanol was completely evaporated. The obtained spanlastics were stored at 4 °C for 24 h to guarantee the complete annealing of the formed bilayer. Those results are consistent with a previous report that revealed that olive extract is appropriate only for the low-temperature thermal application for food and cosmetics because of its low thermal stability.
2.6. Characterization of the Prepared Spanlastics
2.6.1. Determination of Vesicle Size, Size Distribution, and Zeta Potential
The OLE-containing spanlastics’ size and polydispersity index were determined with Zetasizer Nano ZSP (Malvern Instruments, Malvern, UK). Spanlastics were analyzed at 25 °C after dilution with deionized water. Procedures were repeated in a triplicate manner and average values were determined [23]. The zeta potential of the Millipore water-diluted spanlastics was evaluated using a Mastersizer (3000 E Malvern Instruments, UK). The average zeta potential was determined [24].
2.6.2. Transmission Electron Microscopy Analysis (TEM)
The morphology and shape of the prepared spanlastics were investigated using electron microscopy (JEM-1400, Jeol, Tokyo, Japan) maintained at 80 kV. For this, a spanlastic suspension was placed onto a carbon-coated copper grid. The carbon-coated grid was kept for 10 min at ambient temperature before examination [12,25].
2.6.3. Entrapment Efficacy (%EE) of the Prepared Liposomes
The entrapped % of the extract, calculated as % of flavonoid entrapment, within the formulation was determined according to Refaat et al., 2019 [12]: washing of the formulation was performed by the centrifugation of 1 mL of spanlastic suspension at 4 °C, 15,000 rpm for 1 h. The separated spanlastics were twice rewashed by dilution with distilled water followed by centrifugation until the complete removal of the unentrapped extract. The prepared spanlastics were destructed by sonication with absolute ethanol and then centrifugation at 15,000 rpm for 30 min. The percentage (%) of trapped flavonoids was determined as described by Refaat et al., 2019 [12]. Briefly, equal volumes of 10% aluminum chloride alcoholic solution and the supernatant were mixed and diluted with absolute alcohol with a final volume of 2 mL. The absorbance was determined against blank at λmax 410 nm using a UV/Vis spectrophotometer (Spectronic Genesys®), with Winspec Software. The mean % of entrapped flavonoids was determined using the equation:
2.6.4. Characterization of the Heat Effect
The relative change in the weight of olive extract and prepared spanlastics relative to the temperature change was studied using Thermogravimetric Analysis (TGA). Twenty milligrams of the dried samples were placed in a platinum pan and heated from 30 to 450 °C with a heat flow rate of 20 °C/min and nitrogen flow rate of 20 mL/min. Fourier-transform infrared (FT-IR) measurements over the wavenumber range from 4000 to 400 cm (Nicollet IS 10 FTIR spectrometer, USA) for the olive extract, blank spanlastics, and olive spanlastics were carried out after dispersion of the samples in KBr discs to study the possible interactions and bonding pattern between Span 60, Tween 80, and olive extract.
2.7. In Vitro Study
2.7.1. In Vitro Antiproliferative Assay
The antiproliferative activity of OLE either free or encapsulated within the spanlastic dosage form was determined by the sulforhodamine B (SRB) assay as mentioned by Skehan et al. 1990 [26], and Vichai and Kirtikara 2006 [27], on the normal fibroblast (WI-32), breast (MCF-7), liver (HepG2), and colorectal (Caco2) cancer cell lines. Cells were cultivated in 96-well microtiter plates at an initial concentration of 1 × 105 cells/well in 150 µL of fresh medium and left for 24 h to adhere to the wells. Concentrations of 0, 5, 12.5, 25, and 50 µg/mL of the respective extract were included. The plates were incubated for 48 h. The cells were fixed with 50 μL of cold trichloroacetic acid (10% final concentration) for 1 h at 4 °C. The plates were cleaned with distilled water (automatic washer Tecan, Germany) and stained with 50 μL of 0.4% SRB dissolved in 1% acetic acid for 30 min, at room temperature. Later, they were washed with 1% acetic acid and air-dried. The dye was solubilized with 100 μL/well of 10 M of tris base (pH 10.5). The optical density of each well was measured spectrophotometrically at 570 nm using an ELISA microplate reader (Sunrise Tecan reader, Germany). Doxorubicin was used as a positive control. The mean background absorbance was automatically subtracted, and mean values of each drug concentration were determined. The experiment was rerun three times, and later, the IC50 values were measured.
2.7.2. Measurement of Intracellular Free Radicals
For the in vitro examination of the potential activity of OLE and OLE-spanlastics as pro-oxidants, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA) dye was applied. Briefly, upon entry into live cells, acetate groups are removed from the H2DCF-DA by the action of the cellular esterases; then, oxidation of the dye to the fluorescent product dichlorofluorescein (DCF) in the presence of free radicals occurs and represents a marker for oxidative stress and pro-oxidant activity. The Caco-2 cells are used here as an example due to their high sensitivity to oxidative stress.
Generally, Caco-2 cells (8 × 105 cells/µL) were treated with the test materials (7 µg/mL) for 1 and 10 min, and then treated with 20 µM of DCFDA for 30 min. After several washes in PBS, the DCF fluorescence was measured using flow cytometric analysis with excitation and emission wavelengths at 495 and 530 nm, respectively. All activity measurements were repeated three times for each test material concentration.
2.7.3. Glutathione Reductase Enzyme Activity
The inhibitory activity of OLE and OLE-spanlastics against glutathione reductase (GR) was measured colorimetrically using the manufacturer protocol (abcam, Cat.No: ab8346). Briefly, the test material was dissolved in dimethyl sulfoxide to produce a stock solution of 1 mg/mL. The reaction mixture consisted of 100 mM of potassium phosphate buffer at pH 7.4, 1 mM of GSSG, 0.1 mM of NADPH, and 25 μL of GR. The test material was added from the stock solution by serial dilutions to give final inhibitor concentrations between 0.05 µg/mL and 50 µg/mL. GR activity was measured at 405 nm at 37 °C. All activity measurements were repeated three times for each test material concentration.
2.7.4. Superoxide Dismutase Activity
The inhibitory activity of OLE and OLE-spanlastics against SOD was measured colorimetrically using the manufacturer’s protocol (abcam, Cat.No: ab65354). Briefly, the test material was dissolved in dimethyl sulfoxide to produce a stock solution of 1 mg/mL. The reaction mixture consisted of 100 mM of potassium phosphate buffer at pH 7.4, 1 mM of formazan dye, 25 μL of xanthine oxidase (to produce superoxide anions), and 25 μL of SOD solution. The test material was added from the stock solution by serial dilutions to give final inhibitor concentrations between 0.05 µg/mL and 50 µg/mL. SOD activity was measured at 450 nm at 37 °C. All activity analyses were repeated three times for each test material concentration.
2.8. In Silico Investigation
2.8.1. Determination of the Potential Protein Targets
Potential protein targets for the OLE identified compounds were proposed by subjecting all these compounds to inverse docking against all proteins hosted in Protein Data Bank (PDB; https://www.rcsb.org/ accessed on 21 October 2021). idTarget platform (http://idtarget.rcas.sinica.edu.tw/ accessed on 21 October 2021) was used for this task. This structural-based screening software applies a unique docking approach called divide-and-conquer docking that adaptively builds small overlapping grids to make the search space on the protein surfaces more constrained, and hence, it can run a huge number of accurate docking experiments in a reduced time. The retrieved results were obtained as a list of binding affinity scores arranged from the highest negative value to the lowest one. We set a binding affinity score of −7 kcal/mol as a cut-off value to select the best targets for each identified compound in OLE. Considering the human proteins related to cancer pathogenesis, GR (PDB: 1BWC) and SOD (PDB: 5YTO) were selected for further modeling study.
2.8.2. Molecular Dynamic Simulation and Binding Free Energy Calculation
The binding free energy calculation (ΔG) and molecular dynamic simulation were conducted as previously described in [28,29,30]. Desmond v. 2.2 software was used for performing MDS experiments. This software applies the OPLS force field. Protein systems were built using the System Builder option, where the protein structure was embedded in an orthorhombic box of TIP3P water together with 0.15 M of Na+ and Cl− ions in 20 Å solvent buffer. Afterward, the prepared systems were energy-minimized and equilibrated for 10 ns. Desmond software automatically parameterizes inputted ligands during the system building step according to the OPLS force field. For simulations performed by NAMD, the parameters and topologies of the compounds were determined either using the Charmm27 force field with the online software Ligand Reader and Modeler (http://www.charmm-gui.org/?doc=input/ligandrm; accessed on: 29 October 2021) or using the VMD plugin Force Field Toolkit (ffTK). Afterward, the generated parameters and topology files were loaded to VMD to readily read the protein–ligand complexes without errors and then to conduct the simulation step.
2.8.3. Binding Free Energy Calculations
Binding free energy calculations (∆G) were performed using the free energy perturbation (FEP) method. Briefly, this method calculates the binding free energy ∆Gbinding according to the following equation: ∆Gbinding = ∆GComplex − ∆GLigand. The value of each ∆G is estimated from a separate simulation using NAMD software. All input files required for simulation by NAMD can be papered by using the online website Charmm-GUI (https://charmm-gui.org/?doc=input/afes.abinding, accessed on 18 May 2021). Subsequently, we can use these files in NAMD to give the expected simulations using the FEP calculation function in NAMD. The equilibration was achieved in the NPT ensemble at 300 K and 1 atm (1.01325 bar) with Langevin piston pressure (for “Complex” and “Ligand”) in the presence of the TIP3P water model. Then, 10 ns of FEP simulations were performed for each compound, and the last 5 ns of the free energy values was measured for the final free energy values. Finally, the generated trajectories were visualized and analyzed using VMD software.
2.9. Statistical Analysis
All in vitro analyses were performed in triplicate. Pooled data were given as the mean ± standard error of the mean (SEM) of at least three separate experiments. The variations among diverse management groups were established by ANOVA, accompanied by Dunnett’s test using PASW Statistics® version 18 (Quarry Bay, Hong Kong). A variation of p < 0.001 was considered statistically significant correlated with a vehicle-treated control group and illustrated by a * symbol. The IC50 values were established utilizing a nonlinear regression curve fitting analysis using GraphPad Prism software version 6 (La Jolla, CA, USA).
