Superoxide Dismutase Multigene Family from a Primitive Chondrostean Sturgeon, Acipenser baerii: Molecular Characterization, Evolution, and Antioxidant Defense during Development and Pathogen Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Ethics Declaration
2.2. Molecular Cloning of SOD cDNAs
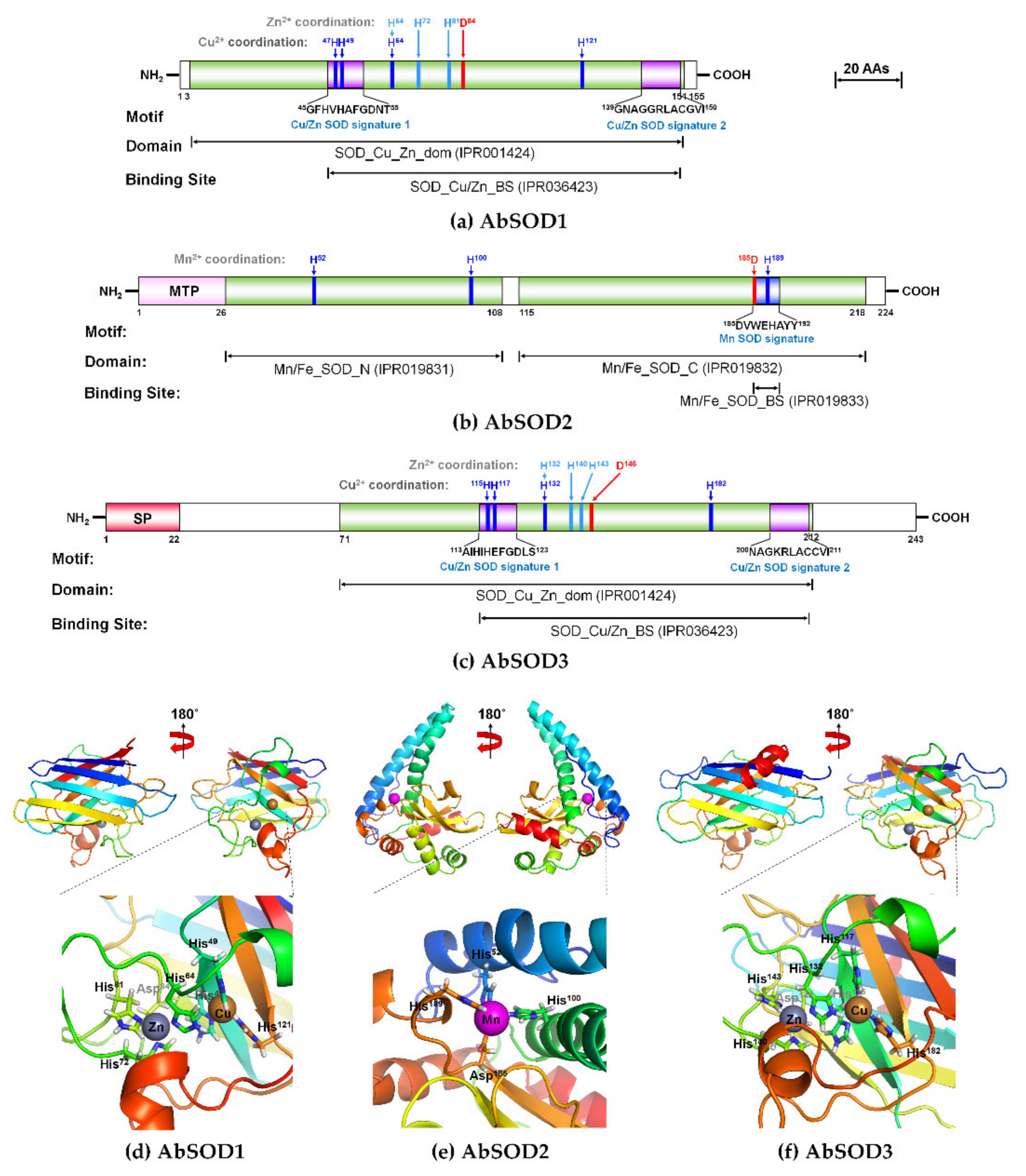
2.3. In Silico Sequence Analysis and Structural Modeling
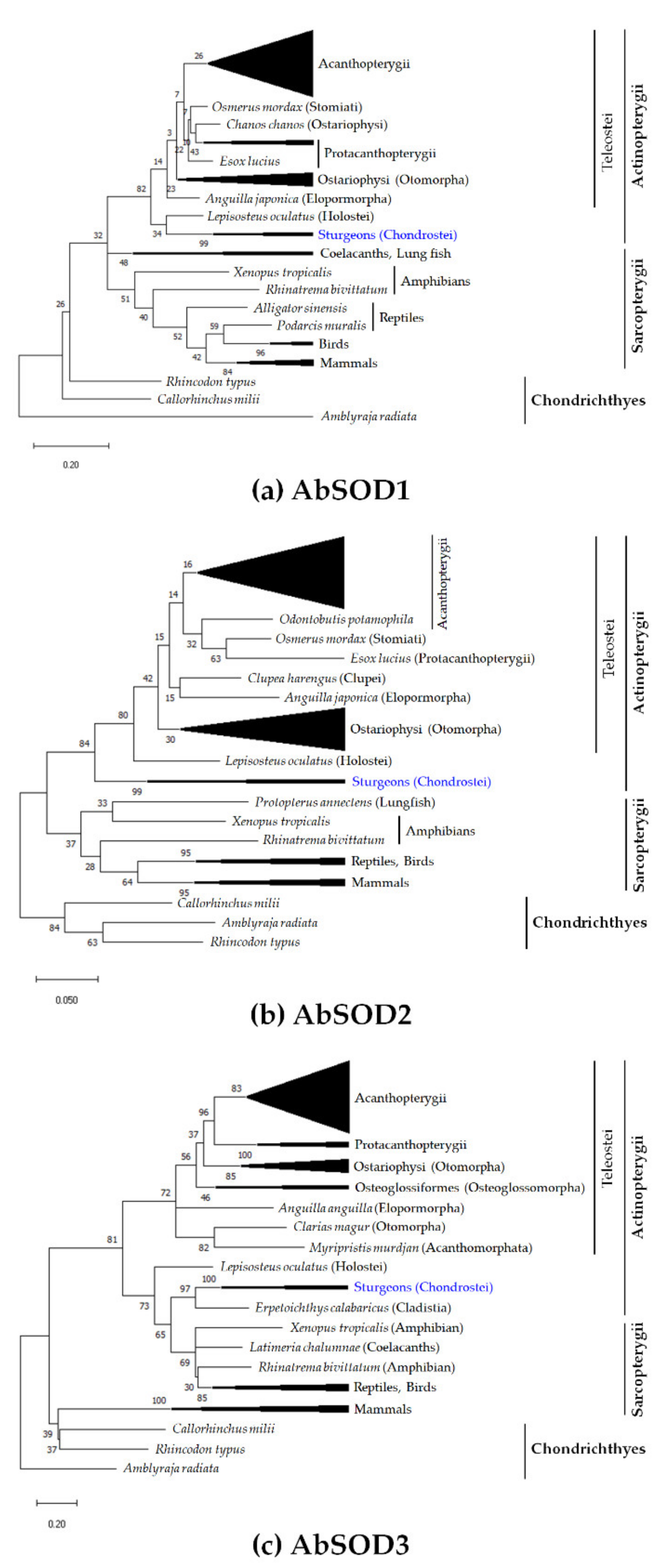
2.4. Molecular Phylogeny and Multiple Sequence Alignment
2.5. Sampling of Tissues, Embryos, and Prelarvae
2.6. In Vivo Immunostimulatory Challenges
2.6.1. Microinjection of LPS or Microbial Pathogens into Prelarvae
2.6.2. A. hydrophila Challenge with Fingerlings
2.7. Nucleic Acid Preparation, RT-qPCR Assay, and Statistics
3. Results
3.1. Characteristics of AbSOD cDNAs and Deduced Amino Acid Sequences
3.2. Assessment of Evolutionary Relationships and Sequence Homology
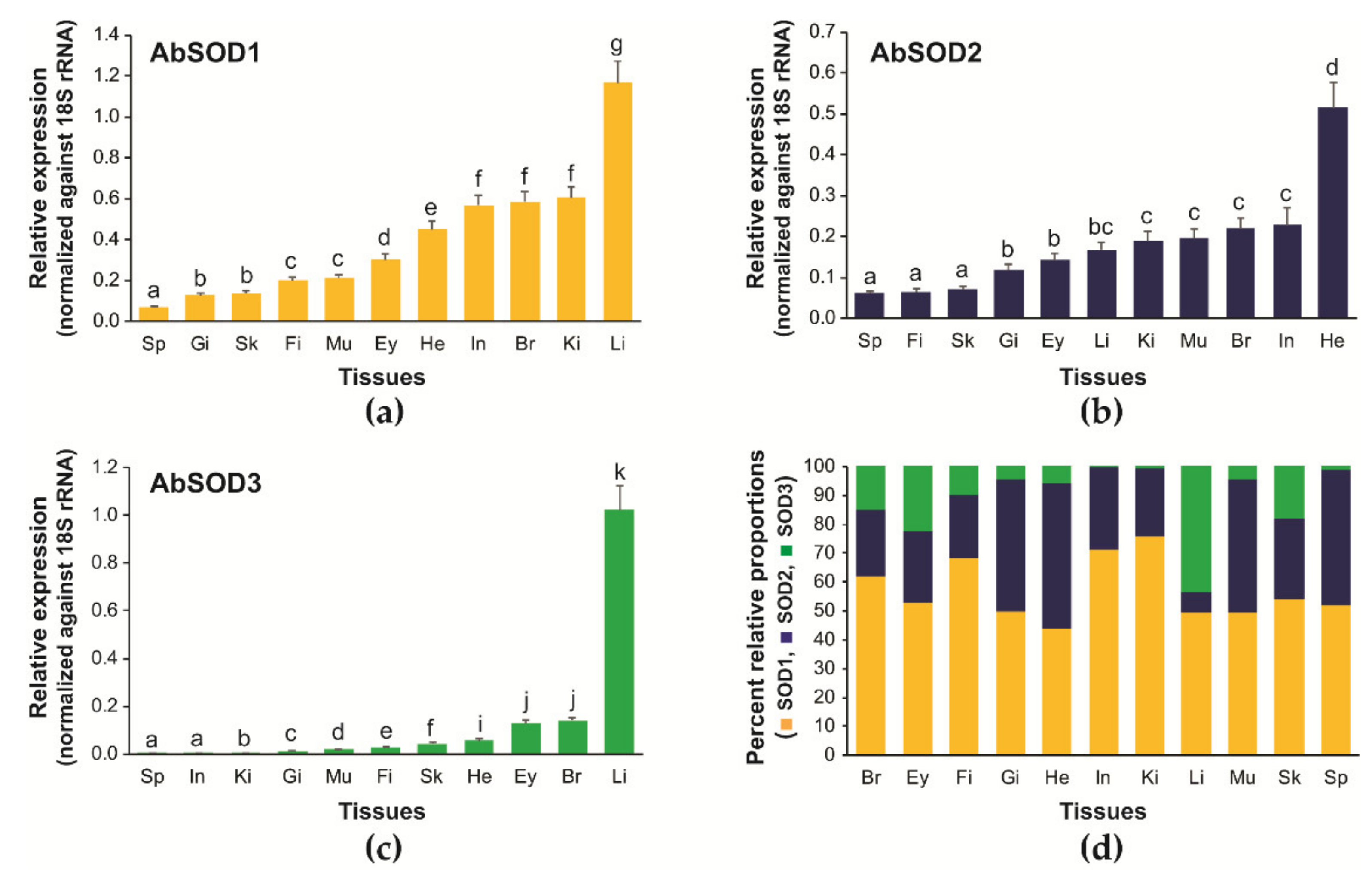
3.3. Tissue Distribution and Relative Expression of A. baerii SOD Isoforms
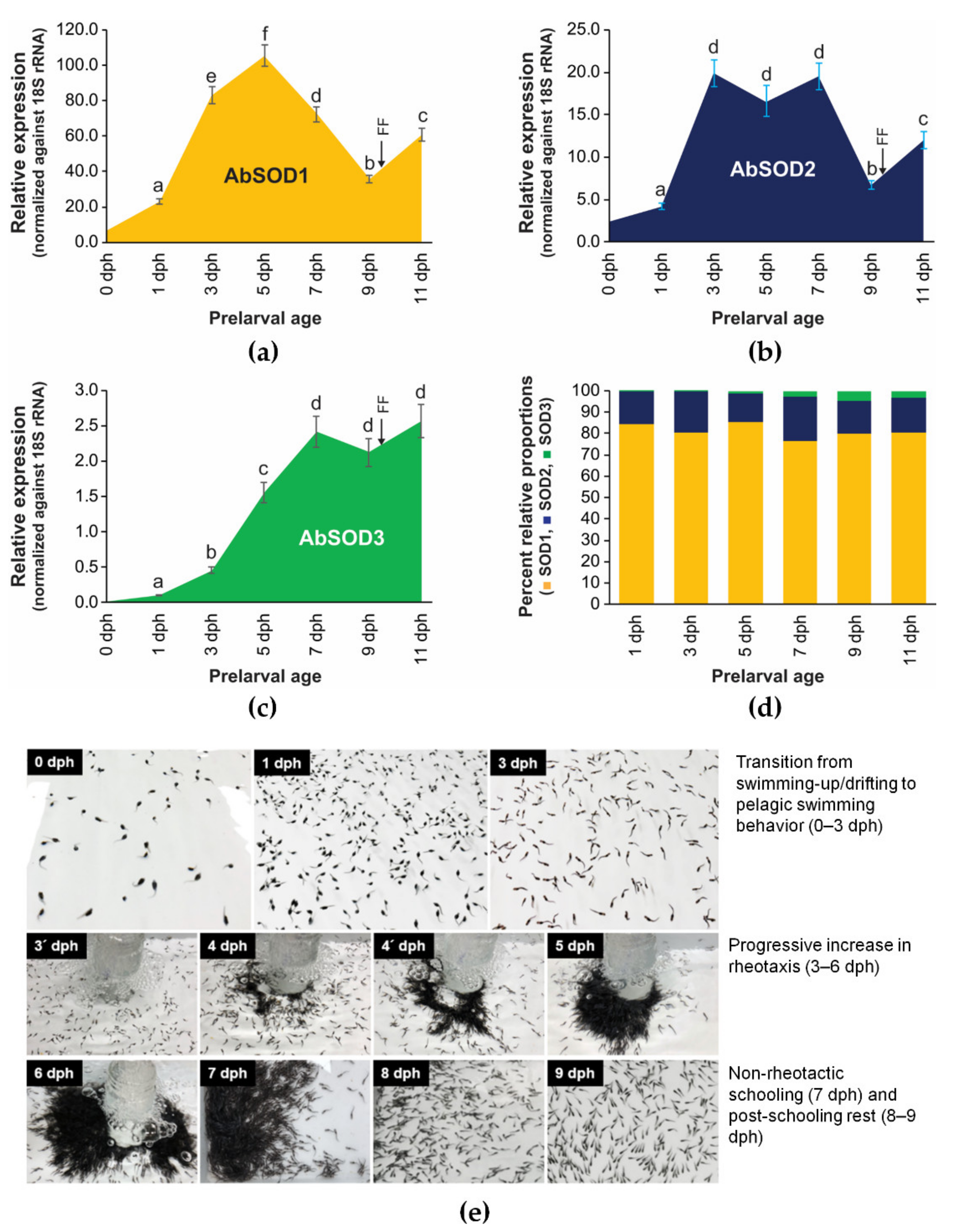
3.4. Expression Patterns of A. baerii SODs during Embryonic and Ontogenic Development
3.4.1. Embryonic Expression
3.4.2. Ontogenic Expression during Prelarval Period
3.5. Transcriptional Responses of A. baerii SODs to Immunostimulatory Treatments
3.5.1. Expression in Prelarvae Microinjected with LPS or A. hydrophila
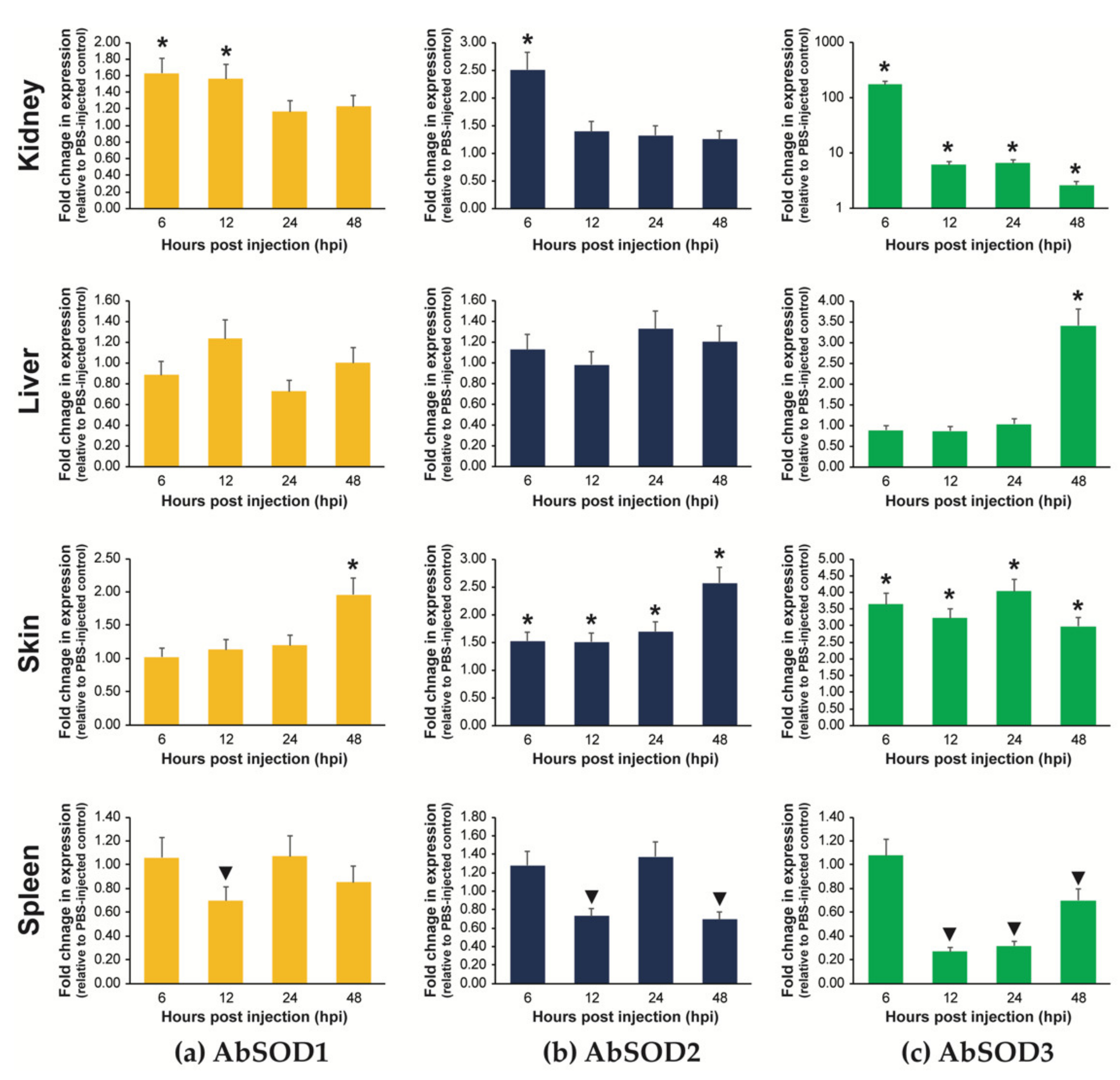
3.5.2. Expression in Fingerlings Injected with A. hydrophila
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davies, K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life 2000, 50, 279–289. [Google Scholar] [CrossRef]
- Auten, R.L.; Davis, J.M. Oxygen toxicity and reactive oxygen species: The devil is in the details. Pediat. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias, L.; Hernández-García, D.; Schnabel, D.; Salas-Vidal, E.; Castro-Obregón, S. Function of reactive oxygen species during animal development: Passive or active? Dev. Biol. 2008, 320, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sies, H. Role of metabolic H2O2 generation redox signaling and oxidative stress. J. Biol. Chem. 2014, 289, 8735–8741. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef]
- Birnie-Gauvin, K.; Costantini, D.; Cooke, S.J.; Willmore, W.G. A comparative and evolutionary approach to oxidative stress in fish: A review. Fish Fish. 2017, 18, 928–942. [Google Scholar] [CrossRef]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef] [PubMed]
- Case, A.J. On the origin of superoxide dismutase: An evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 2017, 6, 82. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.M.; Friedman, D.J.; Ayala, F.J. Superoxide dismutase: An evolutionary puzzle. Proc. Natl. Acad. Sci. USA 1985, 82, 824–828. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.-F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012, 586, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Near, T.J.; Eytan, R.I.; Dornburg, A.; Kuhn, K.L.; Moore, J.A.; Davis, M.P.; Wainwright, P.C.; Friedman, M.; Smith, W.L. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 2012, 109, 13698–13703. [Google Scholar] [CrossRef] [Green Version]
