Resolvin D1 Suppresses H2O2-Induced Senescence in Fibroblasts by Inducing Autophagy through the miR-1299/ARG2/ARL1 Axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Plasmids
2.2. Cell Culture
2.3. siRNA or Plasmid DNA or microRNA Mimics and Inhibitors Transfection
2.4. Cell Viability
2.5. β-Galactosidase Staining
2.6. Acridine Orange Staining
2.7. Western Blot
2.8. Co-Immunoprecipitation
2.9. Confocal Microscopy
2.10. Yeast Two-Hybrid (Y2H) Analysis
2.11. mCherry-GFP-LC3 Adenoviral Infection
2.12. Luciferase Reporter Assay
2.13. Statistical Analysis
3. Results
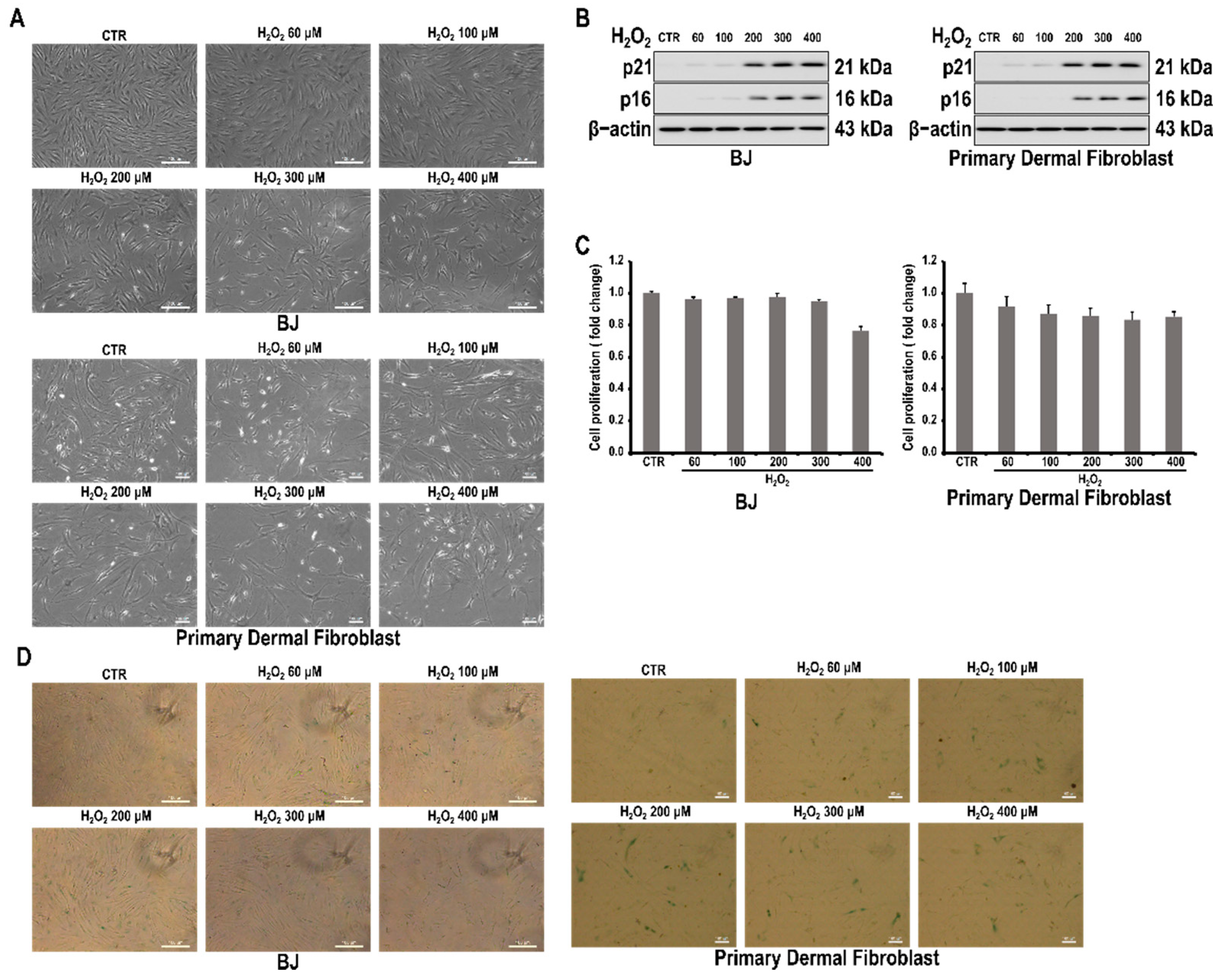
3.1. H2O2 Induces Cell Senescence in Skin Fibroblast Cells
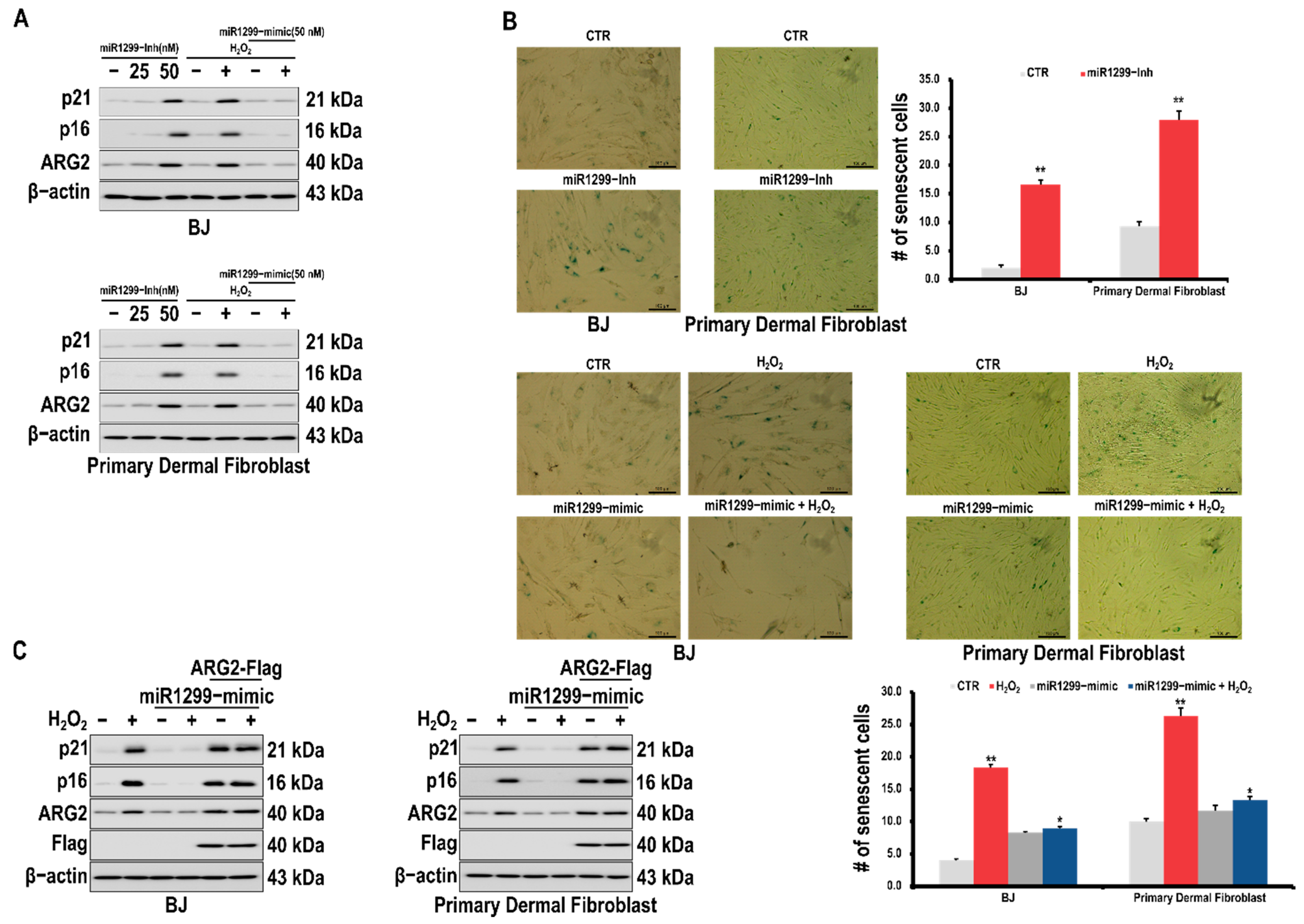
3.2. ARG2 Is Involved in H2O2-Induced Cell Senescence in BJ Skin Fibroblasts
3.3. miR-1299 Is Involved in Cell Senescence by Regulating ARG2 Expression in Dermal Cells
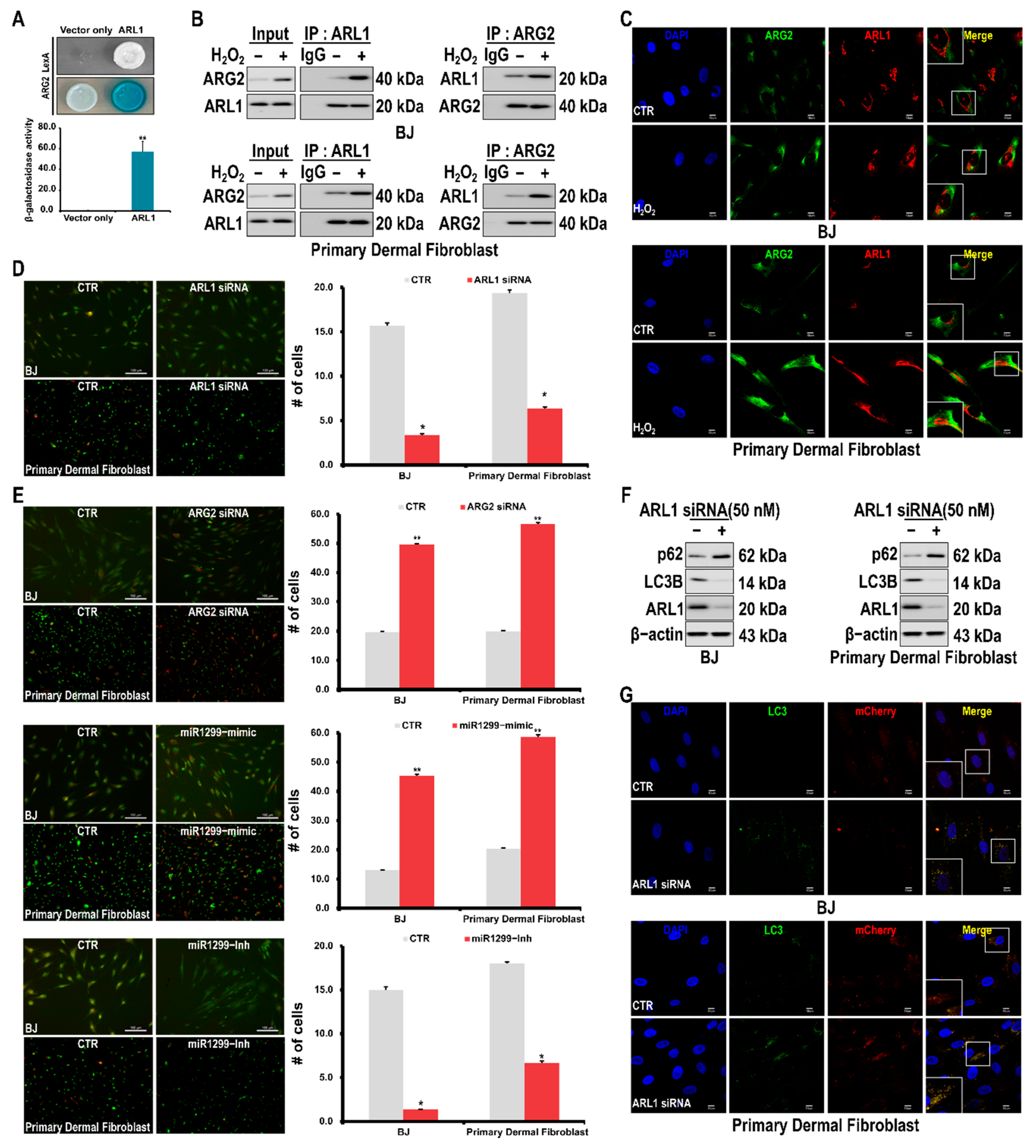
3.4. Interaction of ARG2 with ARL1 Suppresses Autophagy Leading to Cell Senescence
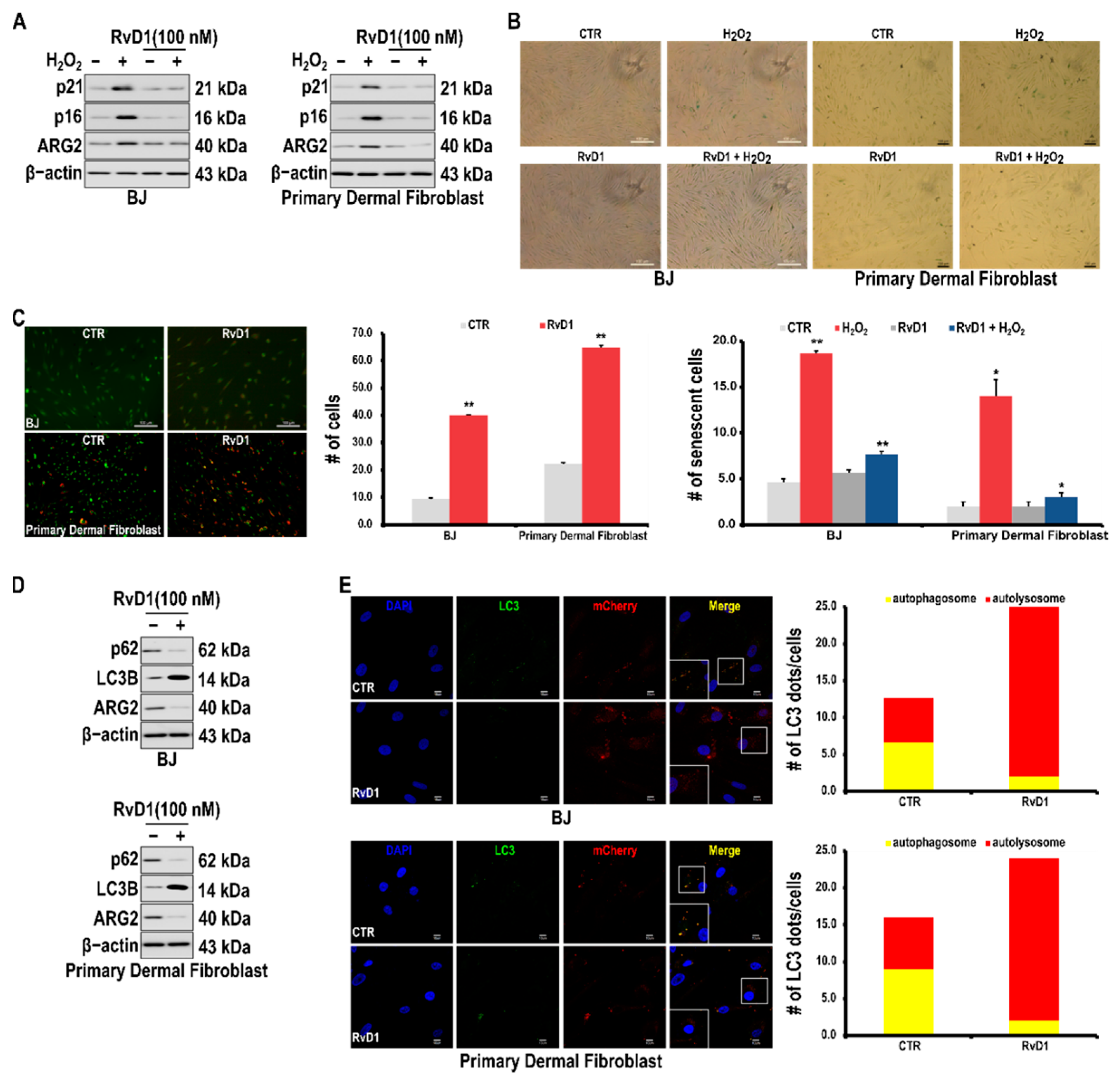
3.5. RvD1 Inhibits H2O2-Induced Cellular Senescence in Fibroblast Cells through the miR-1299/ARG2/ARL1 Axis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, S.; Wang, W.; Tan, H.-Y.; Lu, Y.; Li, Z.; Qu, Y.; Wang, N.; Wang, D. Role of Autophagy in the Maintenance of Stemness in Adult Stem Cells: A Disease-Relevant Mechanism of Action. Front. Cell Dev. Biol. 2021, 9, 715200. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Mariño, G.; Kroemer, G. Autophagy and aging. Cell 2011, 146, 682–695. [Google Scholar] [CrossRef] [Green Version]
