Antioxidant and Neuroprotective Activity of Extra Virgin Olive Oil Extracts Obtained from Quercetano Cultivar Trees Grown in Different Areas of the Tuscany Region (Italy)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. EVOO Sample Extraction
2.3. Analysis of Phenolic Substances
2.4. Cell Culture and Treatments
2.5. MTT and Lactate Dehydrogenase Activity Assays
2.6. Intracellular ROS Production Assay
2.7. Intracellular GSH Levels Assay
2.8. RNA Extraction
2.9. Analysis of mRNA Levels by Reverse Transcriptase Polymerase Chain Reaction
2.10. Proteomic Analysis
2.11. Spot Digestion and Protein Identification
2.12. Bioinformatic Analysis
2.13. Western Blot Analysis
2.14. Statistical Analysis.
3. Results
3.1. Phenolic Content
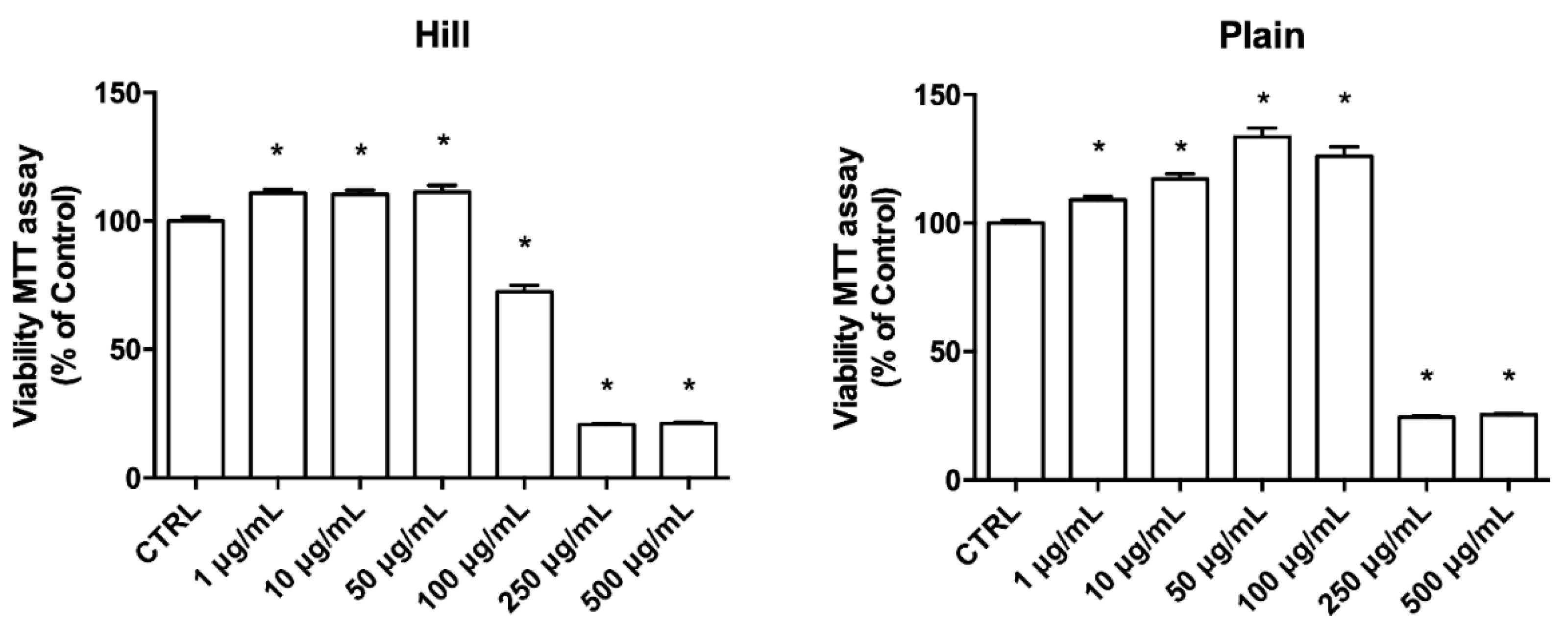
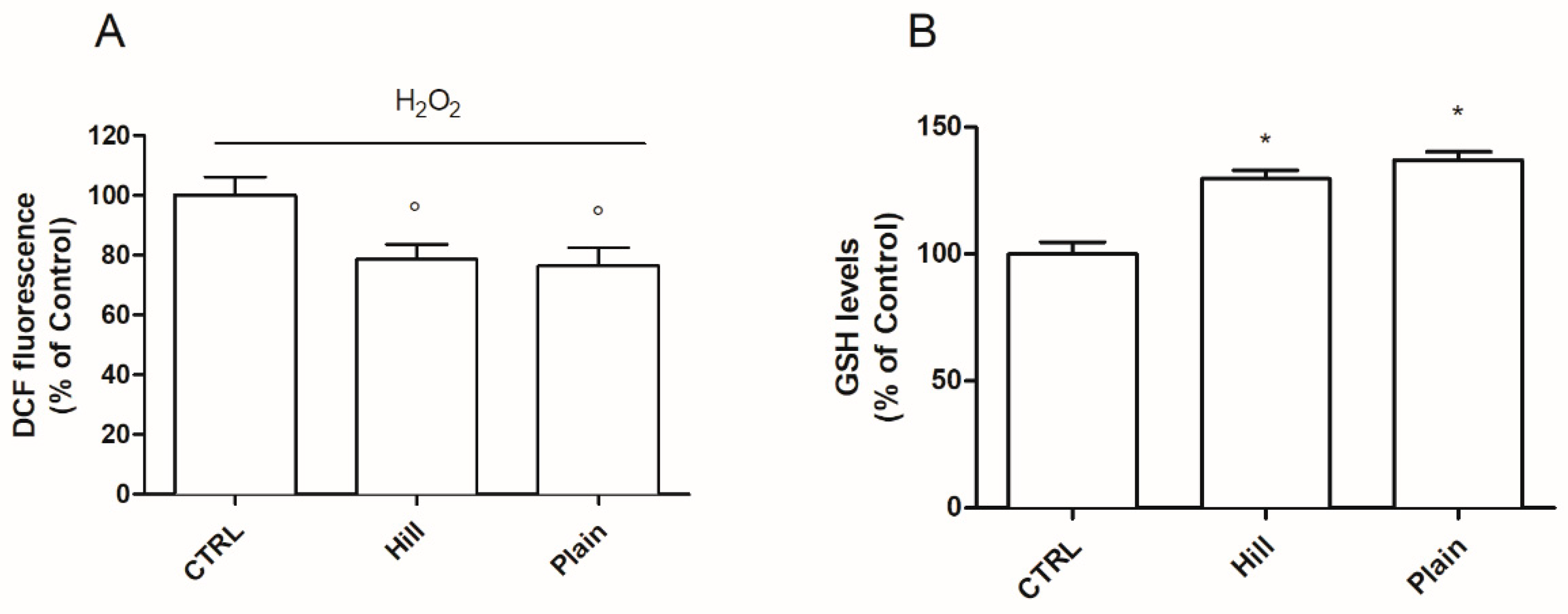
3.2. EVOO Extracts—Mediated Neuroprotection against Oxidative Damage
3.3. Antioxidant Properties of Hill and Plain Extracts
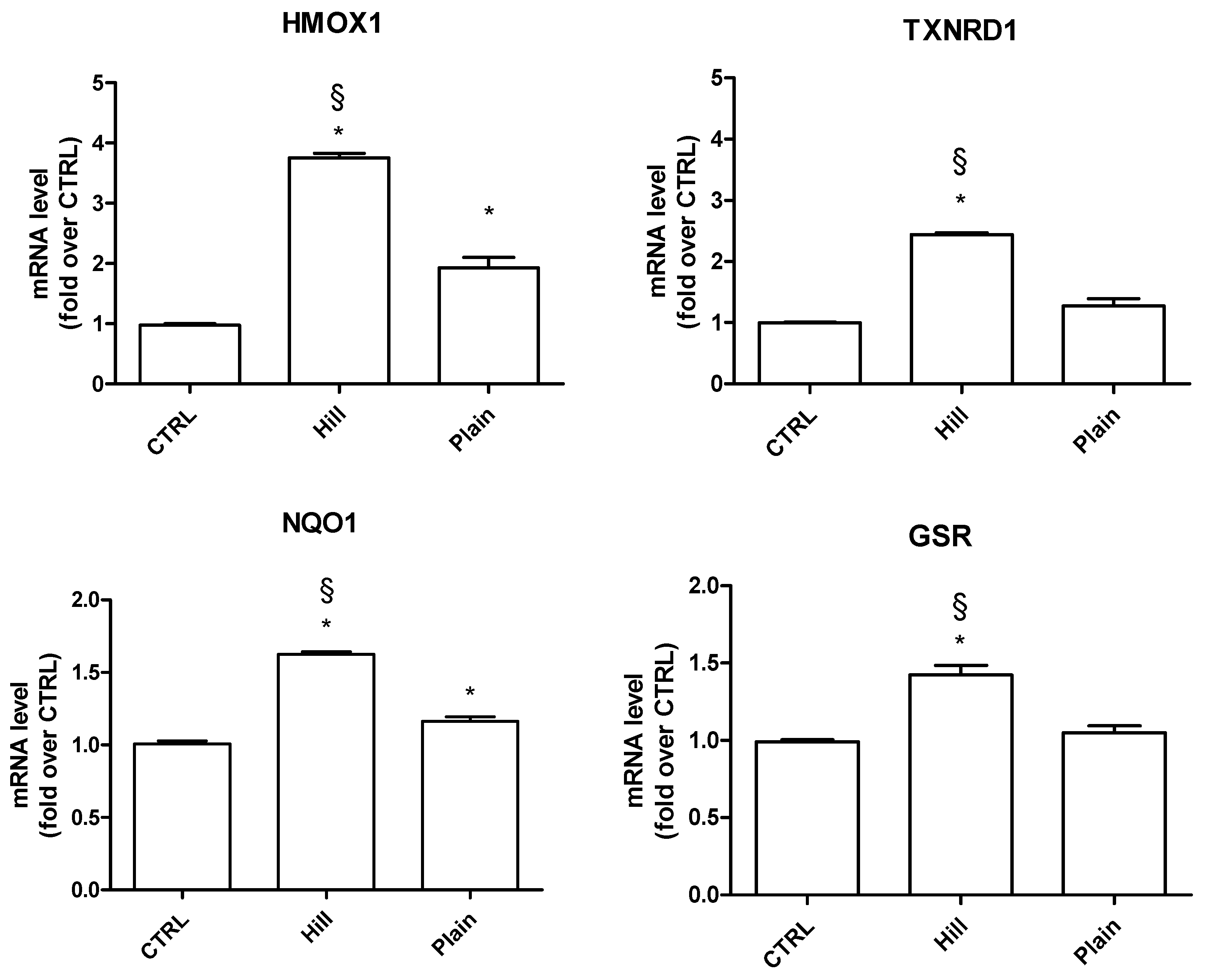
3.4. Modulation of Antioxidant and Pro-Survival Genes by Plain and Hill Extracts
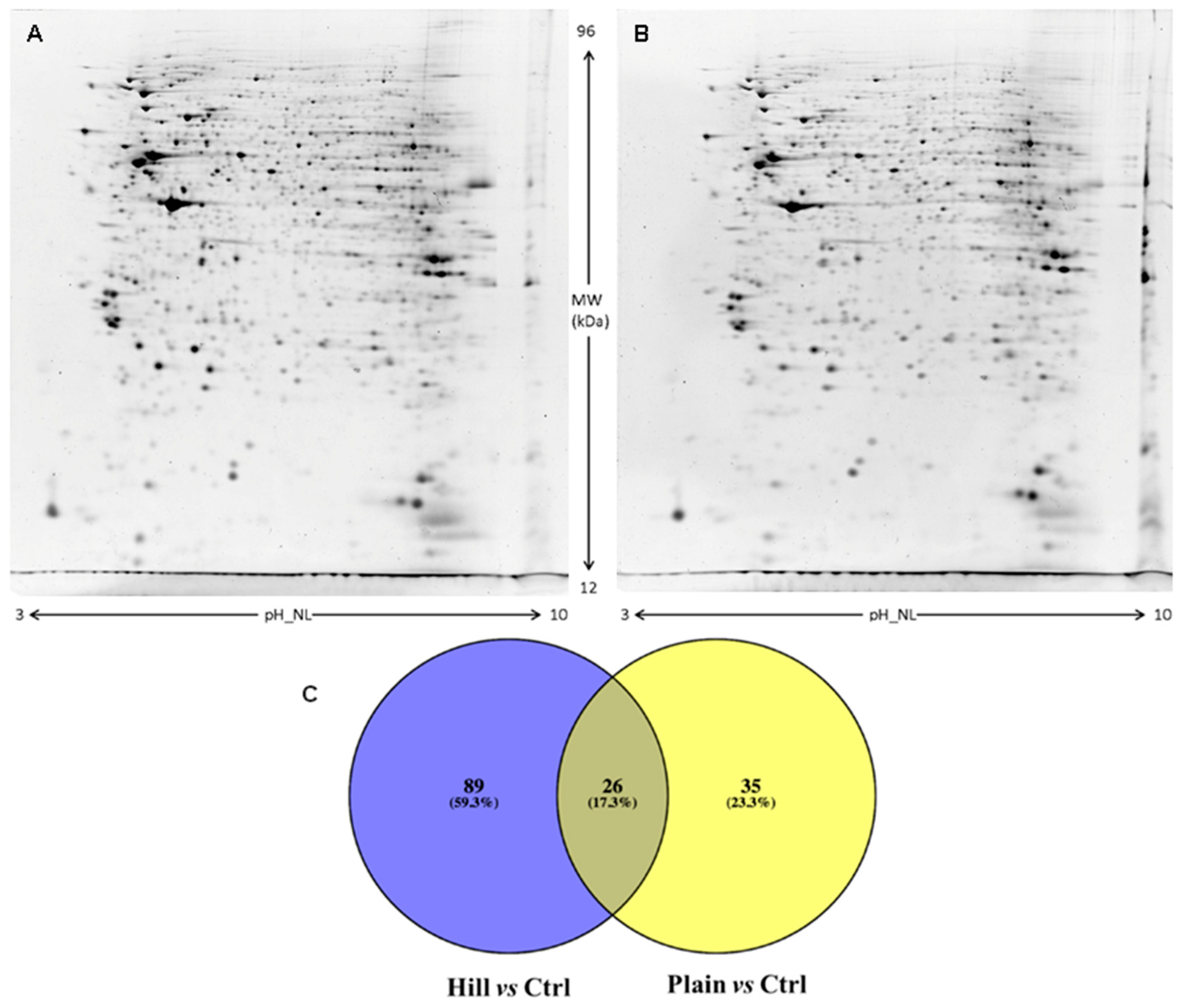
3.5. Protein Expression Analysis
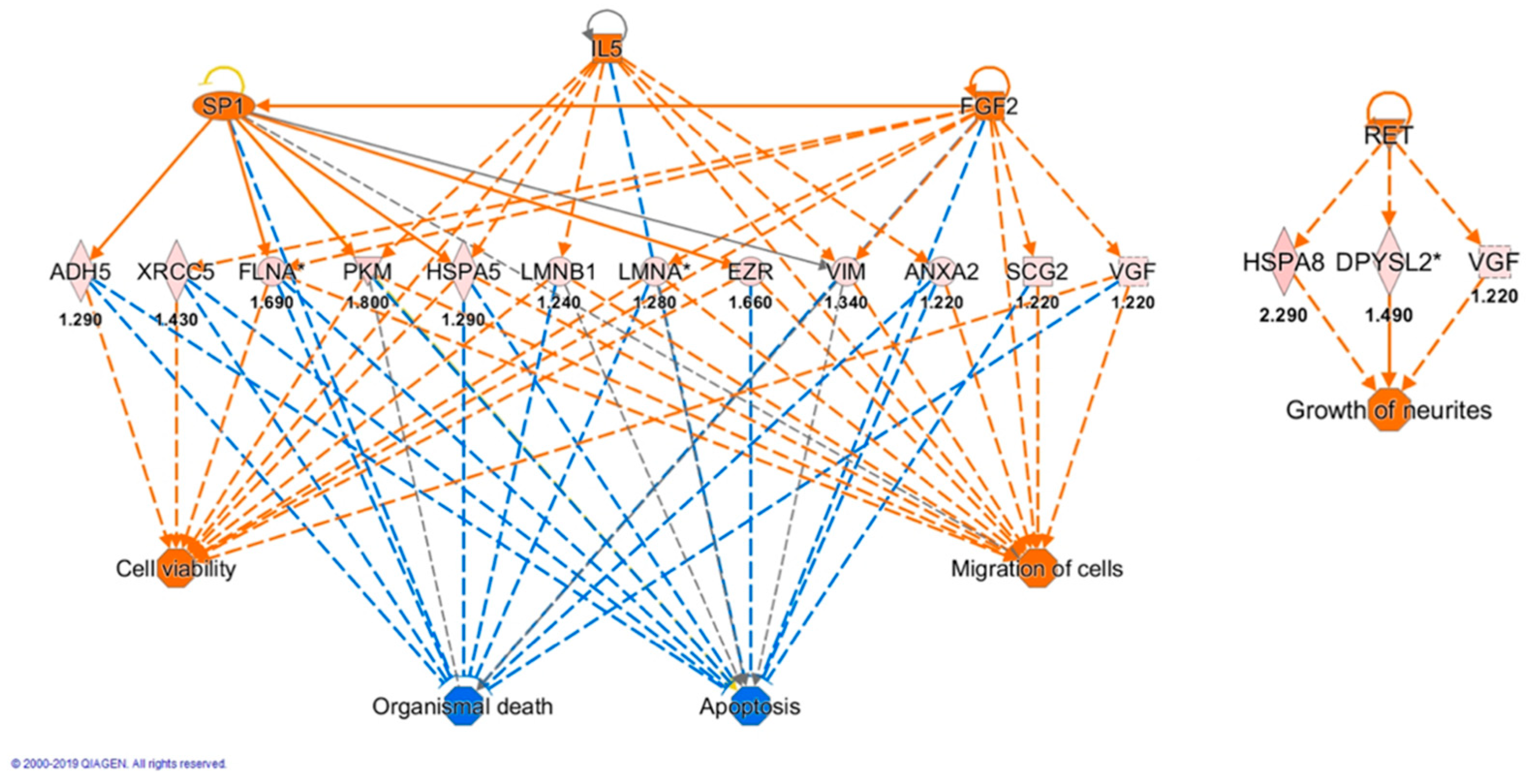
3.6. Bioinformatic Analysis
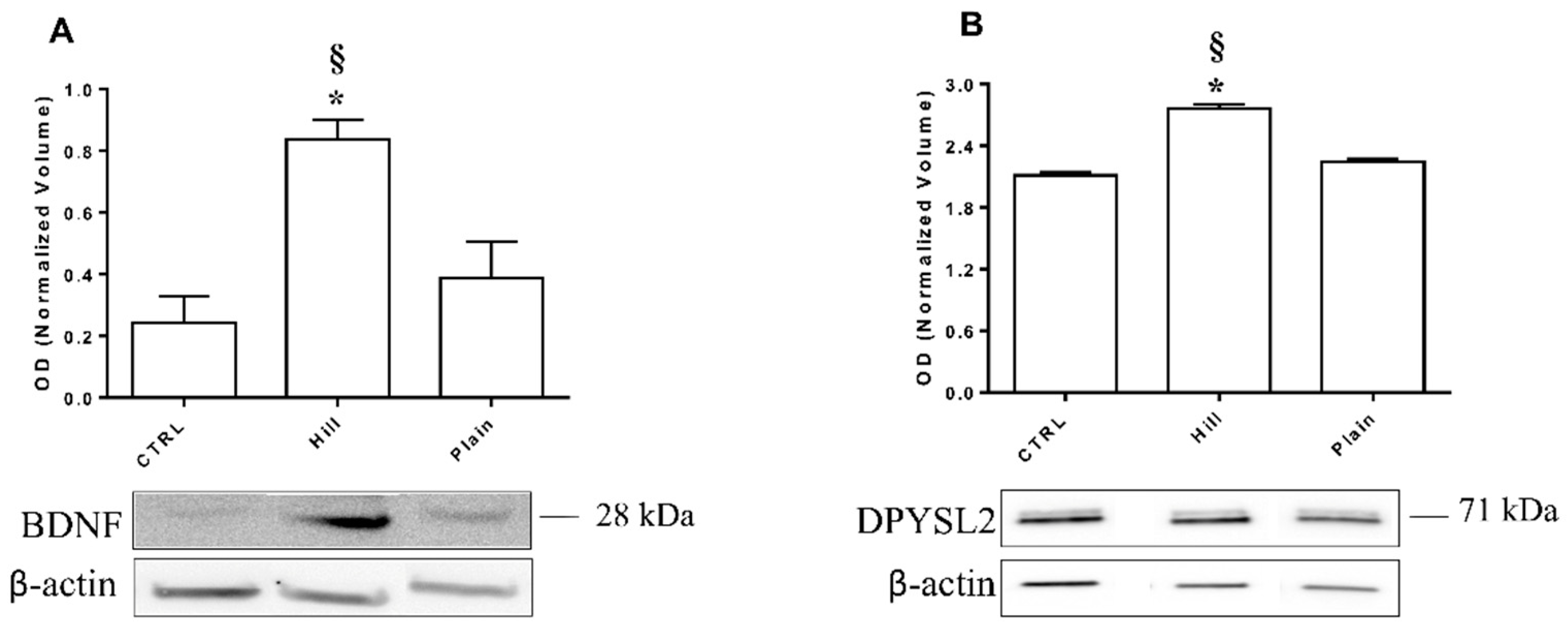
3.7. Validation of BDNF and DPYSL2 Expression in SH-SY5Y Cells Treated with Hill and Plain EVOO Extracts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bennett, J.E.; Stevens, G.A.; Mathers, C.D.; Bonita, R.; Rehm, J.; Kruk, M.E.; Riley, L.M.; Dain, K.; Kengne, A.P.; Chalkidou, K.; et al. NCD Countdown 2030: Worldwide trends in non-communicable disease mortality and progress towards Sustainable Development Goal target 3.4. Lancet 2018, 392, 1072–1088. [Google Scholar] [CrossRef] [Green Version]
- De Simone, A.; Bartolini, M.; Baschieri, A.; Apperley, K.Y.P.; Chen, H.H.; Guardigni, M.; Montanari, S.; Kobrlova, T.; Soukup, O.; Valgimigli, L.; et al. Hydroxy-substituted trans-cinnamoyl derivatives as multifunctional tools in the context of Alzheimer’s disease. Eur. J. Med. Chem. 2017, 139, 378–389. [Google Scholar] [CrossRef]