3. Results and Discussion
3.1. Dereplication of Olea europaea L. cv. Arbosana
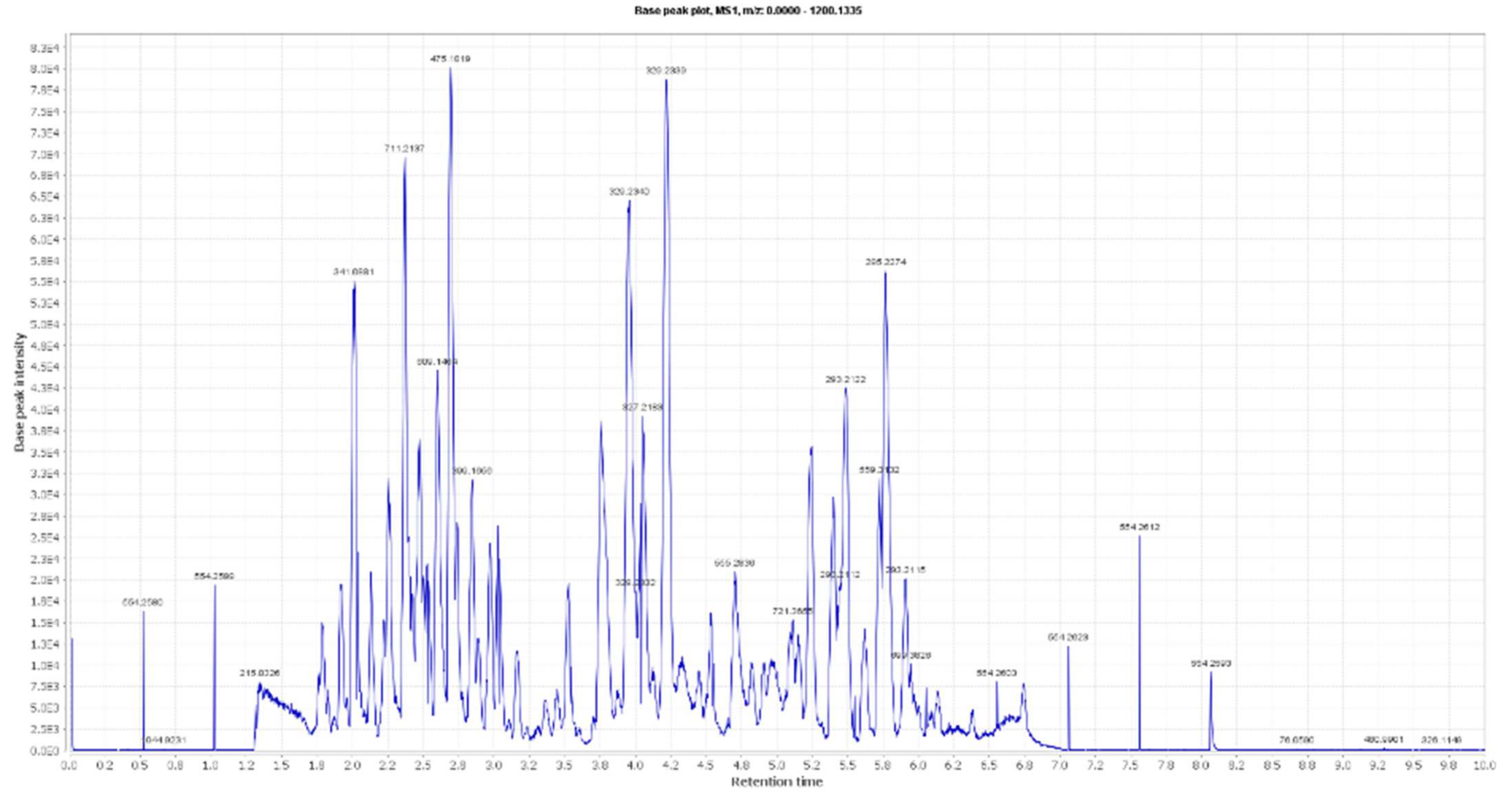
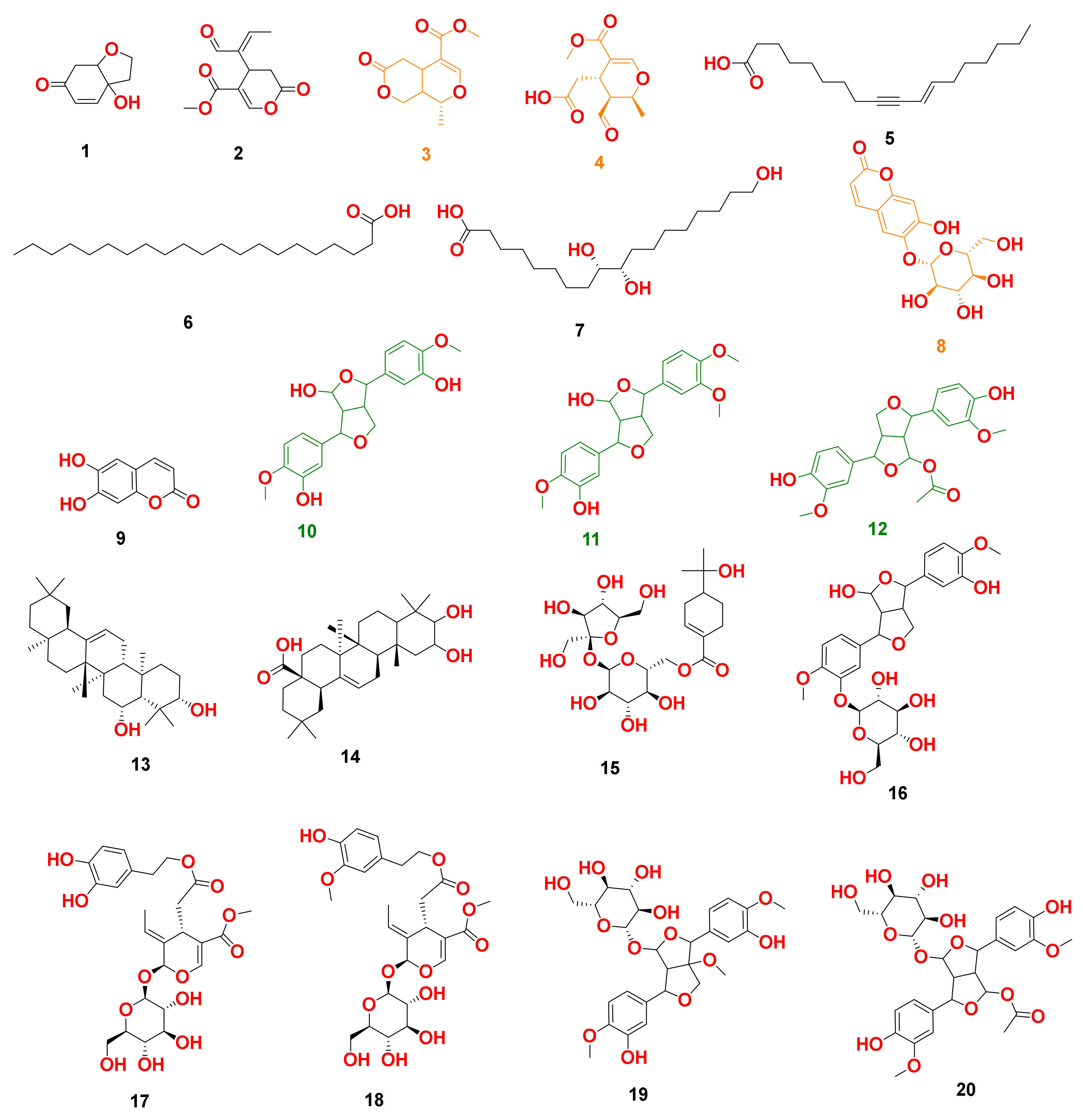
By analyzing OLE, several hits were proposed (Table 1, Figure 1, Figure 2 and Figure 3). The molecular ion mass peaks at m/z 155.0708 and 223.06055 [M + H]+, [M − H]+, respectively, for the predicted molecular formulas C8H10O3 and C11H12O5 gave hits of megaritolactones, and S-(E)-elenolide, halleridone 1, and elenolide 2, respectively, that were formerly isolated from O. europaea [31,32].

Table 1.
Dereplicated metabolites from LC-HRESIMS analysis of Olea europaea L. cv. Arbosana.
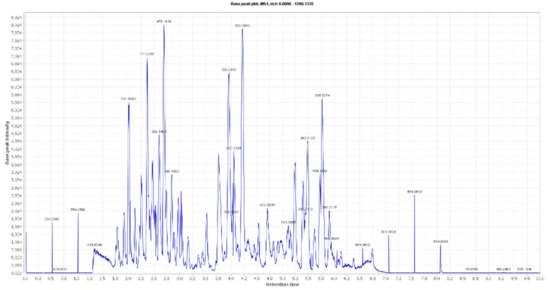
Figure 1.
LC-HRESIMS chromatogram of the dereplicated metabolites of Olea europaea (Positive).
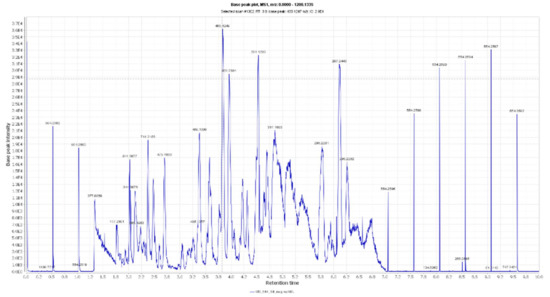
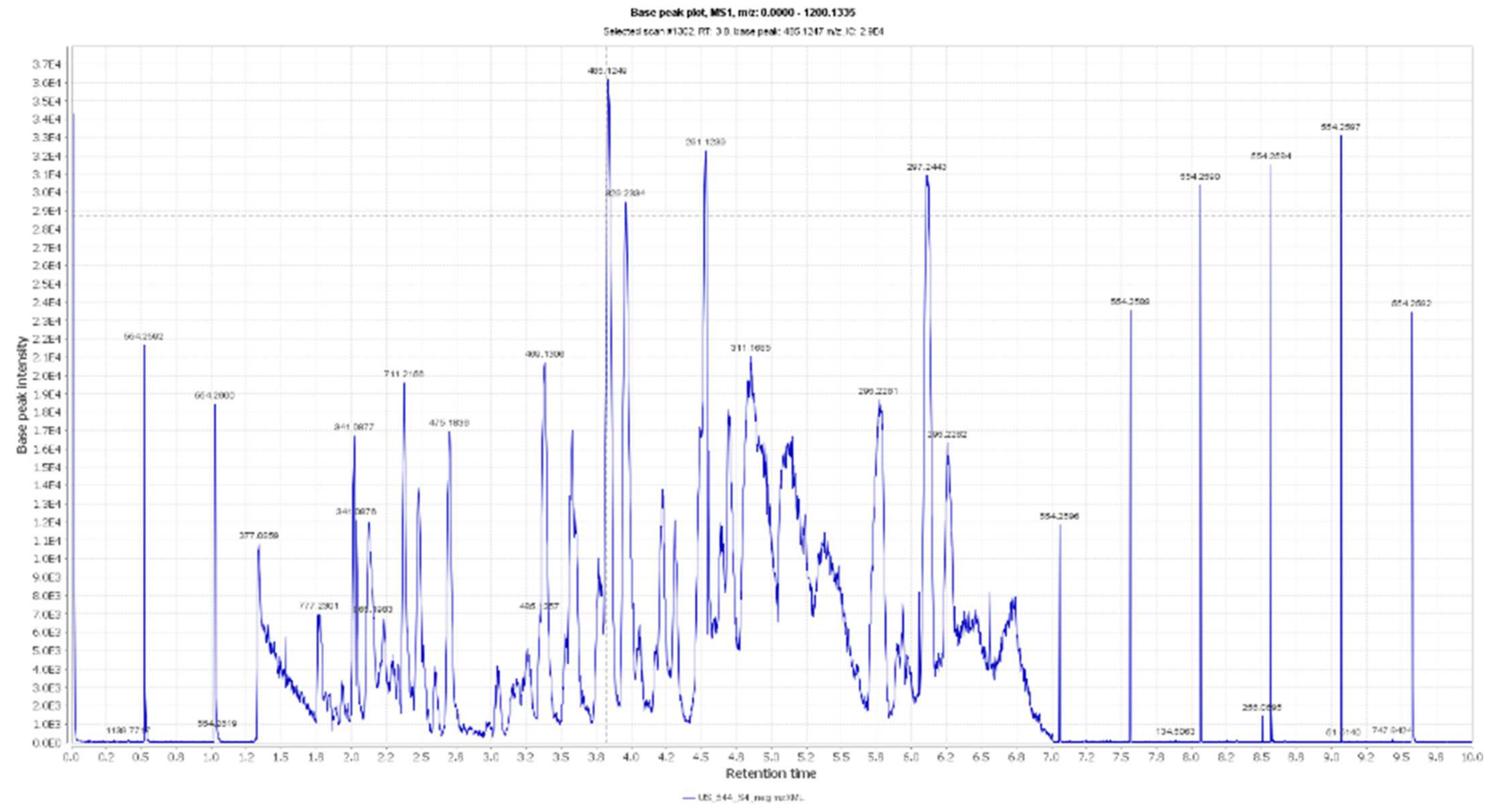
Figure 2.
