Reintroduction of DJ-1 in Müller Cells Inhibits Retinal Degeneration in the DJ-1 Deficient Retina
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Maintenance
2.2. Zebrafish Lines
2.3. Eye Sectioning, Toluidine Blue Staining and Image Analysis
2.4. Cryo-Sectioning
2.5. In Situ Hybridization
2.6. Protein Extraction
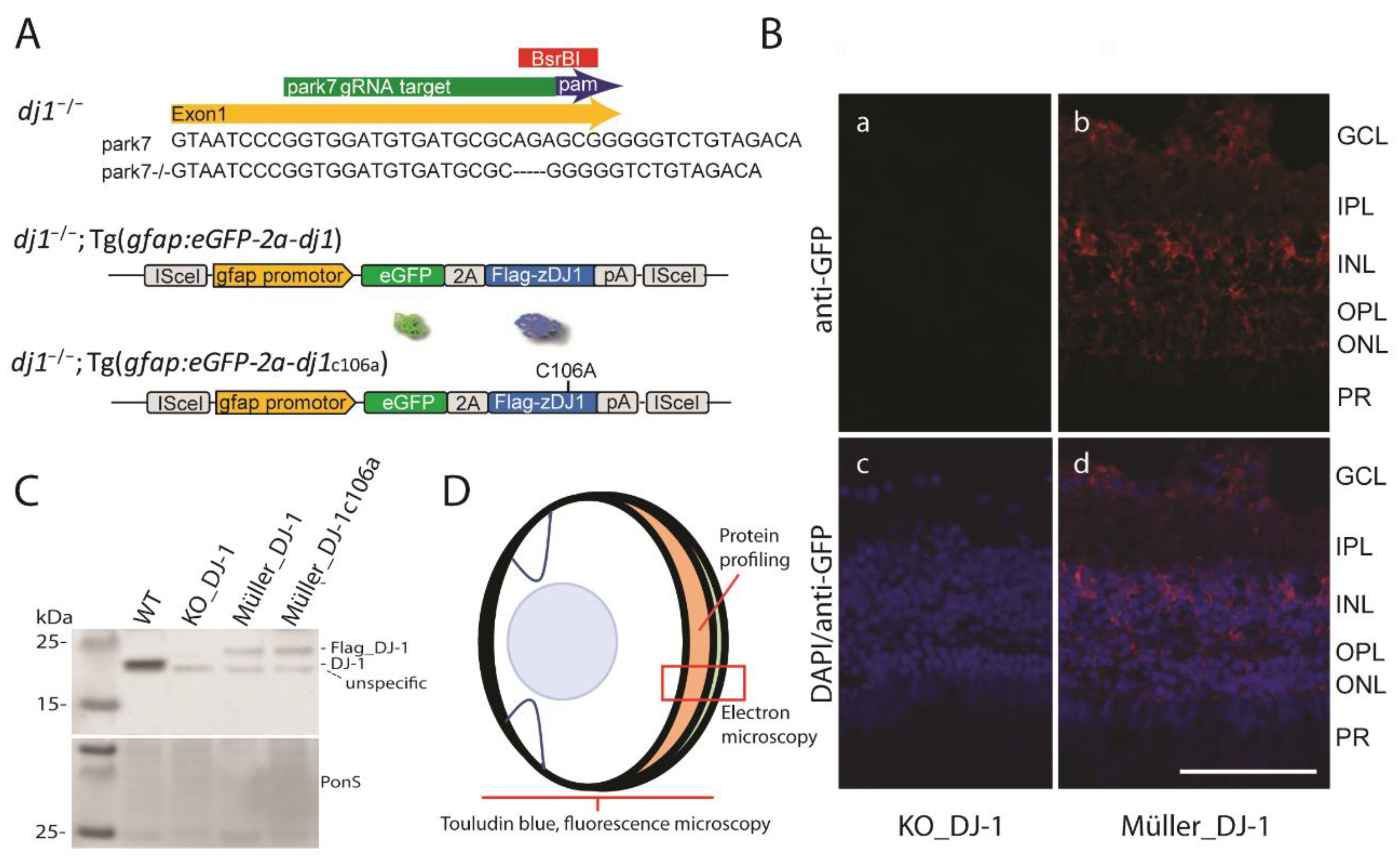
2.7. Verification of Transgenic Line
2.8. Transmission Electron Microscopy
2.9. Isolation of Retina for Mass Spectrometry
2.10. Sample Preparation for Mass Spectrometry
2.11. Label-Free Mass Spectrometry
2.12. Data Interpretation
3. Results
3.1. Generation of Transgenic Zebrafish Lines with Müller Cell Specific Wild Type DJ-1 and DJ-1c106a Expression in a DJ-1 Null Background
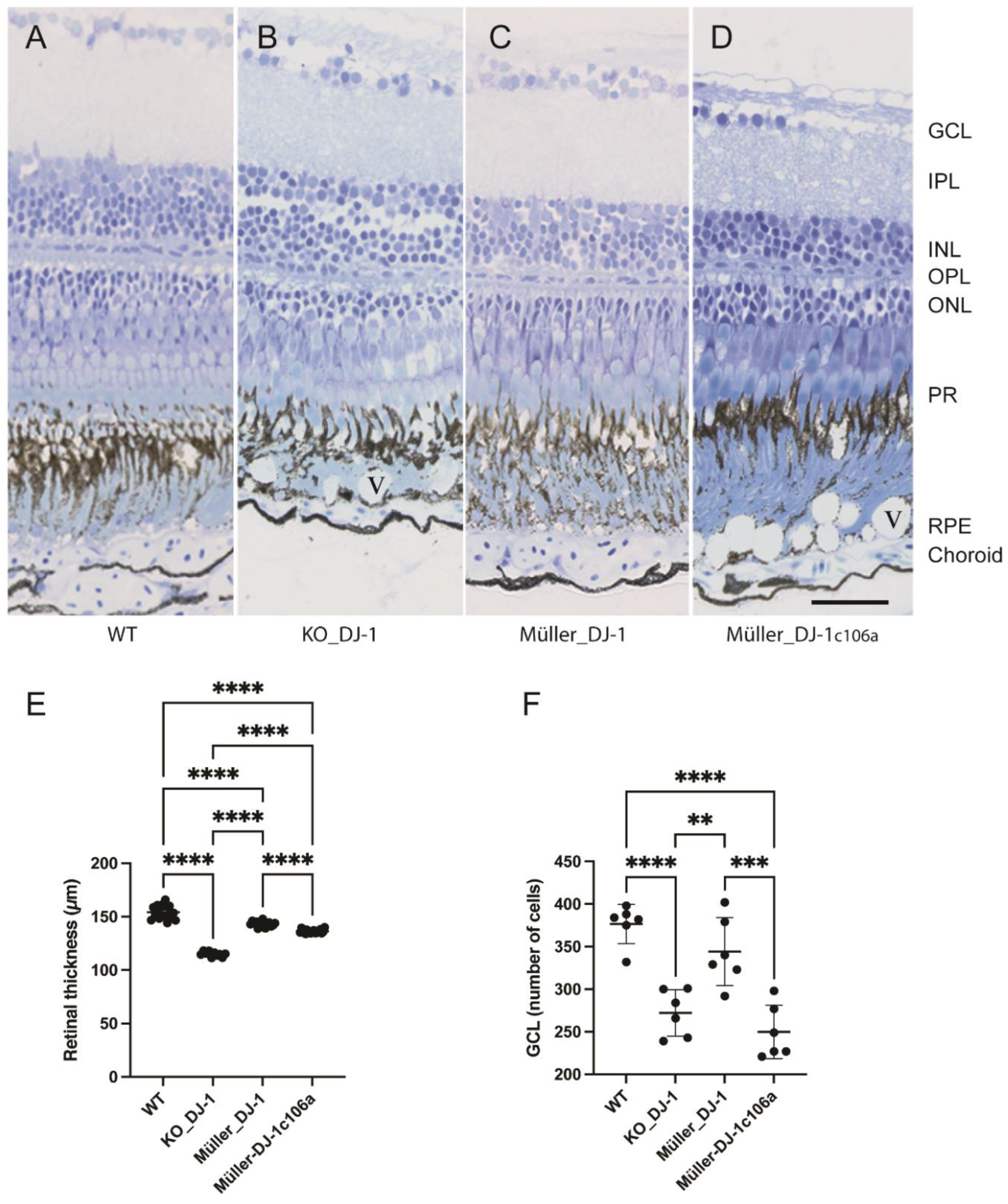
3.2. Retinal Degeneration Induced by the Loss of DJ-1 Can Be Inhibited by Müller Cell Expressed DJ-1
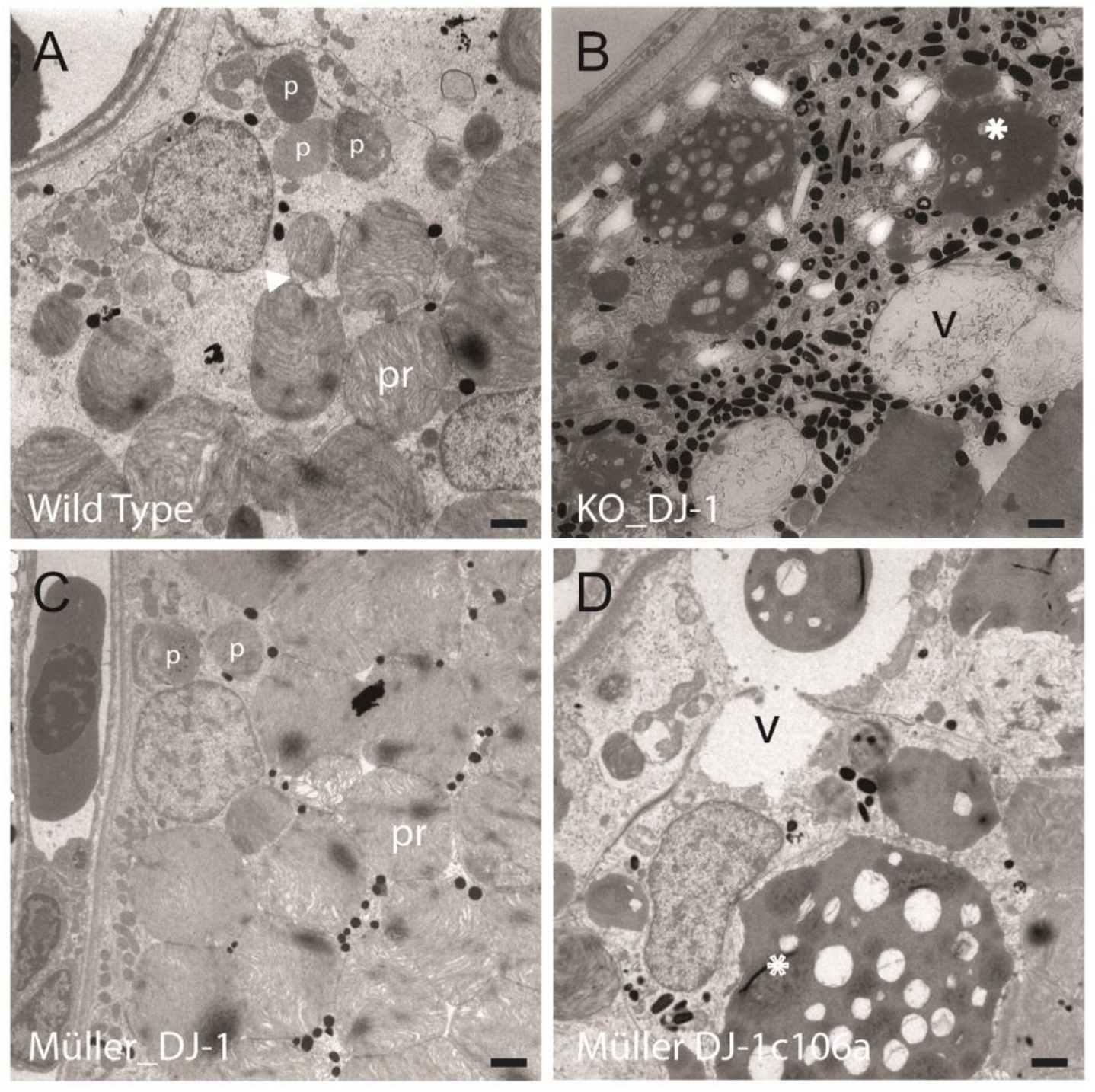
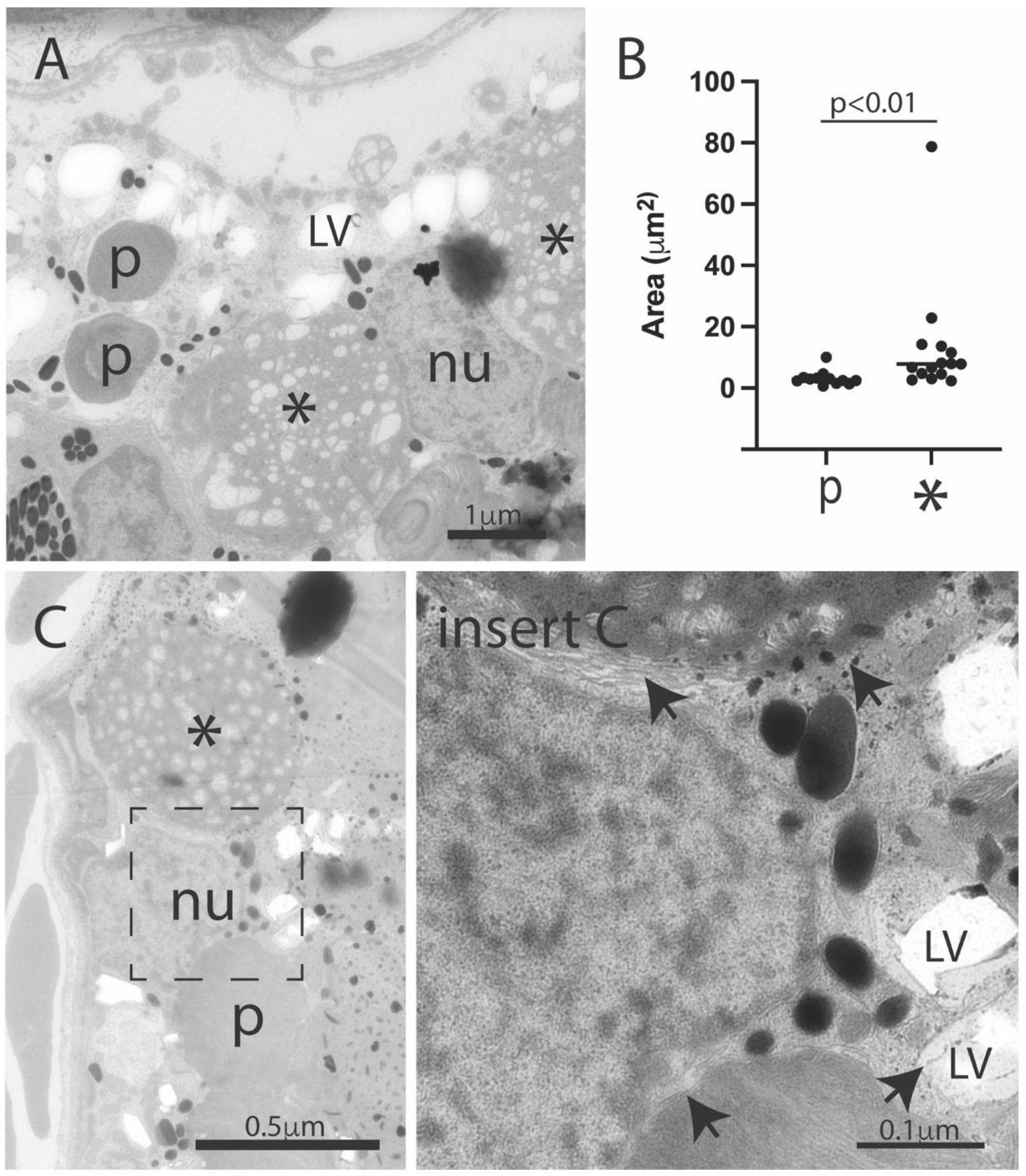
3.3. Introduction of Müller Cell DJ-1 Expression Inhibits DJ-1 Loss-Induced Ultrastructural Changes in the Retinal Pigment Epithelia Cells
3.4. Protein Profiling of Resected Retinas
3.5. Expression of Retinal Cell Markers in Knockout and Transgenic Lines
3.6. Loss of DJ-1 Alters Expression of Proteins Belonging to the Respiratory Complex I and Glycolysis Independently of Reinsertion of Müller Cell DJ-1
3.7. Identification of Retinal Proteins Regulated by the Loss of DJ-1, but with Restored Levels after Introducing Müller Cell DJ-1
3.8. Identification of Proteins with Altered Expression in DJ-1 Knockouts Regardless of Introducing Either Müller Cell DJ-1 or Müller Cell DJ-1c106a
3.9. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ariga, H.; Iguchi-Ariga, S.M.M. DJ-1/PARK7 Protein: Parkinson’s Disease, Cancer and Oxidative Stress-Induced Diseases; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Zhou, W.; Freed, C.R. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J. Biol. Chem. 2005, 280, 43150–43158. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Ren, H.; Jia, N.; Fei, E.; Zhou, T.; Jiang, P.; Wu, M.; Wang, G. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J. Biol. Chem. 2008, 283, 4022–4030. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Petrie, T.G.; Liu, Y.; Liu, J.; Fujioka, H.; Zhu, X. Parkinson’s disease-associated DJ-1 mutations impair mitochondrial dynamics and cause mitochondrial dysfunction. J. Neurochem. 2012, 121, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cai, Z.; Tao, K.; Zeng, W.; Lu, F.; Yang, R.; Feng, D.; Gao, G.; Yang, Q. Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy 2016, 12, 1215–1228. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, X.; Fujioka, H.; Liu, J.; Chen, S.; Zhu, X. DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc. Natl. Acad. Sci. USA 2019, 116, 25322–25328. [Google Scholar] [CrossRef]
