Grape Seeds Proanthocyanidins: Advanced Technological Preparation and Analytical Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Grape Seeds Extract (Ecovitis™) Preparation
2.3. Sample and Standard Preparation
2.4. GPC Analysis
2.5. LC–MS Analysis
2.5.1. LC–MS Conditions
2.5.2. Procyanidin Identification and Quantification by MS
2.6. Statistical Analysis
3. Results
3.1. Preparation of the Ecovitis™
3.2. GPC Results
3.2.1. GPC Procedure and Molecular Weight Calibration
3.2.2. PACs Quantification by GPC Absorbance Profile at 280 nm
3.2.3. GPC Profile Analysis and Determination of Median Degree of Polymerization (mDP)
3.3. LC–MS Results
3.3.1. LC–MS Analysis and Calibration
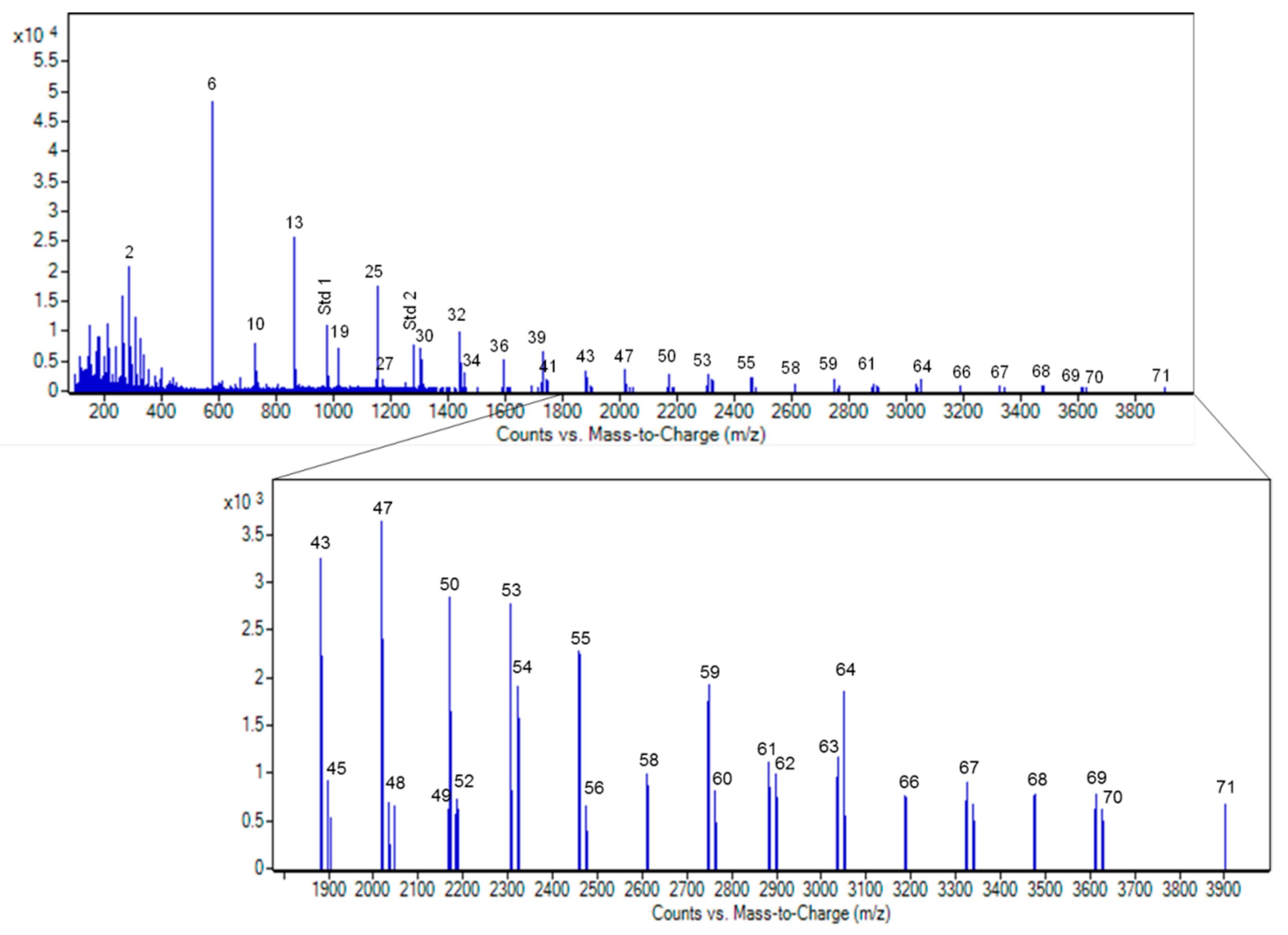
3.3.2. MS Profile of Grape Seed Extracts
3.3.3. Quantitative Analysis of Ecovitis™ Components
3.3.4. Integration of Information from GPC and MS
3.3.5. Application of the Integrated Analytical Procedure to a Benchmark of GSE
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Iahtisham-Ul-Haq; Patel, S.; Pan, X.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A.R. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef] [PubMed]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the Pharmacological Effects of Vitis vinifera (Grape) and its Bioactive Constituents: An update. Phytother. Res. 2016, 30, 1392–1403. [Google Scholar] [CrossRef]
- Cos, P.; De Bruyne, T.; Hermans, N.; Apers, S.; Berghe, D.V.; Vlietinck, A.J. Proanthocyanidins in health care: Current and new trends. Curr. Med. Chem. 2004, 11, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Jain, S.; Bhardwaj, A.; Nagpal, R.; Puniya, M.; Tomar, R.; Singh, V.; Parkash, O.; Prasad, G.B.; Marotta, F.; et al. Biological and medicinal properties of grapes and their bioactive constituents: An update. J. Med. Food 2009, 12, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.Q.; Deng, G.F.; Guo, Y.J.; Li, H.B. Biological activities of polyphenols from grapes. Int. J. Mol. Sci. 2010, 11, 622–646. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, Y.Y. Grape phytochemicals and associated health benefits. Crit. Rev. Food Sci. Nutr. 2013, 53, 1202–1225. [Google Scholar] [CrossRef]