LC-HRESIMS chromatogram of the dereplicated metabolites of Olea europaea (Negative).
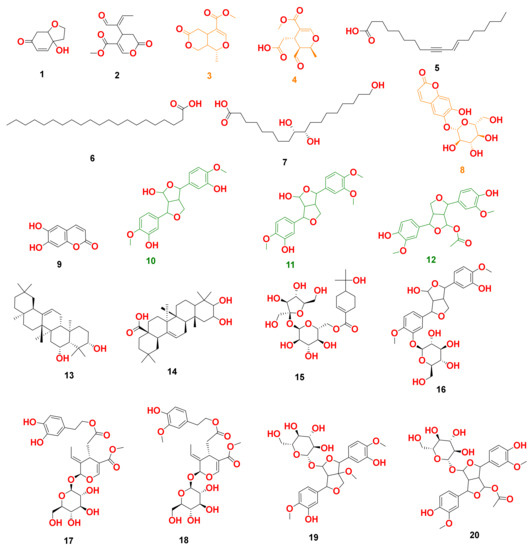
Figure 3.
Dereplicated metabolites from LC-HRESIMS analysis of Olea europaea. Orange-colored structures were predicted to bind to GR’s active site, while the green-colored structures were predicted to bind to SOD’s active site.
The mass ion peaks at m/z 227.09206, 243.0869, 277.2167, 327.3263, and 331.24803 corresponding to the suggested molecular formulas C11H14O5, C11H14O6, C18H30O2, C21H42O2, and C18H36O5 [M + H]+, [M − H]+, respectively, fit a fatty acid, and secoiridoid derivative compounds olenoside A 3, elenaic acid 4, 11-octadecen-9-ynoic acid 5, heneicosanoic acid 6, and 9,10,18-trihydroxyoctadecanoic acid 7, which was previously isolated from O. europaea [33,34,35,36,37]. In addition, the mass ion peaks at m/z 341.08660, 341.0873, 375.1444, 389.1600, and 417.15389 corresponding to the suggested molecular formulas C15H16O9, C15H16O9, C20H22O7, C21H24O7, and C22H24O8 [M + H]+ fit a benzopyrane and tetrahydrofuran lignan derivative compounds 6,7-dihydroxy-2H-1-benzopyran-2-one; 6-O-β-D-glucopyranoside 8, 6,7-dihydroxy-2H-1-benzopyran-2-one; 6-O-β-D-glucopyranoside 9, 7,9′:7′,9-diepoxy-8,8′-lignan-3,3′,4,4′,8-pentol; 3,3′-di-Me ether 10, 7,9′:7′,9-diepoxy-8,8′-lignan-3,3′,4,4′,8-pentol; 3,3′,4′-tri-Me ether 11, and 3,3′,4,4′,8-pentahydroxy-7,9′:7′,9-diepoxylignan; 3,3′-Di-Me ether, 8-Ac 12, which was previously isolated from O. europaea [38,39,40].
Moreover, the molecular ion mass peaks at m/z 443.3889 and 473.36213 [M + H]+, for the predicted molecular formulas C30H50O2 and C30H48O4, gave hits of the triterpenes, 12-oleanene-3,28-diol 13, and 2,3-dihydroxy-13(18)-oleanen-28-oic acid 14, respectively, which were previously isolated from O. europaea [41].
The ion mass peaks at m/z 509.22142, 537.1972, 541.1921, and 555.2078 [M + H]+ for the predicted molecular formulas C22H36O13, C26H32O12, C25H32O13, and C26H34O13, respectively, gave hits of the 6-O-oleuropeoylsucrose 15, which was isolated from O. europaea [4], the tetrahydrofuran lignan nucleus of 7,9′:7′,9-diepoxy-8,8′-lignan-3,3′,4,4′,8-pentol, 3,3′-Di-Me ether, and 4-O-β-D-glucopyranoside 16, which was isolated from O. europaea [35], and secoiridoid derivative compounds oleuropein 17 and oleuropein; 4″-Me ether 18, which were isolated from O. europaea [41,42]. Two main ion peaks with the m/z values of 567.2078 and 579.20783 [M + H]+ with molecular formulas C27H34O13 and C28H34O13, respectively, were detected and dereplicated as 7,9′:7′,9-diepoxy-8,8′-lignan-3,3′,4,4′,5,8-hexol, 3,3′,5-tri-Me ether, 8-O-β-D-glucopyranoside 19, 3,3′,4,4′,8-pentahydroxy-7,9′:7′,9-diepoxylignan, 3,3′-di-Me ether, and 8-Ac, 4-O-β-D-glucopyranoside 20, respectively, which were isolated earlier from O. europaea [4].
3.2. OLE-Containing Spanlastics
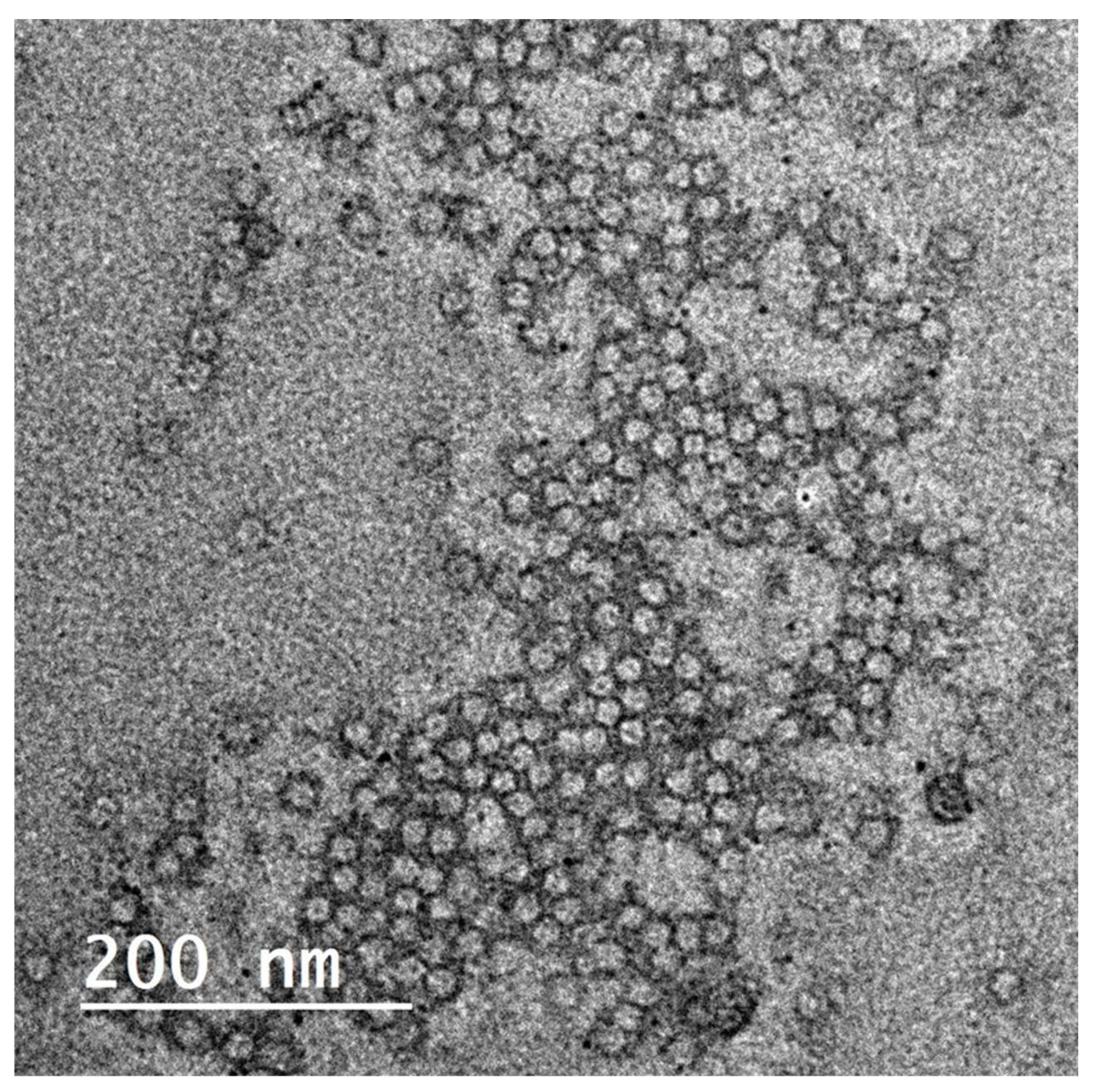
Elastic nanovesicles, so-called spanlastics of OLE, were successfully prepared. The TEM image shows the vesicular structure of the formed spanlastics (Figure 4). The vesicles-obtained particle size of 41 ± 2.4 nm and PDI 0.279. The Zeta potential of the prepared spanlastics was 13.6 ± 2.5. The small size and the narrow size distribution of the formulation were achieved by the adopted spraying method for preparation. Spraying of the organic phase onto the aqueous phase surface provided an enlarged surface area and improved wettability of the sprayed organic phase 13, 44. The formulated spanlastics entrapped 61.43 ± 2.03% of the loaded extract. Adding ethanol during formulation contributed to the enhancement of the drug partitioning and entrapment within the spanlastic vesicles. Moreover, the structure of the spanlastics imparted better inclusion of both the hydrophilic ingredients and the lipophilic ingredients of the extract within the core of the vesicle or the bilayered membrane, respectively.