- Ravi, V.; Venkatesh, B. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 2008, 18, 544–550. [Google Scholar] [CrossRef]
- Almroth, B.C.; Asker, N.; Wassmur, B.; Rosengren, M.; Jutfelt, F.; Gräns, A.; Sundell, K.; Axelsson, M.; Sturve, J. Warmer water temperature results in oxidative damage in an Antarctic fish, the bald notothen. J. Exp. Mar. Biol. Ecol. 2015, 468, 130–137. [Google Scholar] [CrossRef]
- Mazurais, D.; Darias, M.; Zambonino-Infante, J.-L.; Cahu, C. Transcriptomics for understanding marine fish larval development. Can. J. Zool. 2011, 89, 599–611. [Google Scholar] [CrossRef] [Green Version]
- Solnica-Krezel, L. Conserved patterns of cell movements during vertebrate gastrulation. Curr. Biol. 2005, 15, R213–R228. [Google Scholar] [CrossRef] [Green Version]
- Mugoni, V.; Camporeale, A.; Santoro, M.M. Analysis of oxidative stress in zebrafish embryos. J. Vis. Exp. 2014, 89, e51328. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Álvarez, R.M.; Morales, A.E.; Sanz, A. Antioxidant defenses in fish: Biotic and abiotic factors. Rev. Fish Biol. Fish. 2005, 15, 75–88. [Google Scholar] [CrossRef]
- Nozik-Grayck, E.; Suliman, H.B.; Piantadosi, C.A. Extracellular superoxide dismutase. Int. J. Biochem. Cell Biol. 2005, 37, 2466–2471. [Google Scholar] [CrossRef]
- Miriyala, S.; Spasojevic, I.; Tovmasyan, A.; Salvemini, D.; Vujaskovic, Z.; Clair, D.S.; Batinic-Haberle, I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim. Biophys. Acta 2012, 1822, 794–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatzidimitriou, E.; Bisaccia, P.; Corrà, F.; Bonato, M.; Irato, P.; Manuto, L.; Toppo, S.; Bakiu, R.; Santovito, G. Copper/Zinc superoxide dismutase from the crocodile icefish Chionodraco hamatus: Antioxidant defense at constant sub-zero temperature. Antioxidants 2020, 9, 325. [Google Scholar] [CrossRef]
- Liu, H.; He, J.; Chi, C.; Gu, Y. Identification and analysis of icCu/Zn-SOD, Mn-SOD and ecCu/Zn-SOD in superoxide dismutase multigene family of Pseudosciaena crocea. Fish Shellfish Immunol. 2015, 43, 491–501. [Google Scholar] [CrossRef]
- Wang, X.; Song, Q.; Wang, Z.; Xie, Y.; Zhang, D.; Ye, K.; Han, F. Characterizations of intracellular copper/zinc superoxide dismutase from yellow drum (Nibea albiflora, Richardson 1846) and its gene expressions under the ammonia/nitrite stress. Aquat. Toxicol. 2019, 214, 105254. [Google Scholar] [CrossRef]
- Zhang, Z.-W.; Li, Z.; Liang, H.-W.; Li, L.; Luo, X.-Z.; Zou, G.-W. Molecular cloning and differential expression patterns of copper/zinc superoxide dismutase and manganese superoxide dismutase in Hypophthalmichthys molitrix. Fish Shellfish Immunol. 2011, 30, 473–479. [Google Scholar] [CrossRef]
- Sun, S.; Zhu, J.; Jiang, X.; Li, B.; Ge, X. Molecular cloning, tissue distribution and expression analysis of a manganese superoxide dismutase in blunt snout bream Megalobrama amblycephala. Fish Shellfish Immunol. 2014, 38, 340–347. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Ren, Q.; Yin, S.; Liang, F.; Jia, Y. Two superoxide dismutases (SODs) respond to bacterial challenge identified in the marbled eel Anguilla marmorata. Aquaculture 2016, 451, 316–325. [Google Scholar] [CrossRef]