- Feringa, H.H.; Laskey, D.A.; Dickson, J.E.; Coleman, C.I. The effect of grape seed extract on cardiovascular risk markers: A meta-analysis of randomized controlled trials. J. Am. Diet. Assoc. 2011, 111, 1173–1181. [Google Scholar] [CrossRef]
- De La Iglesia, R.; Milagro, F.I.; Campión, J.; Boqué, N.; Martínez, J.A. Healthy properties of proanthocyanidins. Biofactors 2010, 36, 159–168. [Google Scholar] [CrossRef]
- Ma, Z.F.; Zhang, H. Phytochemical Constituents, Health Benefits, and Industrial Applications of Grape Seeds: A Mini-Review. Antioxidants 2017, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Asbaghi, O.; Nazarian, B.; Reiner, Ž.; Amirani, E.; Kolahdooz, F.; Chamani, M.; Asemi, Z. The effects of grape seed extract on glycemic control, serum lipoproteins, inflammation, and body weight: A systematic review and meta-analysis of randomized controlled trials. Phytother. Res. 2020, 34, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Engels, G.; Brinckmann, J. Grape Seed (Vitis vinifera). HerbalGram 2018, 119, 8–14. [Google Scholar]
- Bombardelli, E.; Morazzoni, P. Vitis vinifera L. Fitoterapia 1995, 66, 291–317. [Google Scholar]
- Kupina, A.; Kelm, M.; Monagas, M.J. Scope Grape Seed Extract Laboratory Guidance Document; ABC-AHP-NCNPR Botanical Adulterants Prevention Program: Austin, TX, USA, 2019. [Google Scholar]
- Weber, H.A.; Hodges, A.E.; Guthrie, J.R.; O’Brien, B.M.; Robaugh, D.; Clark, A.P.; Harris, R.K.; Algaier, J.W.; Smith, C.S. Comparison of proanthocyanidins in commercial antioxidants: Grape seed and pine bark extracts. J. Agric. Food Chem. 2007, 55, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Anesi, A.; Mena, P.; Bub, A.; Ulaszewska, M.; Del Rio, D.; Kulling, S.E.; Mattivi, F. Quantification of Urinary Phenyl-γ-Valerolactones and Related Valeric Acids in Human Urine on Consumption of Apples. Metabolites 2019, 9, 254. [Google Scholar] [CrossRef] [Green Version]