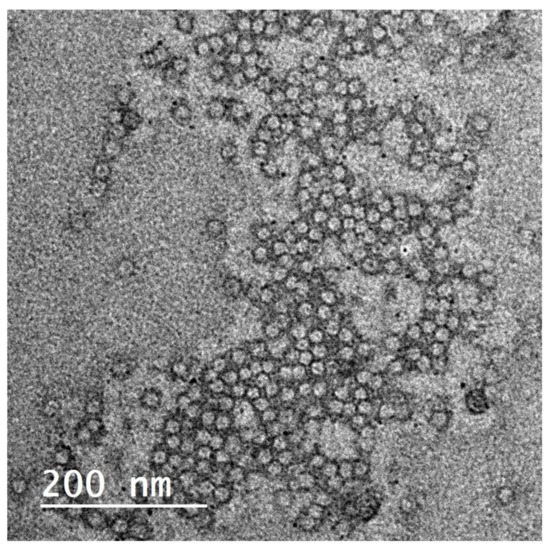
Figure 4.
TEM image for Olea europaea ethanolic extract-containing spanlastics.
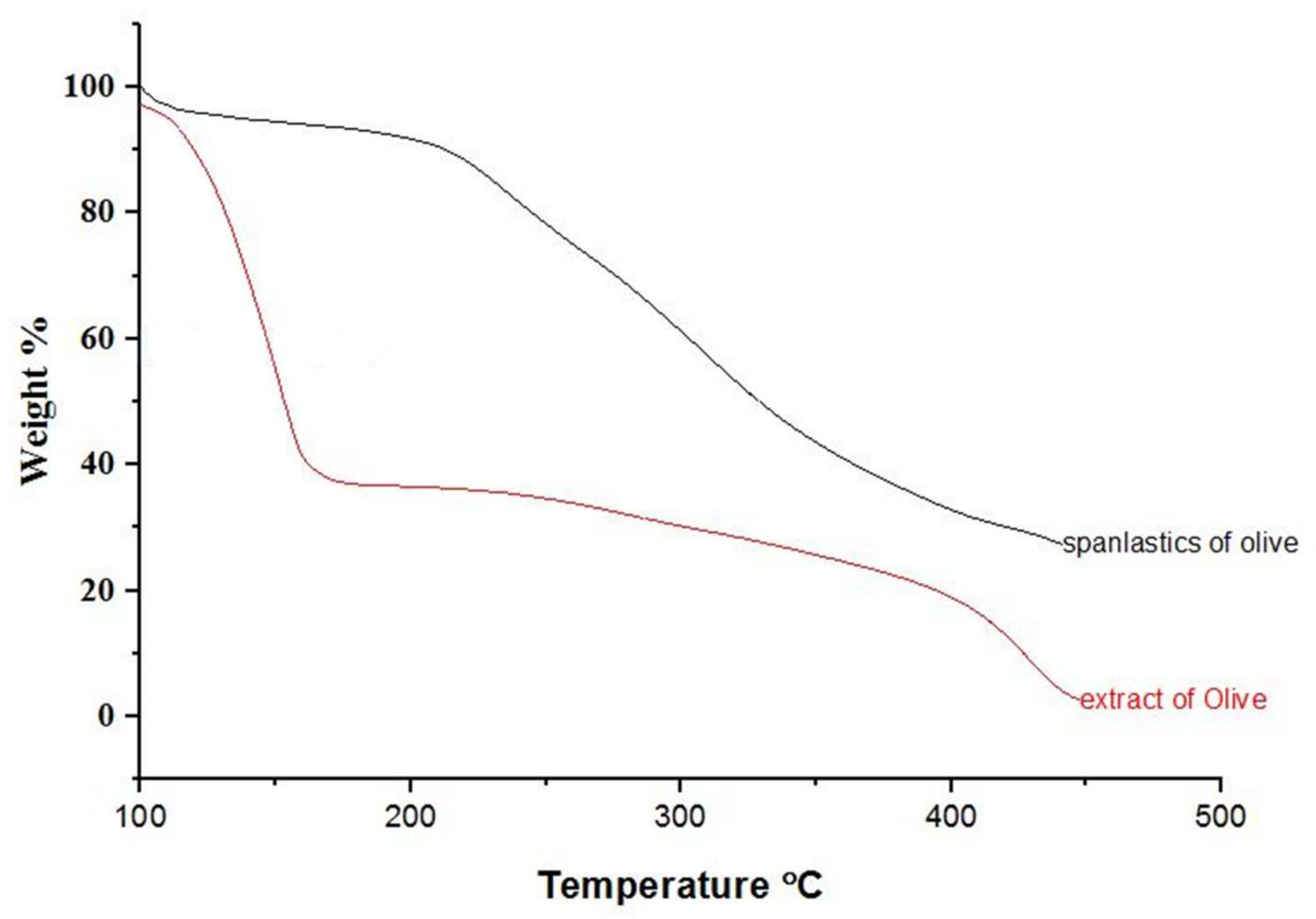
Physicochemical changes in compounds as a function of temperature were evaluated by TGA. TGA curves for OLE and OLE spanlastics are shown in Figure 5. During heating from 30 to 450 °C, about 62 and 13% weight loss was detected at a temperature of 126 °C for OLE and OLE spanlastics, respectively. Weight loss may be due to the phase transition caused by a change in the molecular structure. Results reveal the enhanced thermal balance of the entrapped cargo owing to spanlastic encapsulation.
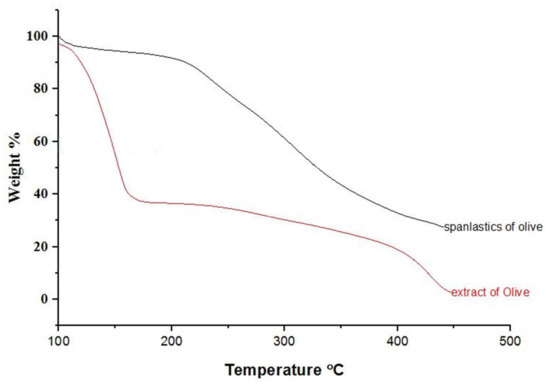
Figure 5.
TGA spectra of olive extract and spanlastics of olive.
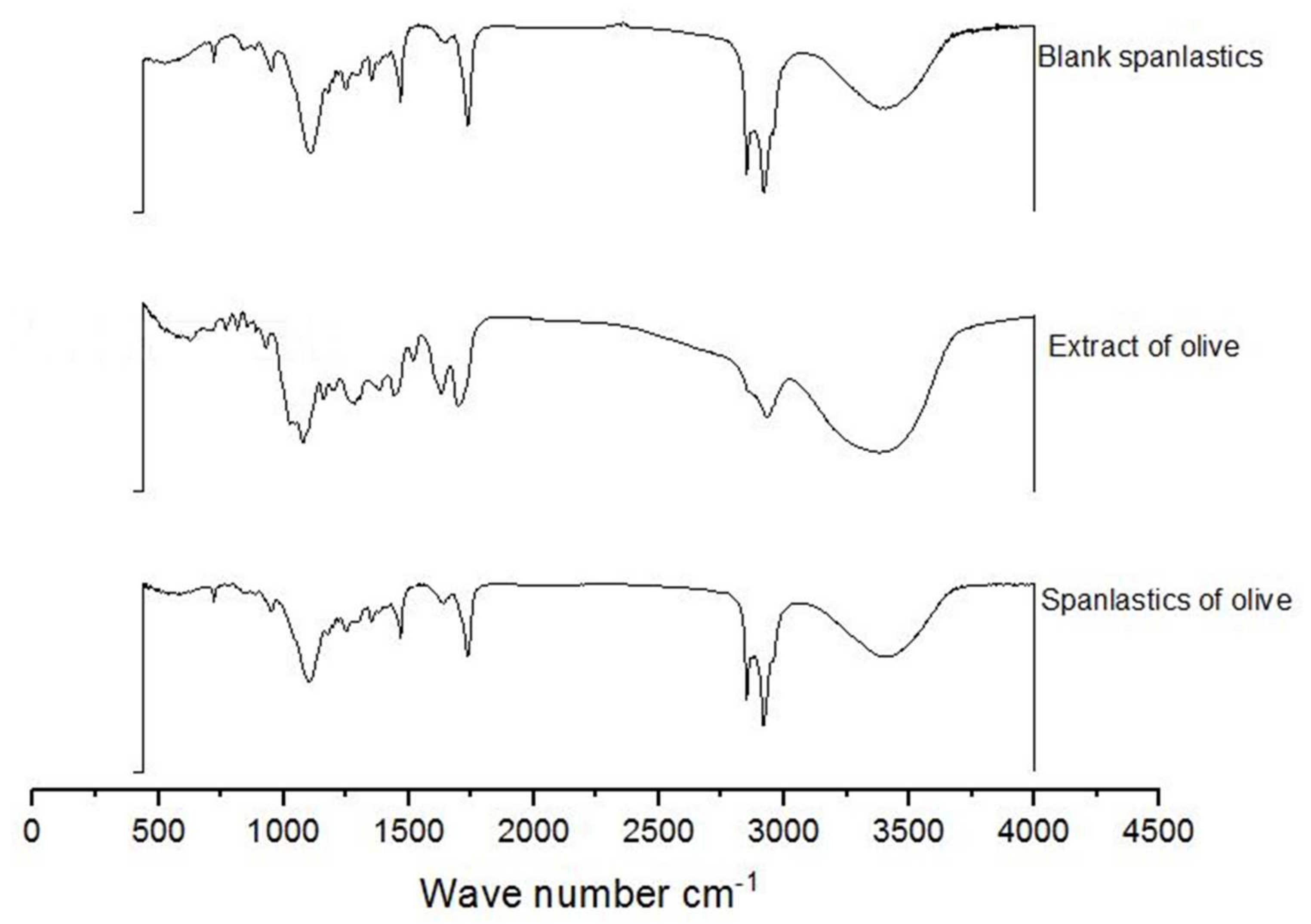
FTIR spectra of OLE, blank spanlastics, and OLE spanlastics were studied to understand the association between components of OLE and components of the spanlastic membrane (Span 60 and Tween 80) (Figure 6). The FTIR spectrum of OLE contains principal bands at 3391, 2942, 1705, 1499, and 1090, and that of Blank spanlastics contains bands at 2928, 2870, 1756, 1481, and 1105. The FTIR spectrum of olive spanlastics contains no extra bands to those involved in both free extract and blank spanlastics spectra, suggesting that the spanlastics encapsulation of OLE within the spanlastics did not set-up new connections.