- Cho, Y.S.; Lee, S.Y.; Bang, I.C.; Kim, D.S.; Nam, Y.K. Genomic organization and mRNA expression of manganese superoxide dismutase (Mn-SOD) from Hemibarbus mylodon (Teleostei, Cypriniformes). Fish Shellfish Immunol. 2009, 27, 571–576. [Google Scholar] [CrossRef]
- Aceto, A.; Amicarelli, F.; Sacchetta, P.; Dragani, B.; Bucciarelli, T.; Masciocco, L.; Miranda, M.; Ilio, C.D. Developmental aspects of detoxifying enzymes in fish (Salmo iridaeus). Free Radic. Res. 1994, 21, 285–294. [Google Scholar] [CrossRef]
- Zengin, H.; Yılmaz, Ö.; Demir, E.; Gökçe, Z. Antioxidant enzymatic defences during embryogenesis of rainbow trout Oncorhynchus mykiss (Walbaum 1792). Turk. J. Fish. Aquat. Sci. 2015, 15, 443–452. [Google Scholar] [CrossRef]
- Díaz, M.E.; Furné, M.; Trenzado, C.E.; García-Gallego, M.; Domezain, A.; Sanz, A. Antioxidant defences in the first life phases of the sturgeon Acipenser naccarii. Aquaculture 2010, 307, 123–129. [Google Scholar] [CrossRef]
- Wagner, J.T.; Knapp, M.J.; Podrabsky, J.E. Antioxidant capacity and anoxia tolerance in Austrofundulus limnaeus embryos. J. Exp. Biol. 2019, 222, jeb204347. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wu, Z.-Q.; Wang, X.-L.; Ren, Q.; Zhang, G.-S.; Liang, F.-F.; Yin, S.-W. Immune responses of two superoxide dismutases (SODs) after lipopolysaccharide or Aeromonas hydrophila challenge in pufferfish, Takifugu obscurus. Aquaculture 2016, 459, 1–7. [Google Scholar] [CrossRef]
- Surai, P.F.; Kochish, I.I.; Fisinin, V.I.; Kidd, M.T. Antioxidant defence systems and oxidative stress in poultry biology: An update. Antioxidants 2019, 8, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birstein, V.J.; Hanner, R.; DeSalle, R. Phylogeny of the Acipenseriformes: Cytogenetic and Molecular Approaches. In Sturgeon Biodiversity and Conservation; Springer: Berlin/Heidelberg, Germany, 1997; pp. 127–155. [Google Scholar]
- Kim, C.-H.; Kim, E.J.; Nam, Y.K. Chondrostean sturgeon hepcidin: An evolutionary link between teleost and tetrapod hepcidins. Fish Shellfish Immunol. 2019, 88, 117–125. [Google Scholar] [CrossRef]
- Kim, C.-H.; Kim, E.J.; Nam, Y.K. Subfunctionalization and evolution of liver-expressed antimicrobial peptide 2 (LEAP2) isoform genes in Siberian sturgeon (Acipenser baerii), a primitive chondrostean fish species. Fish Shellfish Immunol. 2019, 93, 161–173. [Google Scholar] [CrossRef]
- Webb, M.A.; Doroshov, S. Importance of environmental endocrinology in fisheries management and aquaculture of sturgeons. Gen. Comp. Endocrinol. 2011, 170, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, S.Y.; Kim, D.S.; Nam, Y.K. Effects of incubation temperature on egg development, hatching and pigment plug evacuation in farmed Siberian sturgeon Acipenser baerii. Fish. Aquat. Sci. 2013, 16, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Park, C.; Lee, S.Y.; Kim, D.S.; Nam, Y.K. Embryonic development of Siberian sturgeon Acipenser baerii under hatchery conditions: An image guide with embryological descriptions. Fish. Aquat. Sci. 2013, 16, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Park, C.; Nam, Y.K. Effects of incubation temperature on the embryonic viability and hatching time in Russian sturgeon (Acipenser gueldenstaedtii). Fish. Aquat. Sci. 2018, 21, 23. [Google Scholar] [CrossRef] [Green Version]