- Baron, G.; Altomare, A.; Regazzoni, L.; Fumagalli, L.; Artasensi, A.; Borghi, E.; Ottaviano, E.; Del Bo, C.; Riso, P.; Allegrini, P.; et al. Profiling Vaccinium macrocarpon components and metabolites in human urine and the urine ex-vivo effect on Candida albicans adhesion and biofilm-formation. Biochem. Pharmacol. 2020, 173, 113726. [Google Scholar] [CrossRef]
- Takahashi, T.; Kamiya, T.; Hasegawa, A.; Yokoo, Y. Procyanidin oligomers selectively and intensively promote proliferation of mouse hair epithelial cells in vitro and activate hair follicle growth in vivo. J. Invest. Dermatol. 1999, 112, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Gabetta, B.; Fuzzati, N.; Griffini, A.; Lolla, E.; Pace, R.; Ruffilli, T.; Peterlongo, F. Characterization of proanthocyanidins from grape seeds. Fitoterapia 2000, 71, 162–175. [Google Scholar] [CrossRef]
- Bava, M.; Arnoldi, S.; Dell’Acqua, L.; Fontana, S.; La Forgia, F.; Mustich, G.; Roda, G.; Rusconi, C.; Sorrenti, G.; Visconti, G.L.; et al. Quali-quantitative analysis by LC/DAD and GPC of the polyphenols of “Uva di Troia Canosina” grape seeds for the development of an industrial nutraceutical product. J. Chromatogr. Sep. Tech. 2015, 6, 266. [Google Scholar]
- Kennedy, J.A.; Taylor, A.W. Analysis of proanthocyanidins by high-performance gel permeation chromatography. J. Chromatogr. A 2003, 995, 99–107. [Google Scholar] [CrossRef]
- Lin, L.Z.; Harnly, J.M. Quantitation of flavanols, proanthocyanidins, isoflavones, flavanones, dihydrochalcones, stilbenes, benzoic acid derivatives using ultraviolet absorbance after identification by liquid chromatography-mass spectrometry. J. Agric. Food Chem. 2012, 60, 5832–5840. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.