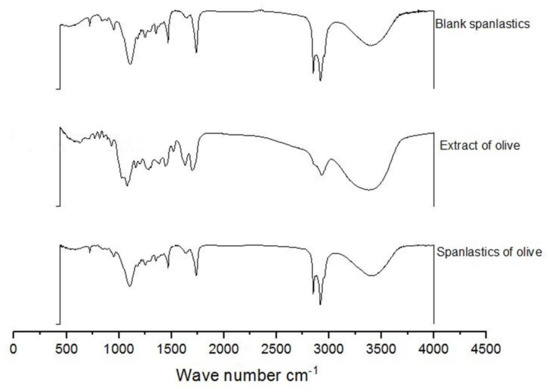
Figure 6.
FTIR spectra of blank spanlastics, olive extract, and spanlastics of olive.
3.3. Antiproliferative Activity of the OLE and OLE-Spanlastics
Many studies [43,44] support our findings in the context of the antiproliferative activity of the OLE. In this study, OLE was screened in vitro for its antiproliferative activity against hepatic, breast, and colorectal cancer cell lines (HepG-2, MCF-7, and Caco-2, respectively) and normal fibroblast cell line (WI-32). Results showed that OLE was able to moderately prohibit the growth of all tested cancer cell lines with IC50 equal to 9.2 ± 0.8, 7.1 ± 0.9, and 6.5 ± 0.7 µg/mL, respectively (Table 2).

Table 2.
In vitro antiproliferative activity of Olea europaea ethanolic crude extract shown as IC50 ± (SD) µM.
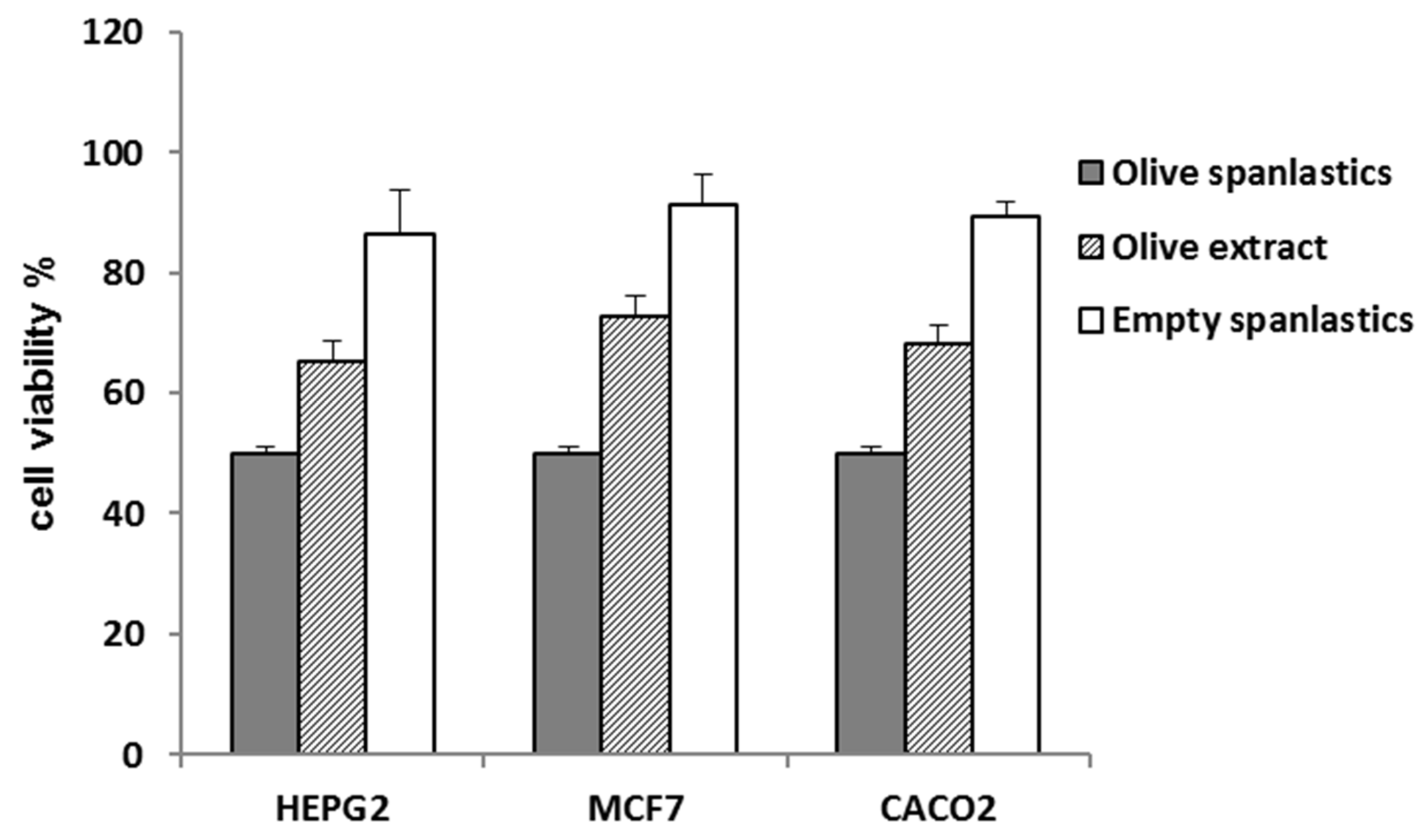
The spanlastic formulation improved the antiproliferative potential of the entrapped extract. IC50 values of the formulated extract were 3.6 ± 0.2, 2.3 ± 0.1, and 1.8 ± 0.1 µg/mL against HepG2, MCF-7, and Caco-2 (Table 2, Figure 7), respectively, p < 0.001.
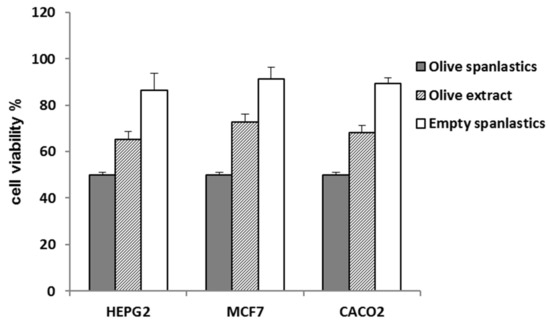
Figure 7.
Cell viability of olive spanlastics and empty spanlastics against HepG-2. MCF-7, and Caco-2.
That may be attributed to the concept of enhanced endocytosis of nanosized particles compared to larger ones [45,46]. Endocytosis of tiny particles needs less energy than that required for larger particles [47]. That means nanosized particles have a better chance for cellular uptake relative to larger counterparts [47]. Herein, cell viability was utilized as a tool to evaluate the extent of extract-containing spanlastic uptake compared to that of the crude extract (Table 2, Figure 7).
3.4. In Silico Study
Target Prediction and Docking Analysis: To propose a probable mode of action for OLE, all annotated compounds (Figure 3) were subject to a comprehensive inverse docking against almost all proteins introduced in the Protein Data Bank using the idTarget docking platform. This online-based molecular modeling software applies a unique virtual screening approach: divide-and-conquer docking. This approach adaptively builds small overlapping grids to improve the speed and accuracy of searching the space on protein surfaces, and thus, it can carry out an enormous number of precise docking experiments in a significantly reduced time [48]. In this docking platform, the query structure can be docked against most of the PDB proteins. Thereafter, the retrieved binding affinity scores of the docked proteins were arranged from the highest negative value to the lowest one.
We set a binding affinity score of −7 kcal/mol as a cut-off value to select the best scoring protein hits. All targets with scores lower than −7 kcal/mol were then carefully investigated to select cancer-related human proteins. Glutathione reductase (GR) showed very interesting binding scores with compounds 3, 4, and 8 (−11.35, −10.63, −9.87 kcal/mol, respectively), while superoxide dismutase (SOD) was a very good binder to compounds 10, 11, and 12 (−10.22, −9.56, and −9.11 kcal/mol, respectively).
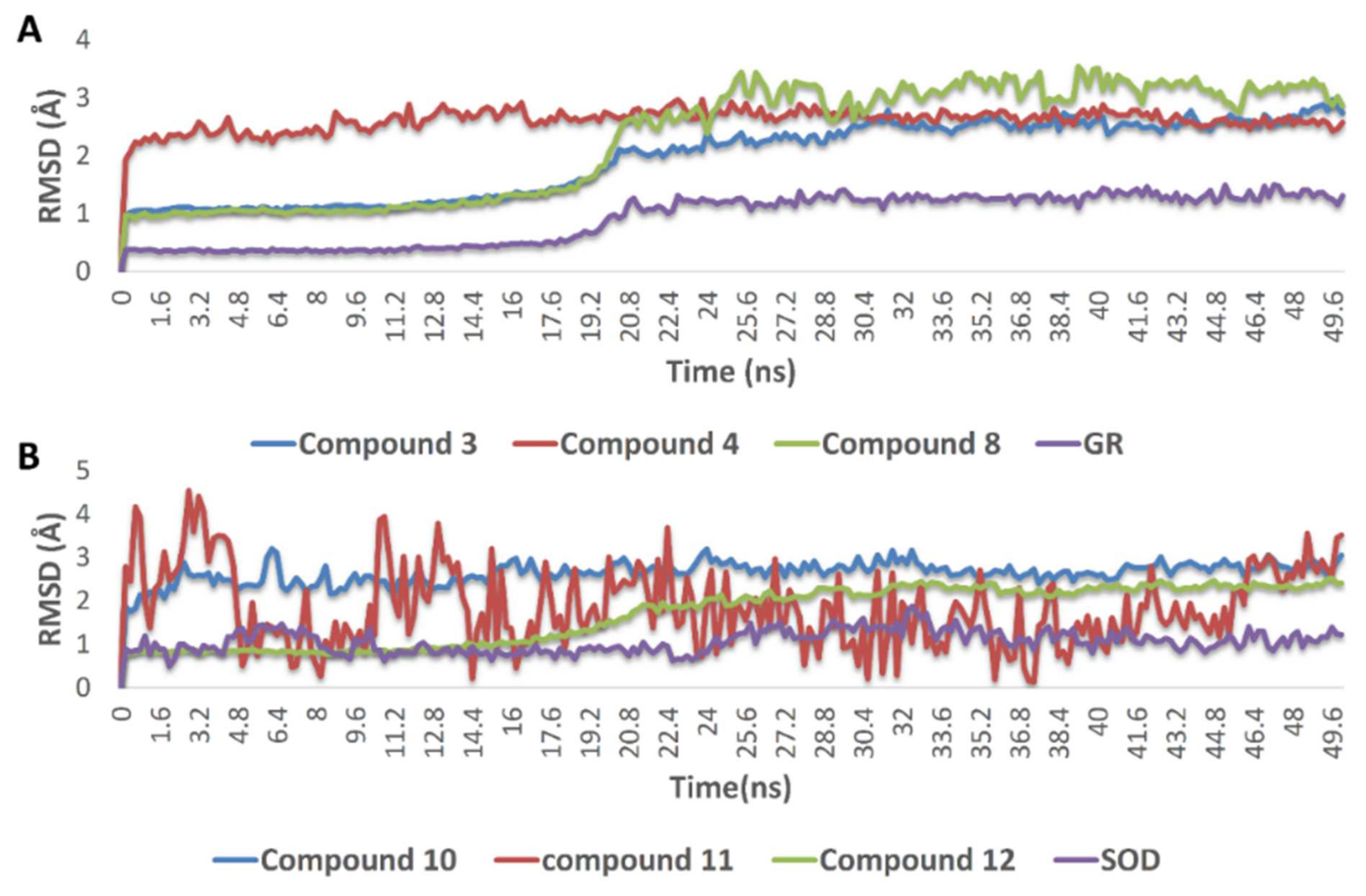
The docking poses of the previously mentioned compounds with GR and SOD were then subjected to 50 ns MDS experiments to study their stability inside the active site of each enzyme and to calculate their binding free energies (ΔG). As shown in Figure 8, compounds 3, 4, 8, 10, 11, and 12 achieved very good binding stabilities inside the active site of both GR and SOD over the simulation time with very low deviations from the starting binding orientations (i.e., docking poses) (RMSD ~ 2.5 Å). Additionally, they achieved very promising ΔG values (−9.45, −8.36, −9.14, −9.76, −10.43, and −8.71 kcal/mol, respectively) as these indicate that they are very good binders to both enzymes. The last snapshot of each MDS was then extracted to investigate the attached mode of each compound inside the effective site of both enzymes.