- Crews, L.; Spencer, B.; Desplats, P.; Patrick, C.; Paulino, A.; Rockenstein, E.; Hansen, L.; Adame, A.; Galasko, D.; Masliah, E. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of α-synucleinopathy. PLoS ONE 2010, 5, e9313. [Google Scholar] [CrossRef] [Green Version]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, D.; Mouchiroud, L.; Andreux, P.A.; Katsyuba, E.; Moullan, N.; Nicolet-dit-Félix, A.A.; Williams, E.G.; Jha, P.; Sasso, G.L.; Huzard, D. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 2016, 22, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Qin, Z.-H. Autophagy, aging and longevity. Autophagy Biol. Dis. 2019, 1026, 509–525. [Google Scholar]
- Gotoh, T.; Sonoki, T.; Nagasaki, A.; Terada, K.; Takiguchi, M.; Mori, M. Molecular cloning of cDNA for nonhepatic mitochondrial arginase (arginase II) and comparison of its induction with nitric oxide synthase in a murine macrophage-like cell line. FEBS Lett. 1996, 395, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, T.; Araki, M.; Mori, M. Chromosomal localization of the human arginase II gene and tissue distribution of its mRNA. Biochem. Biophys. Res. Commun. 1997, 233, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, C.; Ahn, M.; Lee, J.H.; Shin, T. Immunohistochemical study of arginase 1 and 2 in various tissues of rats. Acta Histochem. 2012, 114, 487–494. [Google Scholar] [CrossRef]
- Shin, W.; Berkowitz, D.E.; Ryoo, S. Increased arginase II activity contributes to endothelial dysfunction through endothelial nitric oxide synthase uncoupling in aged mice. Exp. Mol. Med. 2012, 44, 594–602. [Google Scholar] [CrossRef]
- Ryoo, S.; Gupta, G.; Benjo, A.; Lim, H.K.; Camara, A.; Sikka, G.; Lim, H.K.; Sohi, J.; Santhanam, L.; Soucy, K. Endothelial arginase II: A novel target for the treatment of atherosclerosis. Circ. Res. 2008, 102, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef]
- Yang, Z.; Ming, X.-F. Arginase: The emerging therapeutic target for vascular oxidative stress and inflammation. Front. Immunol. 2013, 4, 149. [Google Scholar] [CrossRef] [Green Version]