- Gisbert, E.; Nam, Y.K. Early Ontogeny in the Siberian Sturgeon. In The Siberian Sturgeon (Acipenser baerii, Brandt, 1869) Volume 1—Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 131–157. [Google Scholar]
- Song, W.; Jiang, K.; Zhang, F.; Lin, Y.; Ma, L. RNA-sequencing of the sturgeon Acipenser baeri provides insights into expression dynamics of morphogenic differentiation and developmental regulatory genes in early versus late developmental stages. BMC Genom. 2016, 17, 564. [Google Scholar] [CrossRef] [Green Version]
- Isuev, A.; Ismailova, S.; Ozernyuk, N. Comparative study of superoxide dismutase and glutathione peroxidase activity in embryos and skeletal muscles of adult fish. Biol. Bull. 2008, 35, 494–498. [Google Scholar] [CrossRef]
- Ciulli, S.; Volpe, E.; Sirri, R.; Passalacqua, P.; Bianchi, F.C.; Serratore, P.; Mandrioli, L. Outbreak of mortality in Russian (Acipenser gueldenstaedtii) and Siberian (Acipenser baerii) sturgeons associated with sturgeon nucleo-cytoplasmatic large DNA virus. Vet. Microbiol. 2016, 191, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Ture, M.; Ozcelep, T.; Akbulut, B.; Kutlu, I. Disease of Russian sturgeon (Acipenser gueldenstaedtii) caused by Aeromonas sp. Genet. Aquat. Org. 2018, 2, 43–47. [Google Scholar]
- Santi, M.; Pastorino, P.; Foglini, C.; Righetti, M.; Pedron, C.; Prearo, M. A survey of bacterial infections in sturgeon farming in Italy. J. Appl. Ichthyol. 2019, 35, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Brunak, S. Prediction of N-glycosylation across the human proteome and the correlation to protein function. In Proceedings of the Pacific Symposium on Biocomputing, Kauai, HI, USA, 3–7 January 2002; World Scientific: Kauai, HI, USA, 2002; pp. 310–322. [Google Scholar]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wass, M.N.; Kelley, L.A.; Sternberg, M.J. 3DLigandSite: Predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010, 38, W469–W473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. The clustal omega multiple alignment package. Methods Mol. Biol. 2021, 2231, 3–16. [Google Scholar]
- Gisbert, E.; Solovyev, M. Behaviour of Early Life Stages in the Siberian Sturgeon. In The Siberian Sturgeon (Acipenser baerii, Brandt, 1869) Volume 1—Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 159–172. [Google Scholar]
- Kim, E.J.; Nam, Y.K. Anesthetic protocol for microinjection-related handling of Siberian sturgeon (Acipenser baerii; Acipenseriformes) prolarvae. PLoS ONE 2018, 13, e0209928. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Tainer, J.A.; Getzoff, E.D.; Beem, K.M.; Richardson, J.S.; Richardson, D.C. Determination and analysis of the 2 Å structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982, 160, 181–217. [Google Scholar] [CrossRef]
- Petersen, S.V.; Oury, T.D.; Valnickova, Z.; Thøgersen, I.B.; Højrup, P.; Crapo, J.D.; Enghild, J.J. The dual nature of human extracellular superoxide dismutase: One sequence and two structures. Proc. Natl. Acad. Sci. USA 2003, 100, 13875–13880. [Google Scholar] [CrossRef] [Green Version]
- Azadmanesh, J.; Trickel, S.R.; Borgstahl, G.E. Substrate-analog binding and electrostatic surfaces of human manganese superoxide dismutase. J. Struct. Biol. 2017, 199, 68–75. [Google Scholar] [CrossRef]
- Kim, E.J.; Park, C.; Nam, Y.K. Behavioral characteristics of a chondrostean sturgeon species Acipenser baerii prelarvae in response to different environmental light intensities in a diel photoperiodic cycle. J. Anim. Reprod. Biotechnol. 2020, 35, 245–257. [Google Scholar] [CrossRef]
- Azadmanesh, J.; Borgstahl, G.E. A review of the catalytic mechanism of human manganese superoxide dismutase. Antioxidants 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakhit, R.; Chakrabartty, A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2006, 1762, 1025–1037. [Google Scholar] [CrossRef] [Green Version]
- Borgstahl, G.E.; Parge, H.E.; Hickey, M.J.; Beyer, W.F., Jr.; Hallewell, R.A.; Tainer, J.A. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell 1992, 71, 107–118. [Google Scholar] [CrossRef]
- Antonyuk, S.V.; Strange, R.W.; Marklund, S.L.; Hasnain, S.S. The structure of human extracellular copper–zinc superoxide dismutase at 1.7 Å resolution: Insights into heparin and collagen binding. J. Mol. Biol. 2009, 388, 310–326. [Google Scholar] [CrossRef]
- Tibell, L.A.; Skärfstad, E.; Jonsson, B.-H. Determination of the structural role of the N-terminal domain of human extracellular superoxide dismutase by use of protein fusions. Biochim. Biophys. Acta 1996, 1292, 47–52. [Google Scholar] [CrossRef]
- Carlsson, L.; Marklund, S.; Edlund, T. The rat extracellular superoxide dismutase dimer is converted to a tetramer by the exchange of a single amino acid. Proc. Natl. Acad. Sci. USA 1996, 93, 5219–5222. [Google Scholar] [CrossRef] [Green Version]
- Santovito, G.; Cassini, A.; Piccinni, E. Cu, Zn superoxide dismutase from Trematomus bernacchii: Functional conservation and erratic molecular evolution in Antarctic teleosts. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 143, 444–454. [Google Scholar] [CrossRef]
- Betancur-R, R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Orti, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [Green Version]
- Hughes, L.C.; Ortí, G.; Huang, Y.; Sun, Y.; Baldwin, C.C.; Thompson, A.W.; Arcila, D.; Betancur-R, R.; Li, C.; Becker, L. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl. Acad. Sci. USA 2018, 115, 6249–6254. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Zhao, W.; Jia, L.; Lu, J.; Qiao, T.; Qu, Q. The oldest articulated osteichthyan reveals mosaic gnathostome characters. Nature 2009, 458, 469–474. [Google Scholar] [CrossRef]
- Fink, R.C.; Scandalios, J.G. Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Yin, S. Effects of salinity change on two superoxide dismutases (SODs) in juvenile marbled eel Anguilla marmorata. PeerJ 2016, 4, e2149. [Google Scholar] [CrossRef] [Green Version]
- Hansen, B.; Rømma, S.; Garmo, Ø.; Olsvik, P.; Andersen, R. Antioxidative stress proteins and their gene expression in brown trout (Salmo trutta) from three rivers with different heavy metal levels. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006, 143, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhang, X.; Shu, L.; Chen, L.; Sun, L.; Qian, H.; Liu, W.; Fu, Z. Oxidative stress response and gene expression with atrazine exposure in adult female zebrafish (Danio rerio). Chemosphere 2010, 78, 846–852. [Google Scholar] [CrossRef]