- Canet-Aviles, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozawa, K.; Tsumoto, H.; Miura, Y.; Yamaguchi, J.; Iguchi-Ariga, S.M.M.; Sakuma, T.; Yamamoto, T.; Uchiyama, Y. DJ-1 is indispensable for the S-nitrosylation of Parkin, which maintains function of mitochondria. Sci. Rep. 2020, 10, 4377. [Google Scholar] [CrossRef] [Green Version]
- Moscovitz, O.; Ben-Nissan, G.; Fainer, I.; Pollack, D.; Mizrachi, L.; Sharon, M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015, 6, 6609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junn, E.; Jang, W.H.; Zhao, X.; Jeong, B.S.; Mouradian, M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009, 87, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonilha, V.L.; Bell, B.A.; Rayborn, M.E.; Samuels, I.S.; King, A.; Hollyfield, J.G.; Xie, C.; Cai, H. Absence of DJ-1 causes age-related retinal abnormalities in association with increased oxidative stress. Free Radic. Biol. Med. 2017, 104, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Bonilha, V.L.; Bell, B.A.; Rayborn, M.E.; Yang, X.; Kaul, C.; Grossman, G.H.; Samuels, I.S.; Hollyfield, J.G.; Xie, C.; Cai, H.; et al. Loss of DJ-1 elicits retinal abnormalities, visual dysfunction, and increased oxidative stress in mice. Exp. Eye Res. 2015, 139, 22–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Nieto, J.; Uribe, M.L.; Esteve-Rudd, J.; Herrero, M.T.; Campello, L. A role for DJ-1 against oxidative stress in the mammalian retina. Neurosci. Lett. 2019, 708, 134361. [Google Scholar] [CrossRef]
- Bringmann, A.; Pannicke, T.; Grosche, J.; Francke, M.; Wiedemann, P.; Skatchkov, S.N.; Osborne, N.N.; Reichenbach, A. Muller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 2006, 25, 397–424. [Google Scholar] [CrossRef] [PubMed]
- Eastlake, K.; Luis, J.; Limb, G.A. Potential of Muller Glia for Retina Neuroprotection. Curr. Eye Res. 2020, 45, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karl, M.O.; Hayes, S.; Nelson, B.R.; Tan, K.; Buckingham, B.; Reh, T.A. Stimulation of neural regeneration in the mouse retina. Proc. Natl. Acad. Sci. USA 2008, 105, 19508–19513. [Google Scholar] [CrossRef] [Green Version]
- Frøyset, A.K.; Edson, A.J.; Gharbi, N.; Khan, E.A.; Dondorp, D.; Bai, Q.; Tiraboschi, E.; Suster, M.L.; Connolly, J.B.; Burton, E.A.; et al. Astroglial DJ-1 over-expression up-regulates proteins involved in redox regulation and is neuroprotective in vivo. Redox Biol. 2018, 16, 237–247. [Google Scholar] [CrossRef]
- De Miranda, B.R.; Rocha, E.M.; Bai, Q.; Ayadi, A.; Hinkle, D.; Burton, E.A.; Greenamyre, T.J. Astrocyte-specific DJ-1 overexpression protects against rotenone-induced neurotoxicity in a rat model of Parkinson’s disease. Neurobiol. Dis. 2018, 115, 101–114. [Google Scholar] [CrossRef]
- Edson, A.J.; Hushagen, H.A.; Froyset, A.K.; Elda, I.; Khan, E.A.; Di Stefano, A.; Fladmark, K.E. Dysregulation in the Brain Protein Profile of Zebrafish Lacking the Parkinson’s Disease-Related Protein DJ-1. Mol. Neurobiol. 2019, 56, 8306–8322. [Google Scholar] [CrossRef]
- Koke, J.R.; Mosier, A.L.; Garcia, D.M. Intermediate filaments of zebrafish retinal and optic nerve astrocytes and Muller glia: Differential distribution of cytokeratin and GFAP. BMC Res. Notes 2010, 3, 50. [Google Scholar] [CrossRef] [Green Version]
- Kuzmanovic, M.; Dudley, V.J.; Sarthy, V.P. GFAP promoter drives Muller cell-specific expression in transgenic mice. Invest. Ophthalmol. Vis. Sci. 2003, 44, 3606–3613. [Google Scholar] [CrossRef] [Green Version]
- Bonifati, V.; Rizzu, P.; Squitieri, F.; Krieger, E.; Vanacore, N.; van Swieten, J.C.; Brice, A.; van Duijn, C.M.; Oostra, B.; Meco, G.; et al. DJ-1( PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 2003, 24, 159–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekker, M.S.; Janssen, S.; Seppi, K.; Poewe, W.; de Vries, N.M.; Theelen, T.; Nonnekes, J.; Bloem, B.R. Ocular and visual disorders in Parkinson’s disease: Common but frequently overlooked. Parkinsonism. Relat. Disord. 2017, 40, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Lee, J.Y.; Kim, T.W.; Yoon, E.J.; Oh, S.; Kim, Y.K.; Kim, J.M.; Woo, S.J.; Kim, K.W.; Jeon, B. Retinal thinning associates with nigral dopaminergic loss in de novo Parkinson disease. Neurology 2018, 91, e1003–e1012. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, X.; Qi, J.; Pant, O.P.; Lu, C.W.; Hao, J. DJ-1 in Ocular Diseases: A Review. Int. J. Med. Sci. 2018, 15, 430–435. [Google Scholar] [CrossRef] [Green Version]
- Legland, D.; Arganda-Carreras, I.; Andrey, P. MorphoLibJ: Integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 2016, 32, 3532–3534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharbi, N.; Zhao, X.F.; Ellingsen, S.; Fjose, A. Zebrafish enhancer trap line showing maternal and neural expression of kctd15a. Dev. Growth Differ. 2012, 54, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Aranguren-Abadia, L.; Donald, C.E.; Eilertsen, M.; Gharbi, N.; Tronci, V.; Sorhus, E.; Mayer, P.; Nilsen, T.O.; Meier, S.; Goksoyr, A.; et al. Expression and localization of the aryl hydrocarbon receptors and cytochrome P450 1A during early development of Atlantic cod (Gadus morhua). Aquat. Toxicol. 2020, 226, 105558. [Google Scholar] [CrossRef] [PubMed]
- Froyset, A.K.; Khan, E.A.; Fladmark, K.E. Quantitative proteomics analysis of zebrafish exposed to sub-lethal dosages of beta-methyl-amino-L-alanine (BMAA). Sci. Rep. 2016, 6, 29631. [Google Scholar] [CrossRef] [Green Version]
- Hoang, T.; Wang, J.; Boyd, P.; Wang, F.; Santiago, C.; Jiang, L.; Yoo, S.; Lahne, M.; Todd, L.J.; Jia, M.; et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science 2020, 370. [Google Scholar] [CrossRef]
- Kennedy, C.J.; Rakoczy, P.E.; Constable, I.J. Lipofuscin of the retinal pigment epithelium: A review. Eye 1995, 9, 763–771. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, M.; Mohi, A.; Dettbarn, M.C.; Miura, Y.; Aherrahrou, Z.; Ranjbar, M.; Mutus, B.; Knobloch, J.K. Detection of esterified cholesterol in murine Bruch’s membrane wholemounts with a perfringolysin O-based cholesterol marker. Invest. Ophthalmol. Vis. Sci. 2014, 55, 4759–4767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.L.; Djajadi, H.; Molday, R.S. Cell-Specific Markers for the Identification of Retinal Cells by Immunofluorescence Microscopy. In Retinal Degeneration. Methods in Molecular Biology (Methods and Protocols); Weber, B.L.T., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 935. [Google Scholar]