- Minarini, A.; Milelli, A.; Tumiatti, V.; Rosini, M.; Simoni, E.; Bolognesi, M.L.; Andrisano, V.; Bartolini, M.; Motori, E.; Angeloni, C.; et al. Cystamine-tacrine dimer: A new multi-target-directed ligand as potential therapeutic agent for Alzheimer’s disease treatment. Neuropharmacology 2012, 62, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Féart, C.; Samieri, C.; Barberger-Gateau, P. Mediterranean diet and cognitive function in older adults. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Feart, C.; Samieri, C.; Barberger-Gateau, P. Mediterranean diet and cognitive health: An update of available knowledge. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Motori, E.; Puyal, J.; Toni, N.; Ghanem, A.; Angeloni, C.; Malaguti, M.; Cantelli-Forti, G.; Berninger, B.; Conzelmann, K.K.; Götz, M.; et al. Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 2013, 18, 844–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beal, M.F. Energetics in the pathogenesis of neurodegenerative diseases. Trends Neurosci. 2000, 23, 298–304. [Google Scholar] [CrossRef]
- Epstein, F.H.; Lipton, S.A.; Rosenberg, P.A. Excitatory Amino Acids as a Final Common Pathway for Neurologic Disorders. N. Engl. J. Med. 1994, 330, 613–622. [Google Scholar] [CrossRef]
- Merad-Boudia, M.; Nicole, A.; Santiard-Baron, D.; Saillé, C.; Ceballos-Picot, I. Mitochondrial impairment as an early event in the process of apoptosis induced by glutathione depletion in neuronal cells: Relevance to Parkinson’s disease. Biochem. Pharmacol. 1998, 56, 645–655. [Google Scholar] [CrossRef]
- Hensley, K.; Carney, J.M.; Mattson, M.P.; Aksenova, M.; Harris, M.; Wu, J.F.; Floyd, R.A.; Butterfield, D.A. A model for β-amyloid aggregation and neurotoxicity based on free radical generation by the peptide: Relevance to Alzheimer disease. Proc. Natl. Acad. Sci. USA 1994, 91, 3270–3274. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhou, T.; Ziegler, A.C.; Dimitrion, P.; Zuo, L. Oxidative Stress in Neurodegenerative Diseases: From Molecular Mechanisms to Clinical Applications. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.G.; Zhu, X. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1240–1247. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, D.A.; Di Domenico, F.; Barone, E. Elevated risk of type 2 diabetes for development of Alzheimer disease: A key role for oxidative stress in brain. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1693–1706. [Google Scholar] [CrossRef] [Green Version]
- Angeloni, C.; Malaguti, M.; Barbalace, M.C.; Hrelia, S. Bioactivity of olive oil phenols in neuroprotection. Int. J. Mol. Sci. 2017, 18, 2230. [Google Scholar] [CrossRef] [Green Version]
- Hrelia, P.; Sita, G.; Ziche, M.; Ristori, E.; Marino, A.; Cordaro, M.; Molteni, R.; Spero, V.; Malaguti, M.; Morroni, F.; et al. Common Protective Strategies in Neurodegenerative Disease: Focusing on Risk Factors to Target the Cellular Redox System. Oxid. Med. Cell. Longev. 2020, 2020. [Google Scholar] [CrossRef]
- Kobayashi, M.; Yamamoto, M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, R.; Jaiswal, A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 1996, 93, 14960–14965. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, T.B.; Savall, A.S.; Gutierrez, M.E.Z.; Pinton, S. Neurotrophic factors in Alzheimer’s and parkinson’s diseases: Implications for pathogenesis and therapy. Neural Regen. Res. 2017, 12, 549–557. [Google Scholar] [PubMed]
- Holsinger, R.M.D.; Schnarr, J.; Henry, P.; Castelo, V.T.; Fahnestock, M. Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: Decreased levels in Alzheimer’s disease. Mol. Brain Res. 2000, 76, 347–354. [Google Scholar] [CrossRef]
- Phillips, H.S.; Hains, J.M.; Armanini, M.; Laramee, G.R.; Johnson, S.A.; Winslow, J.W. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron 1991, 7, 695–702. [Google Scholar] [CrossRef]
- Michalski, B.; Fahnestock, M. Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer’s disease. Mol. Brain Res. 2003, 111, 148–154. [Google Scholar] [CrossRef]
- Scarmeas, N.; Stern, Y.; Tang, M.X.; Mayeux, R.; Luchsinger, J.A. Mediterranean diet and risk for Alzheimer’s disease. Ann. Neurol. 2006, 59, 912–921. [Google Scholar] [CrossRef] [Green Version]
- Scarmeas, N.; Stern, Y.; Mayeux, R.; Luchsinger, J.A. Mediterranean diet, alzheimer disease, and vascular mediation. Arch. Neurol. 2006, 63, 1709–1717. [Google Scholar] [CrossRef] [Green Version]
- Corona, G.; Spencer, J.; Dessì, M. Extra virgin olive oil phenolics: Absorption, metabolism, and biological activities in the GI tract. Toxicol. Ind. Health 2009, 25, 285–293. [Google Scholar] [CrossRef]
- Bach-Faig, A.; Berry, E.M.; Lairon, D.; Reguant, J.; Trichopoulou, A.; Dernini, S.; Medina, F.X.; Battino, M.; Belahsen, R.; Miranda, G.; et al. Mediterranean diet pyramid today. Science and cultural updates. Public Health Nutr. 2011, 14, 2274–2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitler, C.M.; Viale, T.M.; Damaj, B.; Crea, R. Hydrolyzed olive vegetation water in mice has anti-inflammatory activity. J. Nutr. 2005, 135, 1475–1479. [Google Scholar] [CrossRef]
- Tripoli, E.; Giammanco, M.; Tabacchi, G.; Di Majo, D.; Giammanco, S.; La Guardia, M. The phenolic compounds of olive oil: Structure, biological activity and beneficial effects on human health. Nutr. Res. Rev. 2005, 18, 98–112. [Google Scholar] [CrossRef]
- Giusti, L.; Angeloni, C.; Barbalace, M.C.; Lacerenza, S.; Ciregia, F.; Ronci, M.; Urbani, A.; Manera, C.; Digiacomo, M.; Macchia, M.; et al. A proteomic approach to uncover neuroprotective mechanisms of oleocanthal against oxidative stress. Int. J. Mol. Sci. 2018, 19, 2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cicerale, S.; Conlan, X.A.; Sinclair, A.J.; Keast, R.S.J. Chemistry and health of olive oil phenolics. Crit. Rev. Food Sci. Nutr. 2009, 49, 218–236. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Pancorbo, A.; Cerretani, L.; Bendini, A.; Segura-Carretero, A.; Gallina-Toschi, T.; Fernández-Gutiérrez, A. Analytical determination of polyphenols in olive oils. J. Sep. Sci. 2005, 28, 837–858. [Google Scholar] [CrossRef]
- Parkinson, L.; Cicerale, S. The health benefiting mechanisms of virgin olive oil phenolic compounds. Molecules 2016, 21, 1734. [Google Scholar] [CrossRef] [Green Version]
- Kalogeropoulos, N.; Tsimidou, M.Z. Antioxidants in greek virgin olive oils. Antioxidants 2014, 3, 387–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Lecce, G.; Piochi, M.; Pacetti, D.; Frega, N.G.; Bartolucci, E.; Scortichini, S.; Fiorini, D. Eleven monovarietal extra virgin olive oils from olives grown and processed under the same conditions: Effect of the cultivar on the chemical composition and sensory traits. Foods 2020, 9, 904. [Google Scholar] [CrossRef]
- Pacetti, D.; Boarelli, M.C.; Giovannetti, R.; Ferraro, S.; Conti, P.; Alfei, B.; Caprioli, G.; Ricciutelli, M.; Sagratini, G.; Fedeli, D.; et al. Chemical and sensory profiling of monovarietal extra virgin olive oils from the italian marche region. Antioxidants 2020, 9, 330. [Google Scholar] [CrossRef] [Green Version]
- Polini, B.; Digiacomo, M.; Carpi, S.; Bertini, S.; Gado, F.; Saccomanni, G.; Macchia, M.; Nieri, P.; Manera, C.; Fogli, S. Oleocanthal and oleacein contribute to the in vitro therapeutic potential of extra virgin oil-derived extracts in non-melanoma skin cancer. Toxicol. In Vitro 2018, 52, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palla, M.; Digiacomo, M.; Cristani, C.; Bertini, S.; Giovannetti, M.; Macchia, M.; Manera, C.; Agnolucci, M. Composition of health-promoting phenolic compounds in two extra virgin olive oils and diversity of associated yeasts. J. Food Compos. Anal. 2018, 74, 27–33. [Google Scholar] [CrossRef]
- Ricciutelli, M.; Marconi, S.; Boarelli, M.C.; Caprioli, G.; Sagratini, G.; Ballini, R.; Fiorini, D. Olive oil polyphenols: A quantitative method by high-performance liquid-chromatography-diode-array detection for their determination and the assessment of the related health claim. J. Chromatogr. A 2017, 1481, 53–63. [Google Scholar] [CrossRef]
- Caprioli, G.; Boarelli, M.C.; Ricciutelli, M.; Sagratini, G.; Fiorini, D. Micro-scaled Quantitative Method to Analyze Olive Oil Polyphenols. Food Anal. Methods 2019, 12, 1133–1139. [Google Scholar] [CrossRef]
- Nzekoue, F.K.; Angeloni, S.; Navarini, L.; Angeloni, C.; Freschi, M.; Hrelia, S.; Vitali, L.A.; Sagratini, G.; Vittori, S.; Caprioli, G. Coffee silverskin extracts: Quantification of 30 bioactive compounds by a new HPLC-MS/MS method and evaluation of their antioxidant and antibacterial activities. Food Res. Int. 2020, 133, 109128. [Google Scholar] [CrossRef]
- Antognoni, F.; Potente, G.; Mandrioli, R.; Angeloni, C.; Freschi, M.; Malaguti, M.; Hrelia, S.; Lugli, S.; Gennari, F.; Muzzi, E.; et al. Fruit quality characterization of new sweet cherry cultivars as a good source of bioactive phenolic compounds with antioxidant and neuroprotective potential. Antioxidants 2020, 9, 677. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, C.; Teti, G.; Barbalace, M.C.; Malaguti, M.; Falconi, M.; Hrelia, S. 17β-Estradiol enhances sulforaphane cardioprotection against oxidative stress. J. Nutr. Biochem. 2017, 42, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, C.; Malaguti, M.; Rizzo, B.; Barbalace, M.C.; Fabbri, D.; Hrelia, S. Neuroprotective Effect of Sulforaphane against Methylglyoxal Cytotoxicity. Chem. Res. Toxicol. 2015, 28, 1234–1245. [Google Scholar] [CrossRef]
- Ciregia, F.; Bugliani, M.; Ronci, M.; Giusti, L.; Boldrini, C.; Mazzoni, M.R.; Mossuto, S.; Grano, F.; Cnop, M.; Marselli, L.; et al. Palmitate-induced lipotoxicity alters acetylation of multiple proteins in clonal β cells and human pancreatic islets. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciregia, F.; Giusti, L.; Da Valle, Y.; Donadio, E.; Consensi, A.; Giacomelli, C.; Sernissi, F.; Scarpellini, P.; Maggi, F.; Lucacchini, A.; et al. A multidisciplinary approach to study a couple of monozygotic twins discordant for the chronic fatigue syndrome: A focus on potential salivary biomarkers. J. Transl. Med. 2013, 11. [Google Scholar] [CrossRef] [Green Version]
- Ciregia, F.; Giusti, L.; Ronci, M.; Bugliani, M.; Piga, I.; Pieroni, L.; Rossi, C.; Marchetti, P.; Urbani, A.; Lucacchini, A. Glucagon-like peptide 1 protects INS-1E mitochondria against palmitate-mediated beta-cell dysfunction: A proteomic study. Mol. Biosyst. 2015, 11, 1696–1707. [Google Scholar] [CrossRef]
- Espinosa-Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Sánchez-Pérez, P.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015, 6, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Kubasek, W.L.; Ausubel, F.M.; Shirley, B.W. A light-independent developmental mechanism potentiates flavonoid gene expression in Arabidopsis seedlings. Plant Mol. Biol. 1998, 37, 217–223. [Google Scholar] [CrossRef]
- Azuma, A.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012, 236, 1067–1080. [Google Scholar] [CrossRef]
- Hernández, I.; Alegre, L.; Munné-Bosch, S. Drought-induced changes in flavonoids and other low molecular weight antioxidants in Cistus clusii grown under Mediterranean field conditions. Tree Physiol. 2004, 24, 1303–1311. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Slimestad, R.; Lea, U.S.; Brede, C.; Løvdal, T.; Ruoff, P.; Verheul, M.; Lillo, C. Temperature and nitrogen effects on regulators and products of the flavonoid pathway: Experimental and kinetic model studies. Plant Cell Environ. 2009, 32, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Influence of harvest year, cultivar and geographical origin on Greek extra virgin olive oils composition: A study by NMR spectroscopy and biometric analysis. Food Chem. 2012, 135, 2561–2568. [Google Scholar] [CrossRef]
- Pezzini, F.; Bettinetti, L.; Di Leva, F.; Bianchi, M.; Zoratti, E.; Carrozzo, R.; Santorelli, F.M.; Delledonne, M.; Lalowski, M.; Simonati, A. Transcriptomic Profiling Discloses Molecular and Cellular Events Related to Neuronal Differentiation in SH-SY5Y Neuroblastoma Cells. Cell. Mol. Neurobiol. 2017, 37, 665–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borriello, A.; Cucciolla, V.; Criscuolo, M.; Indaco, S.; Oliva, A.; Giovane, A.; Bencivenga, D.; Iolascon, A.; Zappia, V.; Della Ragione, F. Retinoic acid induces p27 Kip1 nuclear accumulation by modulating its phosphorylation. Cancer Res. 2006, 66, 4240–4248. [Google Scholar] [CrossRef] [Green Version]
- Cheung, Y.T.; Lau, W.K.W.; Yu, M.S.; Lai, C.S.W.; Yeung, S.C.; So, K.F.; Chang, R.C.C. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology 2009, 30, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, R.; Constantinescu, A.T.; Reichmann, H.; Janetzky, B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. In Neuropsychiatric Disorders an Integrative Approach; Springer: Vienna, Austria, 2007; pp. 17–28. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Grewal, R.; Reutzel, M.; Dilberger, B.; Hein, H.; Zotzel, J.; Marx, S.; Tretzel, J.; Sarafeddinov, A.; Fuchs, C.; Eckert, G.P. Purified oleocanthal and ligstroside protect against mitochondrial dysfunction in models of early Alzheimer’s disease and brain ageing. Exp. Neurol. 2020, 328, 113248. [Google Scholar] [CrossRef]
- Murphy, K.E.; Park, J.J. Can co-activation of Nrf2 and neurotrophic signaling pathway slow Alzheimer’s disease? Int. J. Mol. Sci. 2017, 18, 1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, G.; Johnson, J.A. The Nrf2-ARE pathway: A valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat. CNS Drug Discov. 2012, 7, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.S.; Misonou, Y.; Fujiwara, N.; Takahashi, M.; Miyamoto, Y.; Koh, Y.H.; Suzuki, K.; Taniguchi, N. Induction of thioredoxin reductase as an adaptive response to acrolein in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2005, 327, 1058–1065. [Google Scholar] [CrossRef]
- Coppola, G.; Choi, S.H.; Santos, M.M.; Miranda, C.J.; