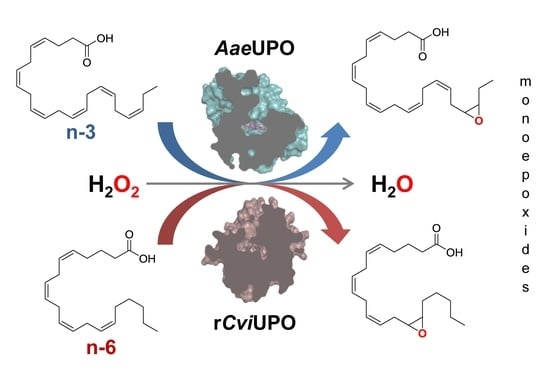
Regioselective and Stereoselective Epoxidation of n-3 and n-6 Fatty Acids by Fungal Peroxygenases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Enzymes
2.3. Enzymatic Reactions
2.4. GC-MS Analyses
2.5. HPLC Analyses
2.6. NMR Analyses
3. Results and Discussion
3.1. UPO Screening for Epoxidation of n-3 and n-6 Fatty Acids
3.2. Regioselective Synthesis of n-3 Fatty-Acid Mono-Epoxides by AaeUPO
3.3. Regioselective Synthesis of n-6 Fatty-Acid Mono-Epoxides by rCviUPO and Two Variants
3.4. Stereoselectivity of Mono-Epoxides from n-3 and n-6 Fatty Acids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sisignano, M.; Steinhilber, D.; Parnham, M.J.; Geisslinger, G. Exploring CYP2J2: Lipid mediators, inhibitors and therapeutic implications. Drug Discov. 2020, 25, 1744–1753. [Google Scholar] [CrossRef]
- Jones, R.D.; Liao, J.; Tong, X.; Xu, D.; Sun, L.; Li, H.; Yang, G.Y. Epoxy-oxylipins and soluble epoxide hydrolase metabolic pathway as targets for NSAID-induced gastroenteropathy and inflammation-associated carcinogenesis. Front. Pharmacol. 2019, 10, 731. [Google Scholar] [CrossRef] [Green Version]
- Darwesh, A.M.; Bassiouni, W.; Adebesin, A.M.; Mohammad, A.S.; Falck, J.R.; Seubert, J.M. A synthetic epoxydocosapentaenoic acid analogue ameliorates cardiac ischemia/reperfusion injury: The involvement of the sirtuin 3-NLRP3 pathway. Int. J. Mol. Sci. 2020, 21, 5261. [Google Scholar] [CrossRef]
- Chen, X.; Li, Z.; Zhang, B.; Hu, R.; Li, J.; Feng, M.; Yao, W.; Zhang, C.; Wan, L.; Zhang, Y. Alleviation of mechanical allodynia by 14,15-epoxyeicosatrienoic acid in a central poststroke pain model: Possible role of allopregnanolone and d-subunit-containing gamma-aminobutyric acid A receptors. J. Pain 2019, 20, 577–591. [Google Scholar] [CrossRef]
- Leineweber, C.G.; Pietzner, A.; Zhang, I.W.; Blessin, U.B.; Rothe, M.; Schott, E.; Schebb, N.H.; Weylandt, K.H. Assessment of the effect of sorafenib on omega-6 and omega-3 epoxyeicosanoid formation in patients with hepatocellular carcinoma. Int. J. Mol. Sci. 2020, 21, 1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imig, J.D. Epoxyeicosatrienoic acids, hypertension, and kidney injury. Hypertension 2015, 65, 476–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliwarga, T.; Evangelista, E.A.; Sotoodehnia, N.; Lemaitre, R.N.; Totah, R.A. Regulation of CYP2J2 and EET levels in cardiac disease and diabetes. Int. J. Mol. Sci. 2018, 19, 1916. [Google Scholar] [CrossRef] [Green Version]
- Imig, J.D.; Jankiewicz, W.K.; Khan, A.H. Epoxy fatty acids: From salt regulation to kidney and cardiovascular therapeutics. Hypertension 2020, 76, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Falck, J.R.; Kumar, P.S.; Reddy, Y.K.; Zou, G.; Capdevila, J.H. Stereospecific synthesis of EET metabolites via Suzuki-Miyaura coupling. Tetrahedron Lett. 2001, 42, 7211–7212. [Google Scholar] [CrossRef]
- Nanba, Y.; Shinohara, R.; Morita, M.; Kobayashi, Y. Stereoselective synthesis of 17,18-epoxy derivative of EPA and stereoisomers of isoleukotoxin diol by ring opening of TMS-substituted epoxide with dimsyl sodium. Org. Biomol. Chem. 2017, 15, 8614–8626. [Google Scholar] [CrossRef]