- Liao, M.L.; Peng, W.H.; Kan, D.; Chien, C.L. Distribution patterns of the zebrafish neuronal intermediate filaments inaa and inab. J. Neurosci. Res. 2019, 97, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Solana-Manrique, C.; Sanz, F.J.; Ripolles, E.; Bano, M.C.; Torres, J.; Munoz-Soriano, V.; Paricio, N. Enhanced activity of glycolytic enzymes in Drosophila and human cell models of Parkinson’s disease based on DJ-1 deficiency. Free Radic. Biol. Med. 2020, 158, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mosienko, V.; Vaccari Cardoso, B.; Prokudina, D.; Huentelman, M.; Teschemacher, A.G.; Kasparov, S. Glio- and neuro-protection by prosaposin is mediated by orphan G-protein coupled receptors GPR37L1 and GPR37. Glia 2018, 66, 2414–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Jin, J.P. Calponin isoforms CNN1, CNN2 and CNN3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene 2016, 585, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilling, D.; Gomer, R.H. The Development of Serum Amyloid P as a Possible Therapeutic. Front. Immunol. 2018, 9, 2328. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Song, E.; Wang, W.; Hsieh, C.H.; Wang, X.; Feng, W.; Wang, X.; Shen, K. Metaxins are core components of mitochondrial transport adaptor complexes. Nat. Commun. 2021, 12, 83. [Google Scholar] [CrossRef]
- Maita, C.; Maita, H.; Iguchi-Ariga, S.M.; Ariga, H. Monomer DJ-1 and its N-terminal sequence are necessary for mitochondrial localization of DJ-1 mutants. PLoS ONE 2013, 8, e54087. [Google Scholar] [CrossRef] [Green Version]
- D’Anna, C.; Cascio, C.; Cigna, D.; Galizzi, G.; Deidda, I.; Bianchi, L.; Russo, D.; Passantino, R.; Bini, L.; Guarneri, P. A retinal proteomics-based study identifies alphaA-crystallin as a sex steroid-regulated protein. Proteomics 2011, 11, 986–990. [Google Scholar] [CrossRef]
- Kaiser, C.J.O.; Peters, C.; Schmid, P.W.N.; Stavropoulou, M.; Zou, J.; Dahiya, V.; Mymrikov, E.V.; Rockel, B.; Asami, S.; Haslbeck, M.; et al. The structure and oxidation of the eye lens chaperone alphaA-crystallin. Nat. Struct. Mol. Biol. 2019, 26, 1141–1150. [Google Scholar] [CrossRef]
- Roskamp, K.W.; Azim, S.; Kassier, G.; Norton-Baker, B.; Sprague-Piercy, M.A.; Miller, R.J.D.; Martin, R.W. Human gammaS-Crystallin-Copper Binding Helps Buffer against Aggregation Caused by Oxidative Damage. Biochemistry 2020, 59, 2371–2385. [Google Scholar] [CrossRef] [PubMed]
- Siintola, E.; Partanen, S.; Stromme, P.; Haapanen, A.; Haltia, M.; Maehlen, J.; Lehesjoki, A.E.; Tyynela, J. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain 2006, 129, 1438–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, J.T.; Shashoua, V.E. Antibodies to ependymin block the sharpening of the regenerating retinotectal projection in goldfish. Brain Res. 1988, 446, 269–284. [Google Scholar] [CrossRef]
- Upadhyay, M.; Milliner, C.; Bell, B.A.; Bonilha, V.L. Oxidative stress in the retina and retinal pigment epithelium (RPE): Role of aging, and DJ-1. Redox Biol 2020, 101623. [Google Scholar] [CrossRef] [PubMed]
- La Morgia, C.; Di Vito, L.; Carelli, V.; Carbonelli, M. Patterns of Retinal Ganglion Cell Damage in Neurodegenerative Disorders: Parvocellular vs Magnocellular Degeneration in Optical Coherence Tomography Studies. Front. Neurol. 2017, 8, 710. [Google Scholar] [CrossRef]
- Bandopadhyay, R.; Kingsbury, A.E.; Cookson, M.R.; Reid, A.R.; Evans, I.M.; Hope, A.D.; Pittman, A.M.; Lashley, T.; Canet-Aviles, R.; Miller, D.W.; et al. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain 2004, 127, 420–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, M.; Muller, V.; Gorner, K.; Kretzschmar, H.A.; Haass, C.; Kahle, P.J. Pathological properties of the Parkinson’s disease-associated protein DJ-1 in alpha-synucleinopathies and tauopathies: Relevance for multiple system atrophy and Pick’s disease. Acta Neuropathol. 2004, 107, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Vecino, E.; Rodriguez, F.D.; Ruzafa, N.; Pereiro, X.; Sharma, S.C. Glia-neuron interactions in the mammalian retina. Prog. Retin. Eye Res. 2016, 51, 1–40. [Google Scholar] [CrossRef] [Green Version]
- Oji, Y.; Hatano, T.; Ueno, S.I.; Funayama, M.; Ishikawa, K.I.; Okuzumi, A.; Noda, S.; Sato, S.; Satake, W.; Toda, T.; et al. Variants in saposin D domain of prosaposin gene linked to Parkinson’s disease. Brain 2020, 143, 1190–1205. [Google Scholar] [CrossRef]
- Chen, H.M.; Lin, C.Y.; Wang, V. Amyloid P component as a plasma marker for Parkinson’s disease identified by a proteomic approach. Clin. Biochem. 2011, 44, 377–385. [Google Scholar] [CrossRef]
- Shi, S.Y.; Lu, S.Y.; Sivasubramaniyam, T.; Revelo, X.S.; Cai, E.P.; Luk, C.T.; Schroer, S.A.; Patel, P.; Kim, R.H.; Bombardier, E.; et al. DJ-1 links muscle ROS production with metabolic reprogramming and systemic energy homeostasis in mice. Nat. Commun. 2015, 6, 7415. [Google Scholar] [CrossRef] [Green Version]
- Weinert, M.; Millet, A.; Jonas, E.A.; Alavian, K.N. The mitochondrial metabolic function of DJ-1 is modulated by 14-3-3beta. FASEB J. 2019, 33, 8925–8934. [Google Scholar] [CrossRef] [PubMed]
- Toft-Kehler, A.K.; Skytt, D.M.; Svare, A.; Lefevere, E.; Van Hove, I.; Moons, L.; Waagepetersen, H.S.; Kolko, M. Mitochondrial function in Muller cells—Does it matter? Mitochondrion 2017, 36, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, H.; Miyagi, M.; Darrow, R.M.; Crabb, J.S.; Hollyfield, J.G.; Organisciak, D.T.; Crabb, J.W. Intense light exposure changes the crystallin content in retina. Exp. Eye Res. 2003, 76, 131–133. [Google Scholar] [CrossRef]