- Yepuri, G.; Velagapudi, S.; Xiong, Y.; Rajapakse, A.G.; Montani, J.P.; Ming, X.F.; Yang, Z. Positive crosstalk between arginase-II and S6K1 in vascular endothelial inflammation and aging. Aging Cell 2012, 11, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Yu, Y.; Montani, J.P.; Yang, Z.; Ming, X.F. Arginase-II induces vascular smooth muscle cell senescence and apoptosis through p66Shc and p53 independently of its L-arginine ureahydrolase activity: Implications for atherosclerotic plaque vulnerability. J. Am. Heart Assoc. 2013, 2, e000096. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Yepuri, G.; Forbiteh, M.; Yu, Y.; Montani, J.-P.; Yang, Z.; Ming, X.-F. ARG2 impairs endothelial autophagy through regulation of MTOR and PRKAA/AMPK signaling in advanced atherosclerosis. Autophagy 2014, 10, 2223–2238. [Google Scholar] [CrossRef] [Green Version]
- Van Neste, C.; Laird, A.; O’Mahony, F.; Van Criekinge, W.; Deforce, D.; Van Nieuwerburgh, F.; Powles, T.; Harrison, D.J.; Stewart, G.D.; De Meyer, T. Epigenetic sampling effects: Nephrectomy modifies the clear cell renal cell cancer methylome. Cell. Oncol. 2017, 40, 293–297. [Google Scholar] [CrossRef] [Green Version]
- Venkatesan, N.; Deepa, P.R.; Khetan, V.; Krishnakumar, S. Computational and in vitro investigation of miRNA-gene regulations in retinoblastoma pathogenesis: MiRNA mimics strategy. Bioinform. Biol. Insights 2015, 9, BBI–S21742. [Google Scholar] [CrossRef] [Green Version]
- dos Santos Schiavinato, J.L.; Haddad, R.; Saldanha-Araujo, F.; Baiochi, J.; Araujo, A.G.; Scheucher, P.S.; Covas, D.T.; Zago, M.A.; Panepucci, R.A. TGF-beta/atRA-induced Tregs express a selected set of microRNAs involved in the repression of transcripts related to Th17 differentiation. Sci. Rep. 2017, 7, 1–17. [Google Scholar]