- Bao, S.; Nie, X.; Ou, R.; Wang, C.; Ku, P.; Li, K. Effects of diclofenac on the expression of Nrf2 and its downstream target genes in mosquito fish (Gambusia affinis). Aquat. Toxicol. 2017, 188, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Nishina, H. Liver development: Lessons from knockout mice and mutant fish. Hepatol. Res. 2009, 39, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Xue, L.; Qi, X.; Bai, Y.; Wu, Y. Gene networks and toxicity/detoxification pathways in juvenile largemouth bass (Micropterus salmoides) liver induced by acute lead stress. Genomics 2020, 112, 20–31. [Google Scholar] [CrossRef]
- Johnson, P.C.; Mikos, A.G.; Fisher, J.P.; Jansen, J.A. Strategic directions in tissue engineering. Tissue Eng. 2007, 13, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Pohjoismäki, J.L.; Goffart, S. The role of mitochondria in cardiac development and protection. Free Radic. Biol. Med. 2017, 106, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Herbers, E.; Kekäläinen, N.J.; Hangas, A.; Pohjoismäki, J.L.; Goffart, S. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion 2019, 44, 85–92. [Google Scholar] [CrossRef]
- Fernández-Vizarra, E.; Enríquez, J.A.; Pérez-Martos, A.; Montoya, J.; Fernández-Silva, P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion 2011, 11, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Folz, R.J.; Guan, J.; Seldin, M.F.; Oury, T.D.; Enghild, J.J.; Crapo, J.D. Mouse extracellular superoxide dismutase: Primary structure, tissue-specific gene expression, chromosomal localization, and lung in situ hybridization. Am. J. Respir. Cell Mol. Biol. 1997, 17, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Peters, L.; Livingstone, D. Antioxidant enzyme activities in embryologic and early larval stages of turbot. J. Fish Biol. 1996, 49, 986–997. [Google Scholar] [CrossRef]
- Kalaimani, N.; Chakravarthy, N.; Shanmugham, R.; Thirunavukkarasu, A.; Alavandi, S.; Santiago, T. Anti-oxidant status in embryonic, post-hatch and larval stages of Asian seabass (Lates calcarifer). Fish Physiol. Biochem. 2008, 34, 151–158. [Google Scholar] [CrossRef]
- Wu, M.; Shariat-Madar, B.; Haron, M.H.; Wu, M.; Khan, I.A.; Dasmahapatra, A.K. Ethanol-induced attenuation of oxidative stress is unable to alter mRNA expression pattern of catalase, glutathione reductase, glutathione-S-transferase (GST1A), and superoxide dismutase (SOD3) enzymes in Japanese rice fish (Oryzias latipes) embryogenesis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 159–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirth, M.; Kirschbaum, F.; Gessner, J.; Krüger, A.; Patriche, N.; Billard, R. Chemical and biochemical composition of caviar from different sturgeon species and origins. Nahrung 2000, 44, 233–237. [Google Scholar] [CrossRef]