- Thanos, S.; Bohm, M.R.; Meyer zu Horste, M.; Prokosch-Willing, V.; Hennig, M.; Bauer, D.; Heiligenhaus, A. Role of crystallins in ocular neuroprotection and axonal regeneration. Prog. Retin. Eye Res. 2014, 42, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, A.; Bringmann, A. New functions of Muller cells. Glia 2013, 61, 651–678. [Google Scholar] [CrossRef]
- Meiser, J.; Delcambre, S.; Wegner, A.; Jager, C.; Ghelfi, J.; d’Herouel, A.F.; Dong, X.; Weindl, D.; Stautner, C.; Nonnenmacher, Y.; et al. Loss of DJ-1 impairs antioxidant response by altered glutamine and serine metabolism. Neurobiol. Dis. 2016, 89, 112–125. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Gillies, M.C.; Madigan, M.C.; Shen, W.; Du, J.; Grunert, U.; Zhou, F.; Yam, M.; Zhu, L. Disruption of De Novo Serine Synthesis in Muller Cells Induced Mitochondrial Dysfunction and Aggravated Oxidative Damage. Mol. Neurobiol. 2018, 55, 7025–7037. [Google Scholar] [CrossRef]
- Kim, J.M.; Cha, S.H.; Choi, Y.R.; Jou, I.; Joe, E.H.; Park, S.M. DJ-1 deficiency impairs glutamate uptake into astrocytes via the regulation of flotillin-1 and caveolin-1 expression. Sci. Rep. 2016, 6, 28823. [Google Scholar] [CrossRef]
- Habas, A.; Hahn, J.; Wang, X.; Margeta, M. Neuronal activity regulates astrocytic Nrf2 signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 18291–18296. [Google Scholar] [CrossRef] [Green Version]
- Clements, C.M.; McNally, R.S.; Conti, B.J.; Mak, T.W.; Ting, J.P. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA 2006, 103, 15091–15096. [Google Scholar] [CrossRef] [Green Version]
- Jablonski, M.M.; Iannaccone, A. Targeted disruption of Muller cell metabolism induces photoreceptor dysmorphogenesis. Glia 2000, 32, 192–204. [Google Scholar] [CrossRef]
- Roesch, K.; Jadhav, A.P.; Trimarchi, J.M.; Stadler, M.B.; Roska, B.; Sun, B.B.; Cepko, C.L. The transcriptome of retinal Muller glial cells. J. Comp. Neurol. 2008, 509, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.E.; Mouradian, M.M. Regulation of Signal Transduction by DJ-1. Adv. Exp. Med. Biol. 2017, 1037, 97–131. [Google Scholar] [CrossRef] [Green Version]
- Imai, Y.; Soda, M.; Inoue, H.; Hattori, N.; Mizuno, Y.; Takahashi, R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 2001, 105, 891–902. [Google Scholar] [CrossRef] [Green Version]
- Sparrow, J.R.; Boulton, M. RPE lipofuscin and its role in retinal pathobiology. Exp. Eye Res. 2005, 80, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Bejarano-Escobar, R.; Sanchez-Calderon, H.; Otero-Arenas, J.; Martin-Partido, G.; Francisco-Morcillo, J. Muller glia and phagocytosis of cell debris in retinal tissue. J. Anat. 2017, 231, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Annesley, S.J.; Jasim, R.A.F.; Fisher, P.R. The Parkinson’s Disease-Associated Protein DJ-1 Protects Dictyostelium Cells from AMPK-Dependent Outcomes of Oxidative Stress. Cells 2021, 10, 1874. [Google Scholar] [CrossRef] [PubMed]
| p-Values (vs. WT) | Max Fold | Average LFQ (log 2) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Gene Name | Protein Name | Total Peptides | Unique Peptides | Score | KO | M_DJ-1 | M_DJ-1c106a | KO/WT | M_DJ-1/WT | M_DJ-1c106a/WT | Wild Type | KO | M_DJ-1 | M_DJ-1c106a |
| Mitochondrial Complex I | |||||||||||||||
| E9QEE8 | ndufb4 | 3 | 3 | 31 | *** | ** | *** | 0.3 | 0.4 | 0.5 | 25.38 ± 0.08 | 23.43 ± 0.29 | 23.89 ± 0.30 | 24.35 ± 0.15 | |
| Q498W6 | ndufa12 | 4 | 4 | 38 | * | ** | * | 0.3 | 0.4 | 0.5 | 26.04 ± 0.20 | 24.41 ± 0.51 | 24.79 ± 0.13 | 25.01 ± 0.4 | |
| Q6P6E5 | ndufb7 | 4 | 4 | 60 | * | ** | * | 0.3 | 0.6 | 0.7 | 26.44 ± 0.10 | 24.89 ± 0.54 | 25.59 ± 0.18 | 25.83 ± 0.18 | |
| Q6DGM9 | ndufa7 | 3 | 3 | 22 | ** | ** | * | 0.4 | 0.5 | 0.5 | 25.61 ± 0.09 | 24.14 ± 0.43 | 24.49 ± 0.20 | 24.66 ± 0.47 | |
| Q6PBJ6 | ndufb6 | 3 | 3 | 23 | *** | * | * | 0.4 | 0.4 | 0.5 | 25.75 ± 0.07 | 24.50 ± 0.05 | 24.56 ± 0.44 | 24.73 ± 0.37 | |
| Q6AZA2 | ndufv1 | 14 | 14 | 156 | * | * | * | 0.5 | 0.6 | 0.6 | 28.20 ± 0.12 | 27.08 ± 0.47 | 27.46 ± 0.27 | 27.42 ± 0.25 | |
| Q6PBX8 | ndufv2 | 5 | 5 | 57 | * | * | * | 0.5 | 0.6 | 0.6 | 26.77 ± 0.21 | 25.67 ± 0.51 | 26.02 ± 0.17 | 26.1 ± 0.24 | |
| Q8AW03 | ndufa6 | 5 | 5 | 65 | ** | ** | * | 0.6 | 0.5 | 0.6 | 26.51 ± 0.12 | 25.68 ± 0.01 | 25.62 ± 0.21 | 25.75 ± 0.24 | |
| F1QHE9 | ndufs7 | 4 | 4 | 37 | * | ** | * | 0.6 | 0.5 | 0.6 | 26.01 ± 0.13 | 25.32 ± 0.14 | 25.05 ± 0.22 | 25.29 ± 0.12 | |
| Q3B7G1 | ndufb5 | 6 | 6 | 43 | * | ** | * | 0.5 | 0.5 | 0.5 | 26.18 ± 0.23 | 25.12 ± 0.11 | 25.16 ± 0.13 | 25.22 ± 0.25 | |
| A0A0U2NDI4 | ND1 | 3 | 3 | 20 | ** | * | ** | 0.6 | 0.7 | 0.6 | 26.00 ± 0.09 | 25.22 ± 0.10 | 25.39 ± 0.19 | 25.34 ± 0.1 | |
| Glycolysis Pathway | |||||||||||||||
| Q9PVK5 | ldha | L-lactate dehydrogenase | 16 | 14 | 185 | * | ** | * | 1.6 | 1.4 | 1.2 | 30.76 ± 0.10 | 31.43 ± 0.29 | 31.22 ± 0.06 | 31.05 ± 0.05 |
| Stress and Inflammatory Response | |||||||||||||||
| F6NYT7 | gpx1a | Glutathione peroxidase | 11 | 11 | 83 | ** | ** | * | 115 | 51 | 47 | 21.65 ± 0.11 | 28.50 ± 0.70 | 27.32 ± 0.54 | 27.21 ± 1.29 |
| Q9DDU5 | gstp1 | Glutathione S-transferase, Pi | 13 | 13 | 175 | * | ** | ** | 1.9 | 1.9 | 1.8 | 31.70 ± 0.14 | 32.58 ± 0.32 | 32.59 ± 0.14 | 32.52 ± 0.07 |
| A0A2R8RU89 | cst14b.2 | Cystatin 14b | 3 | 3 | 23 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | NaN | 24.37 ± 1.19 | 23.82 ± 1.16 | 23.75 ± 1.71 |
| Q5PR64 | hspb1 | Heat shock 27 kDa protein | 9 | 9 | 86 | ** | ** | ** | 0.3 | 0.4 | 0.4 | 27.93 ± 0.21 | 26.33 ± 0.16 | 26.66 ± 0.14 | 26.42 ± 0.17 |
| Lipid Metabolism | |||||||||||||||
| Q5IHX6 | ptges3a | Cytosolic prostaglandin E synthase | 5 | 4 | 48 | *** | *** | *** | 4.4 | 4.0 | 3.6 | 24.01 ± 0.14 | 26.14 ± 0.31 | 26.01 ± 0.13 | 25.76 ± 0.23 |
| F1QVT8 | enpp6 | Choline-specific glycerophosphodiester phosphodiesterase | 8 | 8 | 49 | * | * | ** | 1.8 | 1.5 | 1.6 | 24.97 ± 0.18 | 25.83 ± 0.39 | 25.53 ± 0.12 | 25.65 ± 0.11 |
| p-Value | Max Fold | Average LFQ (log2) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Gene Name | Protein Name | Total pep. | Unique pep. | Score | WT vs. KO | KO vs.M_DJ-1 | DJ-1 vs. DJ-1c106a | KO/WT | M_DJ1/ WT | M_DJ-1c106/WT | WT | KO | M_DJ-1 | M_DJ-1c106a |