-Z.; Sun, J.; Chen, P.; Monagas, M.J.; Harnly, J.M. UHPLC-PDA-ESI/HRMSn profiling method to identify and quantify oligomeric proanthocyanidins in plant products. J. Agric. Food Chem. 2014, 62, 9387–9400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rue, E.A.; Rush, M.D.; Van Breemen, R.B. Procyanidins: A comprehensive review encompassing structure elucidation via mass spectrometry. Phytochem. Rev. 2018, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]



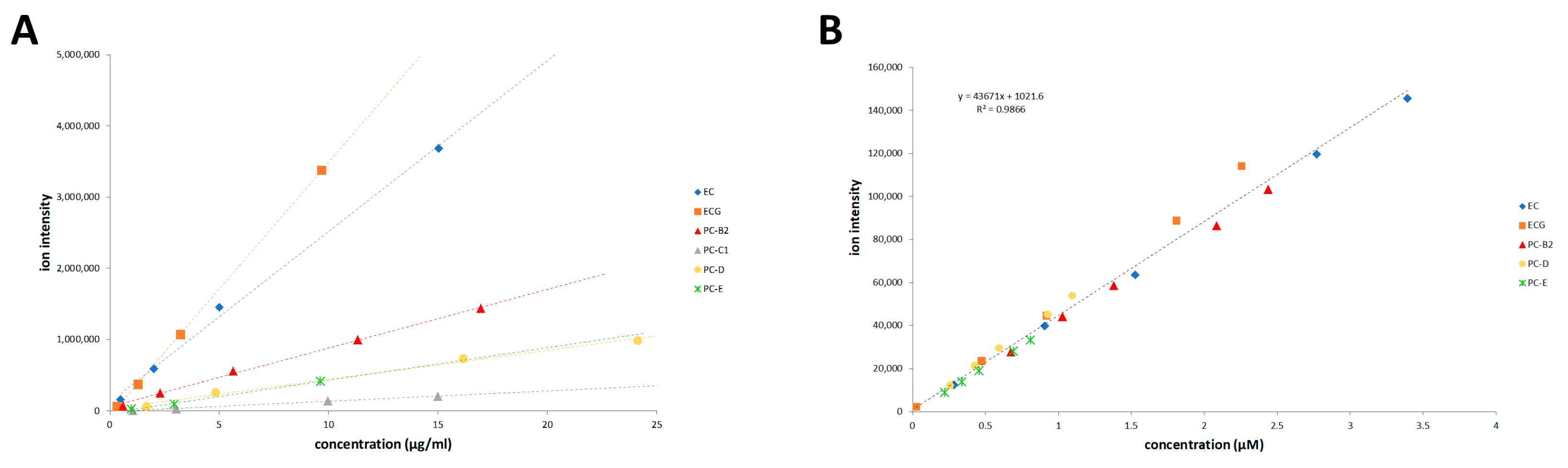

| Standard | MW (g/mol) | DP 1 | λmax (nm) | ε 280 nm 2 (M−1 cm−1) | MRRF 3 | GPC Rt 4 (min) |
|---|---|---|---|---|---|---|
| (+)-Catechin | 290.271 | 1 | 279 | 3950 | 1.00 | 7.09 |
| (−)-Epicatechin | 290.271 | 1 | 279 | 3950 | 1.00 | 7.09 |
| Procyanidin B2 | 578.526 | 2 | 279 | 8050 | 2.00 | 6.78 |
| Procyanidin C1 | 866.778 | 3 | 279 | 12,130 | 3.00 | 6.62 |
| Procyanidin D | 1155.02 | 4 | 279 | 16,075 | 4.00 | 6.48 |
| Procyanidin E | 1443.27 | 5 | 279 | 20,470 | 5.00 | 6.38 |
| EGC/GC | 306.27 | 1 | 279 | 1200 | 0.31 | 7.07 |
| ECG/CG | 442.376 | 2 | 278 | 13,500 | 3.40 | 6.87 |
| EGCG/GCG | 458.375 | 2 | 274 | 10,600 | 2.30 | 6.85 |
| Gallic acid | 170.120 | 1 | 273 | 8500 | 2.15 | 7.38 |
| Ecovitis™ Proanthocyanidins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Compound | MW | Mean | SD | ppm | ID | Compound | MW | Mean | SD | ppm |
| 1 | G | 170.0215 | ND | 37 | PA (A) 4EC 1EGC 1G | 1608.3228 | 1.79 | 1.9 | |||
| 2 | C/EC | 290.079 | 9.51 | 0.23 | −4.3 | 38 | PA (B) 4EC 1EGC 1G | 1610.3385 | 2.59 | 0.67 | −3.0 |
| 3 | EGC | 306.0739 | ND | 39 | PC (B) 6EC | 1730.3960 | 25.12 | 0.66 | 1.4 | ||
| 4 | ECG | 442.0900 | 2.76 | 0.05 | 2.9 | 40 | PA (A) 5EC 1EGC | 1744.3752 | 2.84 | 0.10 | −0.4 |
| 5 | EGCG | 458.0849 | ND | 41 | PC (B) 5EC 2G | 1746.3545 | 10.62 | 0.20 | 3.8 | ||
| 6 | PC (B) 2EC | 578.1424 | 49.85 | 1.89 | −0.7 | 42 | PC (A) 6EC 1G | 1880.3913 | 2.43 | 0.06 | 0.5 |
| 7 | PA (A) 1EC 1EGC | 592.1217 | 1.06 | 0.06 | 8.4 | 43 | PC (B) 6EC 1G | 1882.4070 | 23.57 | 0.59 | 1.8 |