- Capdevila, J.H.; Wei, S.Z.; Helvig, C.; Falck, J.R.; Belosludtsev, Y.; Truan, G.; GrahamLorence, S.E.; Peterson, J.A. The highly stereoselective oxidation of polyunsaturated fatty acids by cytochrome P450BM-3. J. Biol. Chem. 1996, 271, 22663–22671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham-Lorence, S.; Truan, G.; Peterson, J.A.; Falck, J.R.; Wei, S.; Helvig, C.; Capdevila, J.H. An active site substitution, F87V, converts cytochrome P450 BM-3 into a regio- and stereoselective (14S,15R)-arachidonic acid epoxygenase. J. Biol. Chem. 1997, 272, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piazza, G.J.; Nuñez, A.; Foglia, T.A. Epoxidation of fatty acids, fatty methyl esters, and alkenes by immobilized oat seed peroxygenase. J. Mol. Catal. B Enzym. 2003, 21, 143–151. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Paterna, A.; Biondi, D.M.; Patti, A. Lyophilized extracts from vegetable flours as valuable alternatives to purified oxygenases for the synthesis of oxylipins. Bioorg. Chem. 2019, 93, 103325. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.; Schwab, W. Epoxidation, hydroxylation and aromatization is catalyzed by a peroxygenase from Solanum lycopersicum. J. Mol. Catal. B Enzym. 2013, 96, 52–60. [Google Scholar] [CrossRef]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Wambui, V.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer: Cham, Switzerland, 2020; pp. 369–403. [Google Scholar]
- Hofrichter, M.; Ullrich, R. Oxidations catalyzed by fungal peroxygenases. Curr. Opin. Chem. Biol. 2014, 19, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.T.; Ruiz-Dueñas, F.J.; Camarero, S.; Serrano, A.; Linde, D.; Lund, H.; Vind, J.; Tovborg, M.; Herold-Majumdar, O.M.; Hofrichter, M.; et al. Oxidoreductases on their way to industrial biotransformations. Biotechnol. Adv. 2017, 35, 815–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lan, D.; Durrani, R.; Hollmann, F. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Hobisch, M.; Holtmann, D.; Gomez de Santos, P.; Alcalde, M.; Hollmann, F.; Kara, S. Recent developments in the use of peroxygenases- Exploring their high potential in selective oxyfunctionalisations. Biotechnol. Adv. 2020, 51, 107615. [Google Scholar] [CrossRef]
- Grogan, G. Hemoprotein catalyzed oxygenations: P450s, UPOs, and progress toward scalable reactions. JACS Au 2021, 1, 1312–1329. [Google Scholar] [CrossRef]
- Aranda, C.; Carro, J.; González-Benjumea, A.; Babot, E.D.; Olmedo, A.; Linde, D.; Martínez, A.T.; Gutiérrez, A. Advances in enzymatic oxyfunctionalization of aliphatic compounds. Biotechnol. Adv. 2021, 51, 107703. [Google Scholar] [CrossRef]
- González-Benjumea, A.; Marques, G.; Herold-Majumdar, O.M.; Kiebist, J.; Scheibner, K.; del Río, J.C.; Martínez, A.T.; Gutiérrez, A. High epoxidation yields of vegetable oil hydrolyzates and methyl esters by selected fungal peroxygenases. Front. Bioeng. Biotechnol. 2021, 8, 605854. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, R.; Nuske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anh, D.H.; Ullrich, R.; Benndorf, D.; Svatos, A.; Muck, A.; Hofrichter, M. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 2007, 73, 5477–5485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gröbe, G.; Ullrich, M.; Pecyna, M.; Kapturska, D.; Friedrich, S.; Hofrichter, M.; Scheibner, K. High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. AMB Express. 2011, 1, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, R.; Poraj-Kobielska, M.; Scholze, S.; Halbout, C.; Sandvoss, M.; Pecyna, M.J.; Scheibner, K.; Hofrichter, M. Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 2018, 183, 84–93. [Google Scholar] [CrossRef]
- Kiebist, J.; Schmidtke, K.U.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zänder, D.; Hofrichter, M.; Scheibner, K. A peroxygenase from Chaetomium globosum catalyzes the selective oxygenation of testosterone. ChemBioChem 2017, 18, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Babot, E.D.; del Río, J.C.; Kalum, L.; Martínez, A.T.; Gutiérrez, A. Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea. Biotechnol. Bioeng. 2013, 110, 2332. [Google Scholar] [CrossRef] [Green Version]
- Carro, J.; González-Benjumea, A.; Fernández-Fueyo, E.; Aranda, C.; Guallar, V.; Gutiérrez, A.; Martínez, A.T. Modulating fatty acid epoxidation vs hydroxylation in a fungal peroxygenase. ACS Catal. 2019, 9, 6234–6242. [Google Scholar] [CrossRef] [Green Version]
- Linde, D.; Olmedo, A.; González-Benjumea, A.; Renau, C.; Estévez, M.; Carro, J.; Fernández-Fueyo, E.; Gutiérrez, A.; Martínez, A.T. Two new unspecific peroxygenases from heterologous expression of fungal genes in Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e02899-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Espeja, P.; Garcia-Ruiz, E.; Gonzalez-Perez, D.; Ullrich, R.; Hofrichter, M.; Alcalde, M. Directed evolution of unspecific peroxygenase from Agrocybe aegerita. Appl. Environ. Microbiol. 2014, 80, 3496–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Espeja, P.; Ma, S.; Maté, D.M.; Ludwig, R.; Alcalde, M. Tandem-yeast expression system for engineering and producing unspecific peroxygenase. Enzyme Microb. Technol. 2015, 73–74, 29–33. [Google Scholar] [CrossRef]
- Gomez de Santos, P.; Hoang, M.D.; Kiebist, J.; Kellner, H.; Ullrich, R.; Scheibner, K.; Hofrichter, M.; Liers, C.; Alcalde, M. Functional expression of two unusual acidic peroxygenases from Candolleomyces aberdarensis in yeasts by adopting evolved secretion mutations. Appl. Environ. Microbiol. 2021, 87, e00878-21. [Google Scholar]
- Püllmann, P.; Knorrscheidt, A.; Münch, J.; Palme, P.R.; Hoehenwarter, W.; Marillonnet, S.; Alcalde, M.; Westermann, B.; Weissenborn, M.J. A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases. Commun. Biol. 2021, 4, 562. [Google Scholar] [CrossRef] [PubMed]
- Püllmann, P.; Weissenborn, M.J. Improving the heterologous production of fungal peroxygenases through an episomal Pichia pastoris promoter and signal peptide shuffling system. ACS Synth. Biol. 2021, 10, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Molina-Espeja, P.; Gómez de Santos, P.; Alcalde, M. Directed evolution of unspecific peroxygenase. In Directed Enzyme Evolution: Advances and Applications; Alcalde, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 127–143. [Google Scholar]
- Municoy, M.; González-Benjumea, A.; Carro, J.; Aranda, C.; Linde, D.; Renau-Mínguez, C.; Ullrich, R.; Hofrichter, M.; Guallar, V.; Gutiérrez, A.; et al. Fatty-acid oxygenation by fungal peroxygenases: From computational simulations to preparative regio- and stereo-selective epoxidation. ACS Catal. 2020, 10, 13584–13595. [Google Scholar] [CrossRef]
- González-Benjumea, A.; Carro, J.; Renau, C.; Linde, D.; Fernández-Fueyo, E.; Gutiérrez, A.; Martínez, A.T. Fatty acid epoxidation by the new Collariella virescens peroxygenase and heme-channel variants. Catal. Sci. Technol. 2020, 10, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Knorrscheidt, A.; Soler, J.; Hünecke, N.; Püllmann, P.; Garcia-Borràs, M.; Weissenborn, M.J. Accessing chemo- and regioselective benzylic and aromatic oxidations by protein engineering of an unspecific peroxygenase. ACS Catal. 2021, 11, 7327–7338. [Google Scholar] [CrossRef]
- Lund, H.; Kalum, L.; Hofrichter, M.; Peter, S. Epoxidation Using Peroxygenase. US Patent US 9908860 B2, 6 March 2018. [Google Scholar]
- Otey, C.R. High-throughput carbon monoxide binding assay for cytochromes P450. Methods Mol. Biol. 2003, 230, 137–139. [Google Scholar]
- Aranda, C.; Olmedo, A.; Kiebist, J.; Scheibner, K.; del Río, J.C.; Martínez, A.T.; Gutiérrez, A. Selective epoxidation of fatty acids and fatty acid methyl esters by fungal peroxygenases. ChemCatChem 2018, 10, 3964–3968. [Google Scholar] [CrossRef] [Green Version]
- Cinelli, M.A.; Lee, K.S.S. Asymmetric Total Synthesis of 19,20-Epoxydocosapentaenoic Acid, a Bioactive Metabolite of Docosahexaenoic Acid. J. Org. Chem. 2019, 84, 15362–15372. [Google Scholar] [CrossRef]
- Cinelli, M.A.; Yang, J.; Scharmen, A.; Woodman, J.; Karchalla, L.M.; Lee, K.S.S. Enzymatic synthesis and chemical inversion provide both enantiomers of bioactive epoxydocosapentaenoic acids. J. Lipid Res. 2018, 59, 2237–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodman, J.W.; Cinelli, M.A.; Scharmen-Burgdolf, A.; Lee, K.S.S. Enzymatic synthesis of epoxidized metabolites of docosahexaenoic, eicosapentaenoic, and arachidonic acids. J. Vis. Exp. 2019, 148, e59770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, R.; Nagasawa, M.; Sih, C.J. Synthesis and metabolism of(+/−)-eicosa-cis-14,15-epoxy-cis-8,11-dienoic acid. Tetrahedron Lett. 1974, 15, 423–425. [Google Scholar] [CrossRef]
- Xie, F.; Li, B.X.; Alkayed, N.J.; Xiao, X. Synthesis of 14,15-EET from Arachidonic Acid Using Urea-Hydrogen Peroxide as the Oxidant. Synth. Commun. 2015, 45, 105–110. [Google Scholar] [CrossRef]
- Lucas, D.; Goulitquer, S.; Marienhagen, J.; Fer, M.; Dreano, Y.; Schwaneberg, U.; Amet, Y.; Corcos, L. Stereoselective epoxidation of the last double bond of polyunsaturated fatty acids by human cytochromes P450. J. Lipid Res. 2010, 51, 1125–1133. [Google Scholar] [CrossRef] [Green Version]
- Sanfilippo, C.; Patti, A. Biocatalytic regio- and stereoselective access to w-3 endocannabinoid epoxides with peroxygenase from oat flour. Bioorg. Chem. 2021, 113, 105014. [Google Scholar] [CrossRef]
- Falck, J.R.; Wallukat, G.; Puli, N.; Goli, M.; Arnold, C.; Konkel, A.; Rothe, M.; Fischer, R.; Müller, D.N.; Schunck, W.H. 17(R),18(S)-epoxyeicosatetraenoic acid, a potent eicosapentaenoic acid (EPA) derived regulator of cardiomyocyte contraction: Structure-activity relationships and stable analogues. J. Med. Chem. 2011, 54, 4109–4118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauterbach, B.; Barbosa-Sicard, E.; Wang, M.H.; Honeck, H.; Kärgel, E.; Theuer, J.; Schwartzman, M.L.; Haller, H.; Luft, F.C.; Gollasch, M.; et al. Cytochrome P450-dependent eicosapentaenoic acid metabolites are novel BK channel activators. Hypertension 2002, 39, 609–613. [Google Scholar] [CrossRef]
- Saika, A.; Nagatake, T.; Kishino, S.; Park, S.B.; Honda, T.; Matsumoto, N.; Shimojou, M.; Morimoto, S.; Tiwari, P.; Node, E.; et al. 17(S),18(R)-epoxyeicosatetraenoic acid generated by cytochrome P450 BM-3 from Bacillus megaterium inhibits the development of contact hypersensitivity via G-protein-coupled receptor 40-mediated neutrophil suppression. FASEB Bioadv. 2020, 2, 59–71. [Google Scholar] [CrossRef] [Green Version]








| Substrate | Enzyme | Mono- Epoxide Production 1 | Mono- Epoxide Formulae | Other Epoxides 2 | Hydroxy Fatty Acids | Conversion |
|---|---|---|---|---|---|---|
| HTA (1) | AaeUPO | 97% | 12 | - | 3% | >99% |
| SDA (2) | AaeUPO | >99% | 13 | - | - | >99% |
| ETE (3) | AaeUPO | 93% | 14 | 6% | 1% | >99% |
| EPA (4) | AaeUPO | 95% | 15 | 5% | - | >99% |
| HPA (5) | AaeUPO | 96% | 16 | - | 4% | >99% |
| DPA (6) | AaeUPO | 91% | 17 | 9% | - | >99% |
| DHA (7) | AaeUPO | 94% | 18 | 6% | - | >99% |
| Nisinic (8) | AaeUPO | 91% | 19 | 9% | - | >99% |
| DGLA (9) | rCviUPO | 74% | 20 | 19% | 7% | 96% |
| DGLA (9) | F88L | 51% | 20 | 48% | 1% | 95% |
| DGLA (9) | T158F | 92% | 20 | - | 8% | 93% |
| AA (10) | rCviUPO | 92% | 21 | - | 8% | 94% |
| AA (10) | F88L | 74% | 21 | 25% | 1% | 99% |
| AA (10) | T158F | 95% | 21 | - | 5% | 57% |
| AdA (11) | rCviUPO | 74% | 22 | 19% | 7% | >99% |
| AdA (11) | F88L | 20% | 22 | 80% | - | >99% |
| AdA (11) | T158F | 95% | 22 | - | 5% | 94% |
| Substrate | Enzyme | Epoxide Product | Main Enantiomer |
|---|---|---|---|
| ETE (3) | AaeUPO | 17,18-EpEDE (14) | >99% S/R 1 |
| EPA (4) | AaeUPO | 17,18-EpETrE (15) | >99% S/R |
| DPA (6) | AaeUPO | 19,20-EpDTrE (17) | >99% S/R |
| DHA (7) | AaeUPO | 19,20-EpDPE (18) | >99% S/R |
| DGLA (9) | T158F | 14,15-EpEDE (20) | 56% S/R 1 |
| AA (10) | rCviUPO | 14,15-EpETE (21) | 56% R/S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Benjumea, A.; Linde, D.; Carro, J.; Ullrich, R.; Hofrichter, M.; Martínez, A.T.; Gutiérrez, A. Regioselective and Stereoselective Epoxidation of n-3 and n-6 Fatty Acids by Fungal Peroxygenases. Antioxidants 2021, 10, 1888. https://doi.org/10.3390/antiox10121888
González-Benjumea A, Linde D, Carro J, Ullrich R, Hofrichter M, Martínez AT, Gutiérrez A. Regioselective and Stereoselective Epoxidation of n-3 and n-6 Fatty Acids by Fungal Peroxygenases. Antioxidants. 2021; 10(12):1888. https://doi.org/10.3390/antiox10121888
Chicago/Turabian StyleGonzález-Benjumea, Alejandro, Dolores Linde, Juan Carro, René Ullrich, Martin Hofrichter, Angel T. Martínez, and Ana Gutiérrez. 2021. "Regioselective and Stereoselective Epoxidation of n-3 and n-6 Fatty Acids by Fungal Peroxygenases" Antioxidants 10, no. 12: 1888. https://doi.org/10.3390/antiox10121888