| F1R6R2 | apcs | Amyloid P component, serum | 9 | 9 | 171 | * | * | n.s. | 2.6 | 0.6 | 1.2 | 28.87 ± 0.07 | 30.27 ± 0.61 | 28.14 ± 0.40 | 29.17 ± 0.72 |
| Q8UVZ4 | psap | Prosaposin | 6 | 6 | 64 | * | * | n.s. | 1.5 | 1.1 | 1.4 | 27.27 ± 0.11 | 27.88 ± 0.17 | 27.41 ± 0.16 | 27.73 ± 0.35 |
| A4FUN8 | rhol | Rhodopsin-like | 4 | 4 | 49 | * | * | n.s. | 0.5 | 0.8 | 0.5 | 26.63 ± 0.36 | 25.61 ± 0.25 | 26.40 ± 0.24 | 25.73 ± 0.24 |
| Q6PBA5 | cnn2 | Calponin | 4 | 3 | 30 | 25.02 ± 0.74 | 23.90 ± 0.54 | ||||||||
| A0A2R8QT38 | rpl36a | 60S ribosomal protein L36a | 2 | 2 | 14 | 24.47 ± 0.05 | 24.74 ± 0.26 | ||||||||
| E7F0E8 | xpo1a | Exportin 1 | 21 | 4 | 29 | 24.02 ± 0.43 | 23.93 ± 0.09 | ||||||||
| E7F3I2 | gpr37a | G protein-coupled receptor 37a | 2 | 2 | 15 | 24.40 ± 0.02 | 24.48 ± 0.18 | ||||||||
| Q5RKQ2 | mtx1b | Metaxin | 2 | 2 | 11 | 22.31 ± 0.24 | 22.10 ± 0.15 | ||||||||
| A0A0R4 IDW9 | sec23ip | SEC23-interacting protein | 3 | 2 | 24 | 23.38 ± 0.34 | 22.85 ± 0.04 | ||||||||
| p-Value (vs. KO) | Max Fold | Average LFQ Values (log2) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Gene Name | Protein Name | Total Pep. | Unique Pep. | Score | WT | M_DJ-1 | M_DJ-1c106a | KO/WT | M_DJ-1/WT | M_DJ-1c106a /WT | WT | KO | M_DJ-1 | M_DJ-1c106a |
| P17561 | epd | Ependymin | 7 | 7 | 280 | * | ** | * | 6.7 | 0.5 | 0.9 | 29.05 ± 0.37 | 31.80 ± 0.67 | 28.06 ± 0.23 | 28.97 ± 0.91 |
| A3KPR3 | histh1l | Histone-H1-like | 6 | 5 | 38 | * | * | *** | 2.3 | 1.3 | 1.1 | 25.43 ± 0.18 | 26.66 ± 0.17 | 25.80 ± 0.19 | 25.52 ± 0.04 |
| Q8JH28 | ctsd | Cathepsin D | 11 | 11 | 107 | * | * | * | 1.8 | 1.4 | 1.4 | 29.25 ± 0.08 | 30.13 ± 0.17 | 29.70 ± 0.06 | 29.69 ± 0.09 |
| Q502B7 | mcee | Methylmalonyl CoA epim. | 4 | 4 | 30 | * | * | * | 1.4 | 1.2 | 1.2 | 25.55 ± 0.05 | 26.00 ± 0.06 | 25.78 ± 0.08 | 25.77 ± 0.08 |
| Q5XTN6 | crygm1 | Crystallin,γM1 | 7 | 5 | 59 | 2.0 | 23.44 | 24.42 ± 1.68 | n.d. | n.d. | |||||
| Q4ZHG3 | crygm2a | Crystallin,γM2a | 4 | 3 | 39 | 1.7 | 23.42 | 24.21 ± 1.59 | n.d. | n.d. | |||||
| Q6DGJ1 | grifin | Grifin | 4 | 4 | 34 | 1.9 | 24.71 | 25.60 ± 0.87 | 23.87 | n.d. | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharbi, N.; Røise, D.; Førre, J.-E.; Edson, A.J.; Hushagen, H.A.; Tronci, V.; Frøyset, A.-K.; Fladmark, K.E. Reintroduction of DJ-1 in Müller Cells Inhibits Retinal Degeneration in the DJ-1 Deficient Retina. Antioxidants 2021, 10, 1862. https://doi.org/10.3390/antiox10121862
Gharbi N, Røise D, Førre J-E, Edson AJ, Hushagen HA, Tronci V, Frøyset A-K, Fladmark KE. Reintroduction of DJ-1 in Müller Cells Inhibits Retinal Degeneration in the DJ-1 Deficient Retina. Antioxidants. 2021; 10(12):1862. https://doi.org/10.3390/antiox10121862
Chicago/Turabian StyleGharbi, Naouel, Dagne Røise, Jorunn-Elise Førre, Amanda J. Edson, Helena A. Hushagen, Valentina Tronci, Ann-Kristin Frøyset, and Kari E. Fladmark. 2021. "Reintroduction of DJ-1 in Müller Cells Inhibits Retinal Degeneration in the DJ-1 Deficient Retina" Antioxidants 10, no. 12: 1862. https://doi.org/10.3390/antiox10121862