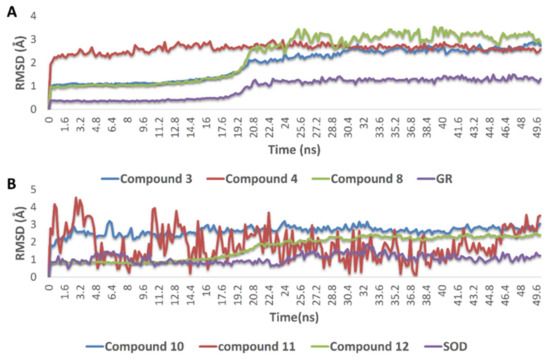
Figure 8.
(A) RMSDs of compounds 3, 4, and 8 inside the active site of GR, and compounds 10, 11, and (B) 12 inside the active site of SOD over 50 ns of MDS.
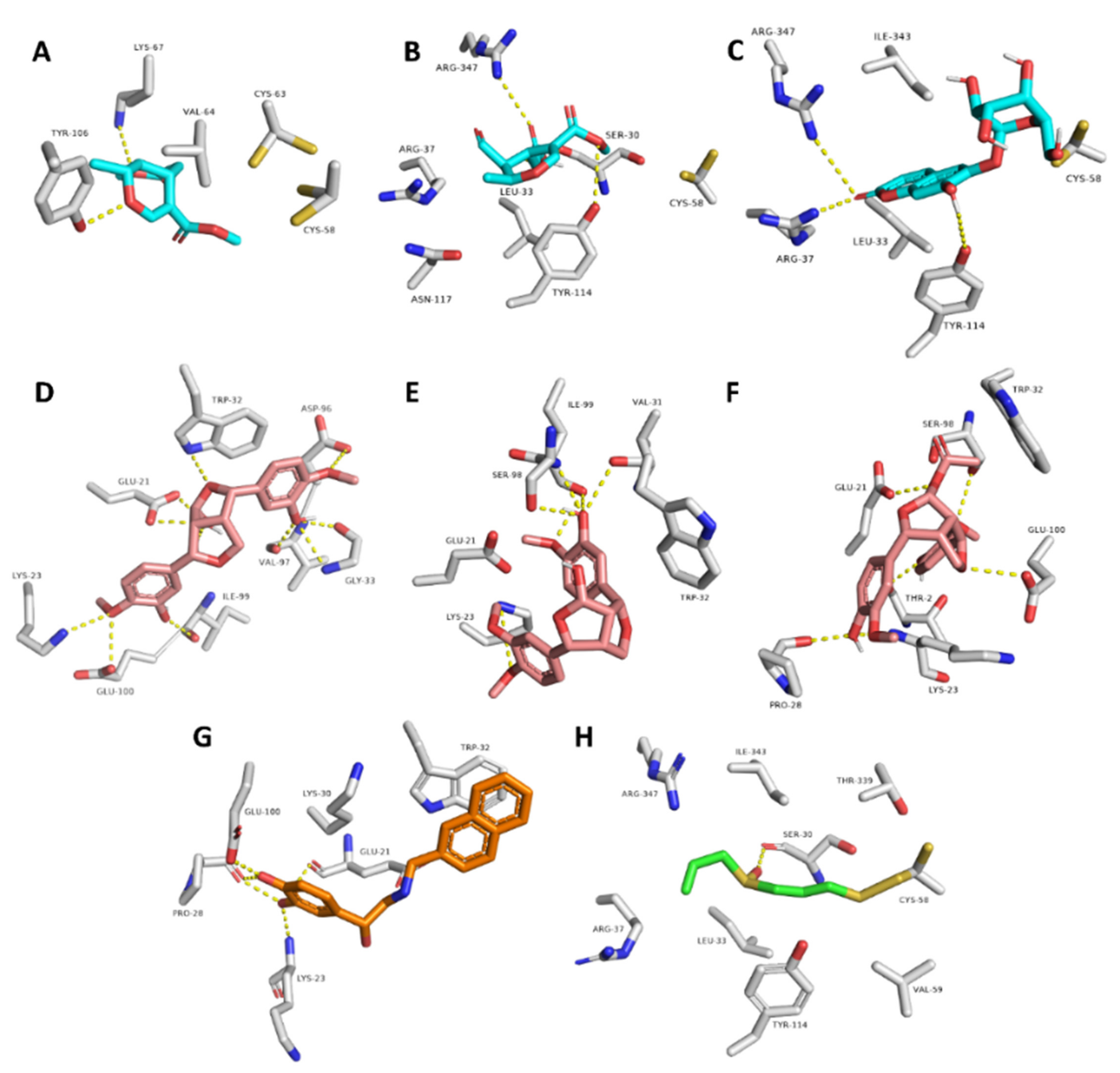
Moreover, compounds 3, 4, and 8 showed multiple stable H-bonds, particularly with ARG-37, LYS-67, TYR-106, TYR-114, and ARG-347 inside the active site of GR, while compounds 10, 11, and 12 exhibited a higher number of H-bonds inside the active site of SOD, where GLU-21, LYS-23, TRP-32, and SER-98 were the common amino acids involved in these interactions (Figure 9).
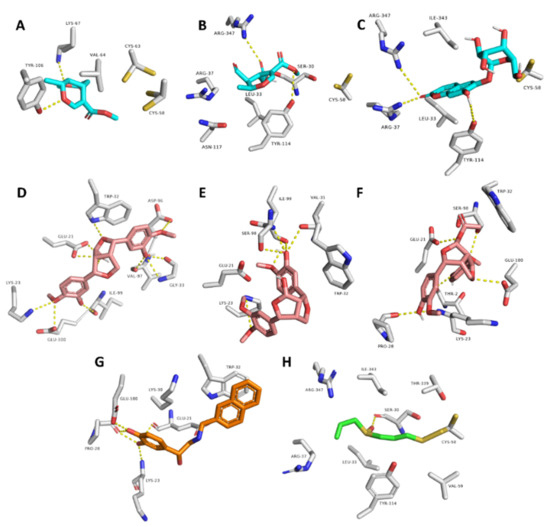
Figure 9.
Binding mode of compounds 3, 4, and 8 inside the active site of GR ((A–C), respectively; brick-red-colored), Scheme 10, 11, and 12 ((D–F), respectively; brick red-colored structures) inside the active site of SOD, in addition to the binding mode of SOD and GR co-crystalized ligands ((G) and (H), respectively).
These two oxidative stress-related enzymes were selected as probable targets for OLE based upon many previous reports that have shown a direct link between the inhibition of these enzymes and the elevated cellular oxidative stress that can eventually lead to cancer cell death. This mechanism of anticancer activity is also very selective because cancer cells are typically highly susceptible to raised oxidative stress in contrast to healthy cells [49,50,51,52,53].
3.5. Pro-Oxidant Activity of OLE and OLE Spanlastic
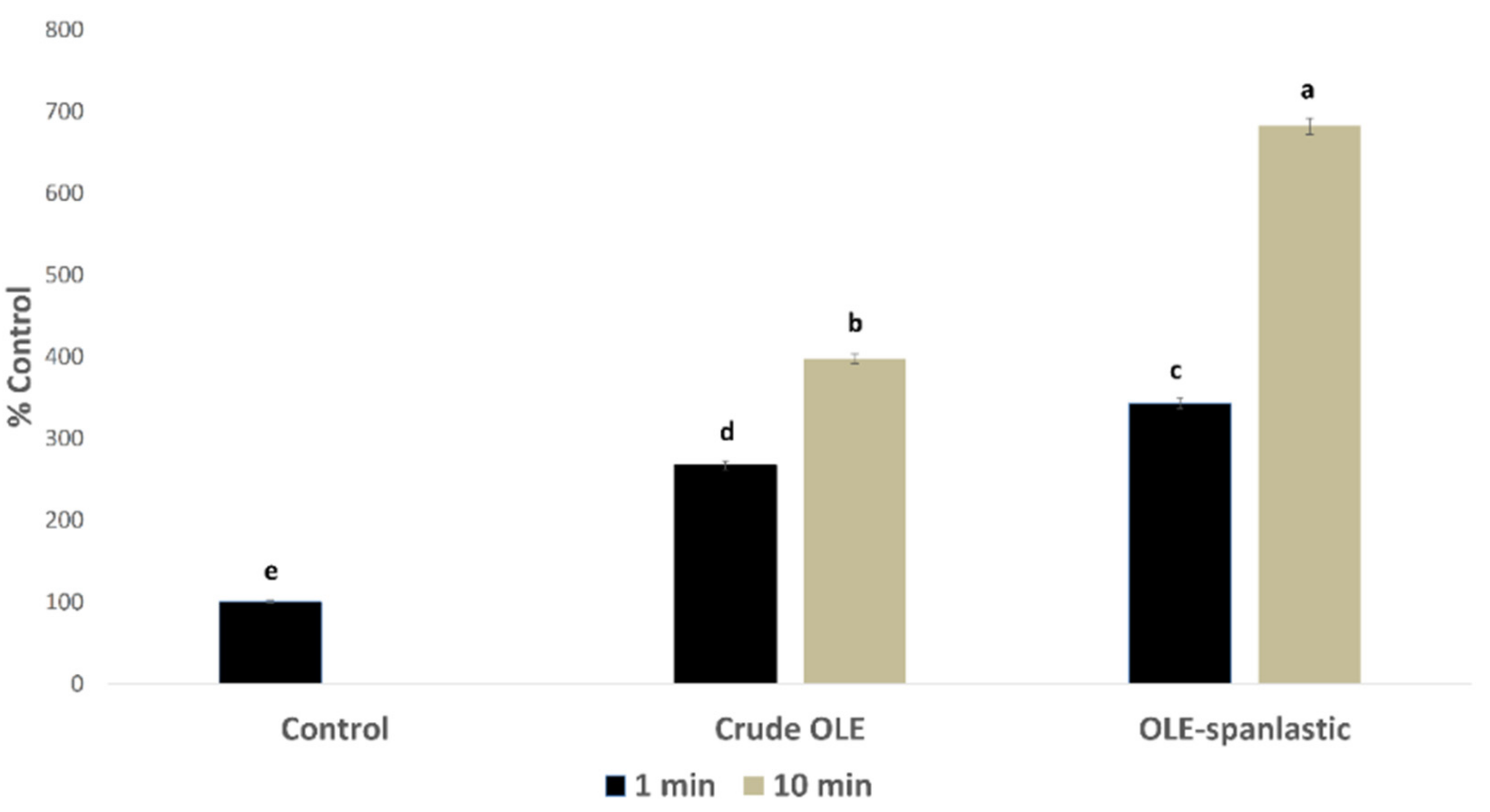
In silico investigation of the OLE revealed its potential to exhibit an intracellular pro-oxidant activity. In this study, OLE and its derived spanlastics were screened in vitro for measuring the percentage of free radicals’ production inside Caco-2 cells using DCFDA assay. Results showed a significant rise in the free radical production in Caco-2 cells upon treatment with 7 µg/mL of both OLE and OLE-spanlastics, with %DCF 267 ± 5.15 and 343 ± 4.95, respectively, after 1 min of treatment, and %DCF 398 ± 5.91 and 682 ± 7.17, respectively, after 10 min, compared to the untreated cells (i.e., normal control) (Figure 10).