- Cao, S.; Li, L.; Li, J.; Zhao, H. MiR-1299 impedes the progression of non-small-cell lung cancer through EGFR/PI3K/AKT signaling pathway. OncoTargets Ther. 2020, 13, 7493. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Choi, S.H.; Yi, N.; Lee, T.R.; Lee, A.Y. Arginase-2, a miR-1299 target, enhances pigmentation in melasma by reducing melanosome degradation via senescence-induced autophagy inhibition. Pigment. Cell Melanoma Res. 2017, 30, 521–530. [Google Scholar] [CrossRef]
- Wang, K.; Liu, R.; Li, J.; Mao, J.; Lei, Y.; Wu, J.; Zeng, J.; Zhang, T.; Wu, H.; Chen, L. Quercetin induces protective autophagy in gastric cancer cells: Involvement of Akt-mTOR-and hypoxia-induced factor 1α-mediated signaling. Autophagy 2011, 7, 966–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rho, S.B.; Lee, K.W.; Lee, S.-H.; Byun, H.J.; Kim, B.-R.; Lee, C.H. Novel Anti-Angiogenic and Anti-Tumour Activities of the N-Terminal Domain of NOEY2 via Binding to VEGFR-2 in Ovarian Cancer. Biomol. Ther. 2021, 29, 506. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Park, M.K.; Kang, G.-J.; Byun, H.J.; Kim, H.J.; Yu, L.; Kim, B.; Chae, H.-S.; Chin, Y.-W.; Shim, J.G. YDJC induces epithelial-mesenchymal transition via escaping from interaction with CDC16 through ubiquitination of PP2A. J. Oncol. 2019, 2019, 3542537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; Nam, H.J.; Han, C.Y.; Joo, M.S.; Jang, K.; Jun, D.W.; Kim, S.G. Liver X Receptor Alpha Activation Inhibits Autophagy and Lipophagy in Hepatocytes by Dysregulating Autophagy-Related 4B Cysteine Peptidase and Rab-8B, Reducing Mitochondrial Fuel Oxidation. Hepatology 2021, 73, 1307–1326. [Google Scholar] [CrossRef]
- Wang, I.H.; Chen, Y.J.; Hsu, J.W.; Lee, F.J.S. The Arl3 and Arl1 GTPases co-operate with Cog8 to regulate selective autophagy via Atg9 trafficking. Traffic 2017, 18, 580–589. [Google Scholar] [CrossRef] [Green Version]