| 8 | PA (B) 1EC 1EGC | 594.1373 | 1.20 | −0.7 | 44 | PA (A) 5EC 1EGC 1G | 1896.3862 | 1.24 | 1.76 | 1.6 | |
| 9 | ECG-Glu | 604.1428 | 1.62 | 0.09 | −0.8 | 45 | PA (B) 5EC 1EGC 1G | 1898.4019 | 4.29 | 0.17 | −2.7 |
| 10 | PC (B) 2EC 1G | 730.1534 | 11.44 | 0.36 | 0.3 | 46 | PC (A) 7EC | 2016.4437 | 2.92 | 0.08 | −1.2 |
| 11 | PA (A) 1EC 1EGC 1G | 744.1326 | ND | 47 | PC (B) 7EC | 2018.4594 | 23.83 | 0.82 | 1.9 | ||
| 12 | PA (B) 1EC 1EGC 1G | 746.1483 | ND | 48 | PC (B) 6EC 2G | 2034.4179 | 7.65 | 0.32 | 3.5 | ||
| 13 | PC (B) 3EC | 866.2058 | 46.36 | 1.77 | 0.7 | 49 | PC (A) 7EC 1G | 2168.4547 | 2.31 | 0.18 | 0.5 |
| 14 | PA (A) 2EC 1EGC | 880.1851 | 1.65 | 7.4 | 50 | PC (B) 7EC 1G | 2170.4703 | 17.51 | 2.72 | 1.9 | |
| 15 | PA (B) 2EC 1EGC | 882.201/882.164 | 1.89 | 0.03 | −17.3/22.7 | 51 | PA (A) 6EC 1EGC 1G | 2184.4496 | ND | ||
| 16 | PA (A) 1EC 2EGC | 896.1800 | 0.90 | 0.03 | 12.9 | 52 | PA (B) 6EC 1EGC 1G | 2186.4653 | 4.12 | 0.15 | −2.5 |
| 17 | PA (B) 1EC 2EGC | 898.1956 | ND | 53 | PC (B) 8EC | 2306.5228 | 11.74 | 4.53 | 0.8 | ||
| 18 | PC (A) 3EC 1G | 1016.2011 | ND | 54 | PC (B) 7EC 2G | 2322.4813 | 13.87 | 0.38 | 2.3 | ||
| 19 | PC (B) 3EC 1G | 1018.2168 | 16.35 | 0.41 | 0.6 | 55 | PC (B) 8EC 1G | 2458.5337 | 8.77 | 0.29 | 1.9 |
| 20 | PA (A) 2EC 1EGC 1G | 1032.1960 | 1.00 | 0.05 | 17.3 | 56 | PA (B) 7EC 1EGC 1G | 2474.5286 | 1.74 | 0.10 | −8.8 |
| 21 | PA (B) 2EC 1EGC 1G | 1034.2117 | ND | 57 | PC (B) 9EC | 2594.5862 | 8.66 | 0.25 | 0.3 | ||
| 22 | PA (A) 1EC 2EGC 1G | 1048.1910 | ND | 58 | PA (B) 8EC 1EGC | 2610.5811 | 11.13 | 0.32 | −1.6 | ||
| 23 | PA (B) 1EC 2EGC 1G | 1050.2066 | ND | 59 | PC (B) 9EC 1G | 2746.5970 | 10.90 | 0.15 | −0.3 | ||
| 24 | PC (A) 4EC | 1152.2536 | 4.50 | 0.23 | −0.8 | 60 | PA (B) 8EC 3G | 2762.5556 | 5.00 | 0.21 | 3.95 |
| 25 | PC (B) 4EC | 1154.2692 | 42.47 | 2.24 | 0.2 | 61 | PC (B) 10EC | 2882.6496 | 9.89 | 0.14 | 0.3 |
| 26 | PA (A) 3EC 1EGC | 1168.2485 | 3.25 | 0.09 | 1.3 | 62 | PC (B) 9EC 2G | 2898.6081 | 7.68 | 0.18 | 1.3 |
| 27 | PA (B) 3EC 2G | 1170.2277 | 5.44 | 0.29 | 6.9 | 63 | PC (B) 10EC 1G | 3034.6605 | 6.05 | 0.16 | 2.7 |
| 28 | PA (B) 2EC 1EGC 2G | 1186.2227 | ND | 64 | PA (B) 9EC 3G | 3050.6190 | 9.89 | 0.24 | 0.62 | ||
| 29 | PC (A) 4EC 1G | 1304.2645 | 1.92 | 0.42 | 0.9 | 65 | PC (B) 11EC | 3170.7129 | 7.99 | 0.31 | 1.3 |
| 30 | PC (B) 4EC 1G | 1306.2802 | 25.12 | 0.69 | 1.2 | 66 | PC (B) 10EC 2G | 3186.6715 | 6.72 | 0.01 | 1.5 |
| 31 | PC (A) 5EC | 1440.3169 | 3.41 | 0.13 | −1.1 | 67 | PC (B) 11EC 1G | 3322.7239 | 10.81 | 1.00 | 1.6 |
| 32 | PC (B) 5EC | 1442.3326 | 42.74 | 1.20 | 1.1 | 68 | PA (B) 11EC 1EGC | 3474.7712 | 6.97 | 1.51 | −1.7 |
| 33 | PA (A) 4EC 1EGC | 1456.3119 | 3.31 | 0.16 | −0.5 | 69 | PC (B) 12EC 1G | 3610.7873 | 6.52 | 1.02 | 0.3 |
| 34 | PC (B) 4EC 2G | 1458.2911 | 6.66 | 0.10 | 4.0 | 70 | PA (B) 11EC 3G | 3626.7458 | 4.50 | 0.47 | 0.2 |
| 35 | PC (A) 5EC 1G | 1592.3279 | 2.32 | 0.10 | 0.4 | 71 | PC (B) 13EC 1G | 3898.8507 | 5.74 | 1.02 | 0.3 |
| 36 | PC (B) 5EC 1G | 1594.3436 | 24.20 | 0.11 | 1.3 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morazzoni, P.; Vanzani, P.; Santinello, S.; Gucciardi, A.; Zennaro, L.; Miotto, G.; Ursini, F. Grape Seeds Proanthocyanidins: Advanced Technological Preparation and Analytical Characterization. Antioxidants 2021, 10, 418. https://doi.org/10.3390/antiox10030418
Morazzoni P, Vanzani P, Santinello S, Gucciardi A, Zennaro L, Miotto G, Ursini F. Grape Seeds Proanthocyanidins: Advanced Technological Preparation and Analytical Characterization. Antioxidants. 2021; 10(3):418. https://doi.org/10.3390/antiox10030418
Chicago/Turabian StyleMorazzoni, Paolo, Paola Vanzani, Sandro Santinello, Antonina Gucciardi, Lucio Zennaro, Giovanni Miotto, and Fulvio Ursini. 2021. "Grape Seeds Proanthocyanidins: Advanced Technological Preparation and Analytical Characterization" Antioxidants 10, no. 3: 418. https://doi.org/10.3390/antiox10030418