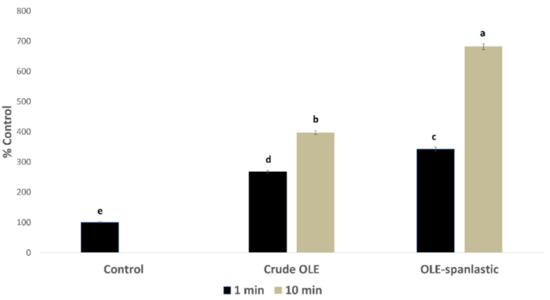
Figure 10.
Free radical production induced by OLE and OLE-spanlastics in Caco-2 cells. The test cells were treated (n = 3) with 7 µg/mL from each test material. Intracellular free radical levels were determined after 1 min and 10 min by using the DCFDA fluorescent method, and the results were expressed as the % produced fluorescence in respect of that of the control (% Control). Bar graphs represent the mean of 3 independent experiments, where the small letters (a, b, c, d, and e) indicate significance between groups (p < 0.001).
Furthermore, we tested OLE for its inhibitory potential toward the enzymatic activity of both GR and SOD to test if these two enzymes that protect cells from elevated free radicals were targeted by OLE chemical components.
Glutathione reductase (GR), or the glutathione-disulfide reductase (GSR) enzyme, catalyzes the reduction of glutathione disulfide (GSSG) to the sulfhydryl form glutathione (GSH), which is a critical molecule in resisting oxidative stress and maintaining the reducing environment of the cell53. Glutathione anions act on a water-soluble formazan dye to produce a color that can be distinguished by the rise in absorbance at 450 nm. The higher the activity of GR in the sample, the lower the amount of formazan dye that is formed. In this study, OLE and its derived spanlastics were screened in vitro for their inhibitory potential toward the enzymatic activity of GR colorimetrically using the manufacturer’s protocol (abcam, Cat.No: ab8346). The results showed that OLE and its derived spanlastics were able to inhibit the activity of the GR enzyme significantly with IC50 values of 1.18 ± 0.12 µg/mL and 0.54 ± 0.11, respectively.
In addition, superoxide dismutase (SOD) is one of the most remarkable antioxidant enzymes. It catalyzes the dismutation of the superoxide anion produced by xanthin oxidase into hydrogen peroxide and molecular oxygen52. As with GR, superoxide anions act on a water-soluble formazan dye to produce a color that can be distinguished by the rise in absorbance at 450 nm. The higher the activity of SOD in the sample, the lower the amount of formazan dye that is formed. In this study, OLE and its derived spanlastics were screened in vitro for their inhibitory potential toward the enzymatic activity of SOD colorimetrically using the manufacturer’s protocol (abcam, Cat.No: ab65354). The results showed that OLE and its derived spanlastics were able to inhibit the activity of the SOD enzyme significantly with IC50 values of 2.33 ± 0.19 µg/mL and 0.73 ± 0.23, respectively.
4. Conclusions
OLE showed mild in vitro antiproliferative activity against HepG-2, MCF-7, and Caco-2, which were enhanced through the entrapment of OLE within spanlastic vesicles. In silico and modeling investigation annotated compounds in OLE to interact with a number of oxidative stress-protecting enzymes (i.e., GR and SOD). In vitro testing supported these findings and uncovered the pro-oxidant capacity of OLE and its derived spanlastics to be able to induce intracellular free radicals in Caco-2 cells and significantly inhibit the enzymatic activity of both GR and SOD. Our findings in the present study highlight the potential of OLE and its nanoformulation (i.e., spanlastics) as promising pro-oxidant anticancer agents.
Author Contributions
Conceptualization, U.R.A., T.S.A., A.M.S., A.H.E., B.F.A., M.M.A.-S., E.M.M., A.M., K.A.Y., H.R., E.M.O., T.D., M.A. and E.A.; methodology, U.R.A., T.S.A., A.M.S., H.R., E.M.O. and T.D.; software, U.R.A., T.S.A., A.M.S., A.H.E., B.F.A., M.M.A.-S. and E.M.M.; formal analysis, U.R.A., T.S.A., A.M.S., A.H.E., B.F.A., M.M.A.-S., E.M.M., A.M., K.A.Y., M.M.G. and H.R.; investigation, U.R.A. and T.S.A.; resources, U.R.A., T.S.A., E.M.O., T.D., A.M.S., A.H.E. and B.F.A.; data curation, U.R.A., T.S.A., A.M.S., A.H.E., B.F.A., M.M.A.-S., E.M.M., A.M., K.A.Y., H.R. and E.A.; writing—original draft, U.R.A., T.S.A., A.M.S. and A.H.E.; writing—review and editing, U.R.A., T.S.A., A.M.S., A.H.E., B.F.A., M.M.A.-S., E.M.M., A.M., K.A.Y., H.R., E.M.O., T.D. and E.A.; supervision, U.R.A. and T.S.A.; project administration: U.R.A., S.A. and T.S.A.; funding acquisition, U.R.A. and T.S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, grant number 375213500.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
Sarah Albogami extends her appreciation to the Taif University Researchers Supporting Project (number TURSP-2020/202), Taif University, Taif, Saudi Arabia. The authors would appreciate presenting their heartfelt acknowledgment to the central laboratory at Jouf University to aid this research.
Conflicts of Interest
The authors claim no conflict of interest.
References
- Hashmi, M.A.; Khan, A.; Hanif, M.; Farooq, U.; Perveen, S. Traditional Uses, Phytochemistry, and Pharmacology of Olea europaea (Olive). Evid.-Based Complement. Altern. Med. 2015, 2015, 541591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanakis, P.; Termentzi, A.; Michel, T.; Gikas, E.; Halabalaki, M.; Skaltsounis, A.-L. From Olive Drupes to Olive Oil. An HPLC-Orbitrap-based Qualitative and Quantitative Exploration of Olive Key Metabolites. Planta Medica 2013, 79, 1576–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibbett, G.S.; Ferguson, L. Olive Production Manual; UCANR Publications: New York, NY, USA, 2005; Volume 3353. [Google Scholar]
- Bendini, A.; Cerretani, L.; Carrasco-Pancorbo, A.; Gómez-Caravaca, A.M.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Lercker, G. Phenolic Molecules in Virgin Olive Oils: A Survey of Their Sensory Properties, Health Effects, Antioxidant Activity and Analytical Methods. An Overview of the Last Decade Alessandra. Molecules 2007, 12, 1679–1719. [Google Scholar] [CrossRef]
- Ghisalberti, E. Biological and pharmacological activity of naturally occurring iridoids and secoiridoids. Phytomedicine 1998, 5, 147–163. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K. Critical Review. Phenolic compounds in olives. Analyst 1998, 123, 31R–44R. [Google Scholar] [CrossRef]
- Erbay, Z.; Icier, F. The importance and potential uses of olive leaves. Food Rev. Int. 2010, 26, 319–334. [Google Scholar] [CrossRef]
- El, S.N.; Karakaya, S. Olive tree (Olea europaea) leaves: Potential beneficial effects on human health. Nutr. Rev. 2009, 67, 632–638. [Google Scholar] [CrossRef]
- Kalua, C.; Allen, M.; Bedgood, D., Jr.; Bishop, A.; Prenzler, P.D.; Robards, K. Olive oil volatile compounds, flavour development and quality: A critical review. Food Chem. 2007, 100, 273–286. [Google Scholar] [CrossRef]
- Ramírez, E.; Brenes, M.; García, P.; Medina, E.; Romero, C. Oleuropein hydrolysis in natural green olives: Importance of the endogenous enzymes. Food Chem. 2016, 206, 204–209. [Google Scholar] [CrossRef] [Green Version]
- Mostafa, M.; Alaaeldin, E.; Aly, U.F.; Sarhan, H.A. Optimization and Characterization of Thymoquinone-Loaded Liposomes with Enhanced Topical Anti-inflammatory Activity. AAPS PharmSciTech 2018, 19, 3490–3500. [Google Scholar] [CrossRef]
- Refaat, H.; Naguib, Y.W.; Elsayed, M.M.A.; Sarhan, H.A.A.; Alaaeldin, E. Modified Spraying Technique and Response Surface Methodology for the Preparation and Optimization of Propolis Liposomes of Enhanced Anti-Proliferative Activity against Human Melanoma Cell Line A375. Pharmaceutics 2019, 11, 558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refaat, H.; Mady, F.M.; Sarhan, H.A.; Rateb, H.S.; Alaaeldin, E. Optimization and evaluation of propolis liposomes as a promising therapeutic approach for COVID-19. Int. J. Pharm. 2021, 592, 120028. [Google Scholar] [CrossRef]
- González-Ortega, R.; Šturm, L.; Skrt, M.; Di Mattia, C.D.; Pittia, P.; Ulrih, N.P. Liposomal Encapsulation of Oleuropein and an Olive Leaf Extract: Molecular Interactions, Antioxidant Effects and Applications in Model Food Systems. Food Biophys. 2021, 16, 84–97. [Google Scholar] [CrossRef]
- Kakkar, S.; Kaur, I.P. Spanlastics—A novel nanovesicular carrier system for ocular delivery. Int. J. Pharm. 2011, 413, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, A.M.; El-Setouhy, D.A.; Habib, B.A.; Tayel, S.A. Enhancement of Transdermal Delivery of Haloperidol via Spanlastic Dispersions: Entrapment Efficiency vs. Particle Size. AAPS PharmSciTech 2019, 20, 95. [Google Scholar] [CrossRef]