- Regulski, M.J. Cellular senescence: What, why, and how. Wounds Compend. Clin. Res. Pract. 2017, 29, 168–174. [Google Scholar]
- Beauséjour, C.M.; Krtolica, A.; Galimi, F.; Narita, M.; Lowe, S.W.; Yaswen, P.; Campisi, J. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 2003, 22, 4212–4222. [Google Scholar] [CrossRef]
- Rodier, F.; Campisi, J. Four faces of cellular senescenceFour faces of senescence. J. Cell Biol. 2011, 192, 547–556. [Google Scholar] [CrossRef]
- Coppé, J.-P.; Patil, C.K.; Rodier, F.; Sun, Y.; Muñoz, D.P.; Goldstein, J.; Nelson, P.S.; Desprez, P.-Y.; Campisi, J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008, 6, e301. [Google Scholar] [CrossRef]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef]
- Bernard, M.; Yang, B.; Migneault, F.; Turgeon, J.; Dieudé, M.; Olivier, M.-A.; Cardin, G.B.; El-Diwany, M.; Underwood, K.; Rodier, F. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy 2020, 16, 2004–2016. [Google Scholar] [CrossRef]
- Baeeri, M.; Bahadar, H.; Rahimifard, M.; Navaei-Nigjeh, M.; Khorasani, R.; Rezvanfar, M.A.; Gholami, M.; Abdollahi, M. α-Lipoic acid prevents senescence, cell cycle arrest, and inflammatory cues in fibroblasts by inhibiting oxidative stress. Pharmacol. Res. 2019, 141, 214–223. [Google Scholar] [CrossRef]
- Yu, Y.; Xiong, Y.; Montani, J.-P.; Yang, Z.; Ming, X.-F. Arginase-II activates mTORC1 through myosin-1b in vascular cell senescence and apoptosis. Cell Death Dis. 2018, 9, 313. [Google Scholar] [CrossRef] [Green Version]
- Hamdan, Y.; Mazini, L.; Malka, G. Exosomes and Micro-RNAs in Aging Process. Biomedicines 2021, 9, 968. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Prattichizzo, F.; Giuliani, A.; Matacchione, G.; Rippo, M.R.; Sabbatinelli, J.; Bonafè, M. miR-21 and miR-146a: The microRNAs of inflammaging and age-related diseases. Ageing Res. Rev. 2021, 70, 101374. [Google Scholar] [CrossRef]
- Ma, Y.; Qi, M.; An, Y.; Zhang, L.; Yang, R.; Doro, D.H.; Liu, W.; Jin, Y. Autophagy controls mesenchymal stem cell properties and senescence during bone aging. Aging Cell 2018, 17, e12709. [Google Scholar] [CrossRef] [Green Version]
- Italiano, A.; Chen, C.L.; Thomas, R.; Breen, M.; Bonnet, F.; Sevenet, N.; Longy, M.; Maki, R.G.; Coindre, J.M.; Antonescu, C.R. Alterations of the p53 and PIK3CA/AKT/mTOR pathways in angiosarcomas: A pattern distinct from other sarcomas with complex genomics. Cancer 2012, 118, 5878–5887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, F.-J.S.; Huang, C.-F.; Yu, W.-L.; Buu, L.-M.; Lin, C.-Y.; Huang, M.-C.; Moss, J.; Vaughan, M. Characterization of an ADP-ribosylation Factor-like 1 Protein in Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 30998–31005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Horstmann, H.; Ng, C.; Hong, W. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 2001, 114, 4543–4555. [Google Scholar] [CrossRef] [PubMed]
- Van Valkenburgh, H.; Shern, J.F.; Sharer, J.D.; Zhu, X.; Kahn, R.A. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: Characterizing ARL1-binding proteins. J. Biol. Chem. 2001, 276, 22826–22837. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Tai, G.; Hong, W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol. Biol. Cell 2004, 15, 4426–4443. [Google Scholar] [CrossRef] [Green Version]
- Price, H.P.; Panethymitaki, C.; Goulding, D.; Smith, D.F. Functional analysis of TbARL1, an N-myristoylated Golgi protein essential for viability in bloodstream trypanosomes. J. Cell Sci. 2005, 118, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, A.; Bieler, B.M.; Harper, D.C.; Cowan, D.A.; Sutterwala, S.; Gay, D.M.; Cole, N.B.; McCaffery, J.M.; Marks, M.S. A role for GRIP domain proteins and/or their ligands in structure and function of the trans Golgi network. J. Cell Sci. 2003, 116, 4441–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setty, S.R.G.; Shin, M.E.; Yoshino, A.; Marks, M.S.; Burd, C.G. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 2003, 13, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Panic, B.; Whyte, J.R.; Munro, S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 2003, 13, 405–410. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H. Role of specialized pro-resolving lipid mediators and their receptors in virus infection: A promising therapeutic strategy for SARS-CoV-2 cytokine storm. Arch. Pharm. Res. 2021, 44, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H. Reversal of Epithelial-Mesenchymal Transition by Natural Anti-Inflammatory and Pro-Resolving Lipids. Cancers 2019, 11, 1841. [Google Scholar] [CrossRef] [Green Version]
- Prieto, P.; Rosales-Mendoza, C.E.; Terrón, V.; Toledano, V.; Cuadrado, A.; López-Collazo, E.; Bannenberg, G.; Martín-Sanz, P.; Fernández-Velasco, M.; Boscá, L. Activation of autophagy in macrophages by pro-resolving lipid mediators. Autophagy 2015, 11, 1729–1744. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H. Epithelial-mesenchymal transition: Initiation by cues from chronic inflammatory tumor microenvironment and termination by anti-inflammatory compounds and specialized pro-resolving lipids. Biochem. Pharmacol. 2018, 158, 261–273. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, K.; Liao, L.; Zhang, T.; Yang, M.; Sun, C. Lipoxin A4 receptor agonist BML-111 induces autophagy in alveolar macrophages and protects from acute lung injury by activating MAPK signaling. Respir. Res. 2018, 19, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Feng, X.; Huang, X.-Y.; Huang, Z.; Huang, Y.; Li, C.; Li, G.; Nong, S.; Wu, R.; Huang, Y. Spautin-1 ameliorates acute pancreatitis via inhibiting impaired autophagy and alleviating calcium overload. Mol. Med. 2016, 22, 643–652. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.J.; Kim, B.; Byun, H.J.; Yu, L.; Nguyen, T.M.; Nguyen, T.H.; Do, P.A.; Kim, E.J.; Cheong, K.A.; Kim, K.S.; et al. Resolvin D1 Suppresses H2O2-Induced Senescence in Fibroblasts by Inducing Autophagy through the miR-1299/ARG2/ARL1 Axis. Antioxidants 2021, 10, 1924. https://doi.org/10.3390/antiox10121924
Kim HJ, Kim B, Byun HJ, Yu L, Nguyen TM, Nguyen TH, Do PA, Kim EJ, Cheong KA, Kim KS, et al. Resolvin D1 Suppresses H2O2-Induced Senescence in Fibroblasts by Inducing Autophagy through the miR-1299/ARG2/ARL1 Axis. Antioxidants. 2021; 10(12):1924. https://doi.org/10.3390/antiox10121924
Chicago/Turabian StyleKim, Hyun Ji, Boram Kim, Hyung Jung Byun, Lu Yu, Tuan Minh Nguyen, Thi Ha Nguyen, Phuong Anh Do, Eun Ji Kim, Kyung Ah Cheong, Kyung Sung Kim, and et al. 2021. "Resolvin D1 Suppresses H2O2-Induced Senescence in Fibroblasts by Inducing Autophagy through the miR-1299/ARG2/ARL1 Axis" Antioxidants 10, no. 12: 1924. https://doi.org/10.3390/antiox10121924
APA StyleKim, H. J., Kim, B., Byun, H. J., Yu, L., Nguyen, T. M., Nguyen, T. H., Do, P. A., Kim, E. J., Cheong, K. A., Kim, K. S., Huy Phùng, H., Rahman, M., Jang, J. Y., Rho, S. B., Kang, G. J., Park, M. K., Lee, H., Lee, K., Cho, J., ... Lee, C. H. (2021). Resolvin D1 Suppresses H2O2-Induced Senescence in Fibroblasts by Inducing Autophagy through the miR-1299/ARG2/ARL1 Axis. Antioxidants, 10(12), 1924. https://doi.org/10.3390/antiox10121924