- Ovissipour, M.; Al-Qadiri, H.M.; Lu, X.; Hu, Y.; Ross, C.F.; Van Eenennaam, J.P.; Doroshov, S.I.; Rasco, B. The effect of white sturgeon (Acipenser transmontanus) ovarian fat deposition on caviar yield and nutritional quality: Introducing image processing method for sturgeon ovary fat determination. Int. Aquat. Res. 2015, 7, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Demchenko, I.T.; Oury, T.D.; Crapo, J.D.; Piantadosi, C.A. Regulation of the brain’s vascular responses to oxygen. Circ. Res. 2002, 91, 1031–1037. [Google Scholar] [CrossRef] [Green Version]
- Wolin, M.S. ecSOD controls the delicate balance of reactive oxygen species in bone marrow and ischemic tissue needed for neovascularization. Circ. Res. 2007, 101, 331–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Reszka, K.J.; Fukai, T.; Weintraub, N.L. Extracellular superoxide dismutase (ecSOD) in vascular biology: An update on exogenous gene transfer and endogenous regulators of ecSOD. Transl. Res. 2008, 151, 68–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.J.; Park, C.; Nam, Y.K. Ontogenetic behavior of farm-bred Russian sturgeon (Acipenser gueldenstaedtii) prelarvae in a diel photoperiodic cycle: Behavioral modifications in response to light intensity. Fish. Aquat. Sci. 2019, 22, 4. [Google Scholar] [CrossRef] [Green Version]
- Harvie, E.A.; Huttenlocher, A. Non-invasive imaging of the innate immune response in a zebrafish larval model of Streptococcus iniae infection. J. Vis. Exp. 2015, 98, e52788. [Google Scholar] [CrossRef] [Green Version]
- Zaccaria, E.; Cao, R.; Wells, J.M.; Van Baarlen, P. A zebrafish larval model to assess virulence of porcine Streptococcus suis strains. PLoS ONE 2016, 11, e0151623. [Google Scholar] [CrossRef] [Green Version]
- Saraceni, P.R.; Romero, A.; Figueras, A.; Novoa, B. Establishment of infection models in zebrafish larvae (Danio rerio) to study the pathogenesis of Aeromonas hydrophila. Front. Microbiol. 2016, 7, 1219. [Google Scholar] [CrossRef] [Green Version]
- Geven, E.J.; Klaren, P.H. The teleost head kidney: Integrating thyroid and immune signalling. Dev. Comp. Immunol. 2017, 66, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Xiao, H.; Zeng, L. Isolation and identification of the hemorrhagic septicemia pathogen from Amur sturgeon, Acipenser schrenckii. J. Appl. Ichthyol. 2011, 27, 799–803. [Google Scholar] [CrossRef]
- Tracz, M.J.; Alam, J.; Nath, K.A. Physiology and pathophysiology of heme: Implications for kidney disease. J. Am. Soc. Nephrol. 2007, 18, 414–420. [Google Scholar] [CrossRef]
- Chiabrando, D.; Vinchi, F.; Fiorito, V.; Mercurio, S.; Tolosano, E. Heme in pathophysiology: A matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 2014, 5, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guardiola, F.A.; Cuesta, A.; Abellán, E.; Meseguer, J.; Esteban, M.A. Comparative analysis of the humoral immunity of skin mucus from several marine teleost fish. Fish Shellfish Immunol. 2014, 40, 24–31. [Google Scholar] [CrossRef]
- Biller, J.D.; Takahashi, L.S. Oxidative stress and fish immune system: Phagocytosis and leukocyte respiratory burst activity. An. Acad. Bras. Ciênc. 2018, 90, 3403–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laith, A.; Najiah, M. Aeromonas hydrophila: Antimicrobial susceptibility and histopathology of isolates from diseased catfish, Clarias gariepinus (Burchell). J. Aquac. Res. Dev. 2014, 5, 215. [Google Scholar]
- Caipang, C.M.A.; Lazado, C.C.; Brinchmann, M.F.; Rombout, J.H.; Kiron, V. Differential expression of immune and stress genes in the skin of Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. D Genom. Proteom. 2011, 6, 158–162. [Google Scholar] [CrossRef]
- Lazado, C.C.; Caipang, C.M.A. Probiotics-pathogen interactions elicit differential regulation of cutaneous immune responses in epidermal cells of Atlantic cod Gadus morhua. Fish Shellfish Immunol. 2014, 36, 113–119. [Google Scholar] [CrossRef]
- Maekawa, S.; Wang, P.-C.; Chen, S.-C. Differential expression of immune-related genes in head kidney and spleen of cobia (Rachycentron canadum) having Streptococcus dysgalactiae infection. Fish Shellfish Immunol. 2019, 92, 842–850. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Rao, J.; Liu, Z.; Chen, Q. Effects of inorganic mercury exposure on histological structure, antioxidant status and immune response of immune organs in yellow catfish (Pelteobagrus fulvidraco). J. Appl. Toxicol. 2018, 38, 843–854. [Google Scholar] [CrossRef]
- Shahi, N.; Ardó, L.; Fazekas, G.; Gócza, E.; Kumar, S.; Rèvèsz, N.; Sándor, Z.J.; Molnár, Z.; Jeney, G.; Jeney, Z. Immunogene expression in head kidney and spleen of common carp (Cyprinus carpio L.) following thermal stress and challenge with Gram-negative bacterium, Aeromonas hydrophila. Aquac. Int. 2018, 26, 727–741. [Google Scholar] [CrossRef]
- Abarike, E.D.; Jia, C.; Jian, J.; Tang, J.; Lu, Y.; Yu, H. Characterization and expression of NADPH oxidase genes in stimulated splenic neutrophils of tilapia, Oreochromis niloticus by a mix of traditional Chinese medicine and Bacillus species (TCMBS). Aquac. Res. 2019, 50, 93–105. [Google Scholar] [CrossRef]
- Xiang, J.; Chen, R.; Xu, D.; Sun, Y.; Liu, H. Characterization of pathological changes and immune-related gene expression in yellow drum (Nibea albiflora) in response to Pseudomonas plecoglossicida and poly I: C challenge. Aquac. Rep. 2020, 17, 100350. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Kim, K.-Y.; Bang, I.-C.; Nam, Y.-K. Characterization of Copper/Zinc-Superoxide dismutase (Cu/Zn-SOD) gene from an endangered freshwater fish species Hemibarbus mylodon (Teleostei; Cypriniformes). Fish. Aquat. Sci. 2011, 14, 43–54. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.-H.; Kim, E.J.; Nam, Y.K. Superoxide Dismutase Multigene Family from a Primitive Chondrostean Sturgeon, Acipenser baerii: Molecular Characterization, Evolution, and Antioxidant Defense during Development and Pathogen Infection. Antioxidants 2021, 10, 232. https://doi.org/10.3390/antiox10020232
Kim C-H, Kim EJ, Nam YK. Superoxide Dismutase Multigene Family from a Primitive Chondrostean Sturgeon, Acipenser baerii: Molecular Characterization, Evolution, and Antioxidant Defense during Development and Pathogen Infection. Antioxidants. 2021; 10(2):232. https://doi.org/10.3390/antiox10020232
Chicago/Turabian StyleKim, Chan-Hee, Eun Jeong Kim, and Yoon Kwon Nam. 2021. "Superoxide Dismutase Multigene Family from a Primitive Chondrostean Sturgeon, Acipenser baerii: Molecular Characterization, Evolution, and Antioxidant Defense during Development and Pathogen Infection" Antioxidants 10, no. 2: 232. https://doi.org/10.3390/antiox10020232