- Griess, B.; Tom, E.; Domann, F.; Teoh-Fitzgerald, M. Extracellular superoxide dismutase and its role in cancer. Free. Radic. Biol. Med. 2017, 112, 464–479. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Yang, C.-X.; Zhang, L.; Yang, C.-Y.; Xu, X.-Q. Baicalein, as a Prooxidant, Triggers Mitochondrial Apoptosis in MCF-7 Human Breast Cancer Cells through Mobilization of Intracellular Copper and Reactive Oxygen Species Generation. OncoTargets Ther. 2019, 12, 10749–10761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephenie, S.; Chang, Y.P.; Gnanasekaran, A.; Esa, N.M.; Gnanaraj, C. An insight on superoxide dismutase (SOD) from plants for mammalian health enhancement. J. Funct. Foods 2020, 68, 103917. [Google Scholar] [CrossRef]
- Shin, J.; Song, M.-H.; Oh, J.-W.; Keum, Y.-S.; Saini, R.K. Pro-oxidant Actions of Carotenoids in Triggering Apoptosis of Cancer Cells: A Review of Emerging Evidence. Antioxidants 2020, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Firczuk, M.; Bajor, M.; Graczyk-Jarzynka, A.; Fidyt, K.; Góral, A.; Zagozdzon, R. Harnessing altered oxidative metabolism in cancer by augmented prooxidant therapy. Cancer Lett. 2020, 471, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alzarea, S.; Elmaidomy, A.; Saber, H.; Musa, A.; Al-Sanea, M.; Mostafa, E.; Hendawy, O.; Youssif, K.; Alanazi, A.; Alharbi, M.; et al. Potential Anticancer Lipoxygenase Inhibitors from the Red Sea-Derived Brown Algae Sargassum cinereum: An In-Silico-Supported In-Vitro Study. Antibiotics 2021, 10, 416. [Google Scholar] [CrossRef]
- New, R.R. Preparation of liposomes. Liposomes Pract. Approach 1990, 13, 33–104. [Google Scholar]
- Ruozi, B.; Tosi, G.; Forni, F.; Fresta, M.; Vandelli, M.A. Atomic force microscopy and photon correlation spectroscopy: Two techniques for rapid characterization of liposomes. Eur. J. Pharm. Sci. 2005, 25, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.M.; Mostafa, M.E.; Alaaeldin, E.; Sarhan, H.A.; Shaykoon, M.S.A.; Allam, S.; Ahmed, A.R.; Elsadek, B.E. Design and Characterisation of Novel Sorafenib-Loaded Carbon Nanotubes with Distinct Tumour-Suppressive Activity in Hepatocellular Carcinoma. Int. J. Nanomed. 2019, 14, 8445–8467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.M.; Alhadrami, H.A.; El-Hawary, S.S.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Abdelmohsen, U.R.; Bakeer, W. Discovery of Two Brominated Oxindole Alkaloids as Staphylococcal DNA Gyrase and Pyruvate Kinase Inhibitors via Inverse Virtual Screening. Microorganisms 2020, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Abouelela, M.E.; Assaf, H.K.; Abdelhamid, R.A.; Elkhyat, E.S.; Sayed, A.M.; Oszako, T.; Belbahri, L.; Zowalaty, A.E.E.; Abdelkader, M.S.A. Identification of Potential SARS-CoV-2 Main Protease and Spike Protein Inhibitors from the Genus Aloe: An In Silico Study for Drug Development. Molecules 2021, 26, 1767. [Google Scholar] [CrossRef]
- Orfali, R.; Rateb, M.; Hassan, H.; Alonazi, M.; Gomaa, M.; Mahrous, N.; GabAllah, M.; Kandeil, A.; Perveen, S.; Abdelmohsen, U.; et al. Sinapic Acid Suppresses SARS CoV-2 Replication by Targeting Its Envelope Protein. Antibiotics 2021, 10, 420. [Google Scholar] [CrossRef]
- Mousouri, E.; Melliou, E.; Magiatis, P. Isolation of Megaritolactones and Other Bioactive Metabolites from ‘Megaritiki’ Table Olives and Debittering Water. J. Agric. Food Chem. 2014, 62, 660–667. [Google Scholar] [CrossRef]
- Rigakou, A.; Diamantakos, P.; Melliou, E.; Magiatis, P. S-(E)-Elenolide: A new constituent of extra virgin olive oil. J. Sci. Food Agric. 2019, 99, 5319–5326. [Google Scholar] [CrossRef] [PubMed]
- Damak, N.; Allouche, N.; Hamdi, B.; Litaudon, M.; Damak, M. New secoiridoid from olive mill wastewater. Nat. Prod. Res. 2011, 26, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Dini, I.; Graziani, G.; Fedele, F.L.; Sicari, A.; Vinale, F.; Castaldo, L.; Ritieni, A. Effects of Trichoderma Biostimulation on the Phenolic Profile of Extra-Virgin Olive Oil and Olive Oil By-Products. Antioxidants 2020, 9, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, F.; Liu, Y.; Hettiarachichi, D.S.; Wang, F.; Li, J.; Sunderland, B.; Li, D. Ximenynic Acid Regulation of n-3 PUFA Content in Liver and Brain. Lifestyle Genom. 2020, 13, 64–73. [Google Scholar] [CrossRef]
- Jimenez-Lopez, C.; Carpena, M.; Lourenço-Lopes, C.; Gallardo-Gomez, M.; Lorenzo, J.M.; Barba, F.J.; Prieto, M.A.; Simal-Gandara, J. Bioactive Compounds and Quality of Extra Virgin Olive Oil. Foods 2020, 9, 1014. [Google Scholar] [CrossRef]
- Olmo-García, L.; Kessler, N.; Neuweger, H.; Wendt, K.; Olmo-Peinado, J.M.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Unravelling the Distribution of Secondary Metabolites in Olea europaea L.: Exhaustive Characterization of Eight Olive-Tree Derived Matrices by Complementary Platforms (LC-ESI/APCI-MS and GC-APCI-MS). Molecules 2018, 23, 2419. [Google Scholar] [CrossRef] [Green Version]
- Mukesi, M.; Iweriebor, B.C.; Obi, L.C.; Nwodo, U.U.; Moyo, S.R.; Okoh, A.I. The activity of commercial antimicrobials, and essential oils and ethanolic extracts of Olea europaea on Streptococcus agalactiae isolated from pregnant women. BMC Complement. Altern. Med. 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Hisada, S.; Nishibe, S. Lignans from bark of the Olea plants. I. Chem. Pharm. Bull. 1984, 32, 2730–2735. [Google Scholar] [CrossRef]
- Owen, R.W.; Mier, W.; Giacosa, A.; Hull, W.E.; Spiegelhalder, B.; Bartsch, H. Identification of Lignans as Major Components in the Phenolic Fraction of Olive Oil. Clin. Chem. 2000, 46, 976–988. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wesemann, S.; Krenn, L.; Ladurner, A.; Heiss, E.; Dirsch, V.; Atanasov, A.G. Erythrodiol, an Olive Oil Constituent, Increases the Half-Life of ABCA1 and Enhances Cholesterol Efflux from THP-1-Derived Macrophages. Front. Pharmacol. 2017, 8, 375. [Google Scholar] [CrossRef]
- Edgecombe, S.C.; Stretch, G.L.; Hayball, P.J. Oleuropein, an Antioxidant Polyphenol from Olive Oil, Is Poorly Absorbed from Isolated Perfused Rat Intestine. J. Nutr. 2000, 130, 2996–3002. [Google Scholar] [CrossRef]
- Benot-Dominguez, R.; Tupone, M.G.; Castelli, V.; D’Angelo, M.; Benedetti, E.; Quintiliani, M.; Cinque, B.; Forte, I.M.; Cifone, M.G.; Ippoliti, R.; et al. Olive leaf extract impairs mitochondria by pro-oxidant activity in MDA-MB-231 and OVCAR-3 cancer cells. Biomed. Pharmacother. 2021, 134, 111139. [Google Scholar] [CrossRef]
- De Stefanis, D.; Scimè, S.; Accomazzo, S.; Catti, A.; Occhipinti, A.; Bertea, C.M.; Costelli, P. Anti-Proliferative Effects of an Extra-Virgin Olive Oil Extract Enriched in Ligstroside Aglycone and Oleocanthal on Human Liver Cancer Cell Lines. Cancers 2019, 11, 1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Wang, H.; He, M.; Chen, B.; Yang, B.; Hu, B. Size-dependent cytotoxicity study of ZnO nanoparticles in HepG2 cells. Ecotoxicol. Environ. Saf. 2019, 171, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef] [PubMed]
- Salatin, S.; Dizaj, S.M.; Khosroushahi, A.Y. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol. Int. 2015, 39, 881–890. [Google Scholar] [CrossRef]
- Wang, J.-C.; Chu, P.-Y.; Chen, C.-M.; Lin, J.-H. idTarget: A web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res. 2012, 40, W393–W399. [Google Scholar] [CrossRef] [Green Version]
- Hwang, G.H.; Ryu, J.M.; Jeon, Y.J.; Choi, J.; Han, H.J.; Lee, Y.-M.; Lee, S.; Bae, J.-S.; Jung, J.-W.; Chang, W.; et al. The role of thioredoxin reductase and glutathione reductase in plumbagin-induced, reactive oxygen species-mediated apoptosis in cancer cell lines. Eur. J. Pharmacol. 2015, 765, 384–393. [Google Scholar] [CrossRef]
- Begum, S.S.; Das, D.; Gour, N.K.; Deka, R.C. Computational modelling of nanotube delivery of anti-cancer drug into glutathione reductase enzyme. Sci. Rep. 2021, 11, 4950. [Google Scholar] [CrossRef]
- Güller, P.; Karaman, M.; Güller, U.; Aksoy, M.; Küfrevioğlu Ömer, I. A study on the effects of inhibition mechanism of curcumin, quercetin, and resveratrol on human glutathione reductase through in vitro and in silico approaches. J. Biomol. Struct. Dyn. 2021, 39, 1744–1753. [Google Scholar] [CrossRef]
- Wilkes, J.G.; Alexander, M.S.; Cullen, J.J. Superoxide Dismutases in Pancreatic Cancer. Antioxidants 2017, 6, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Chen, Y.; Zhao, J.; Shi, J.; Wang, M.; Qiu, S.; Hu, Y.; Xu, Y.; Cui, Y.; Liu, C.; et al. The Specific Inhibition of SOD1 Selectively Promotes Apoptosis of Cancer Cells via Regulation of the ROS Signaling Network. Oxidative Med. Cell. Longev. 2019, 2019, 1–21. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).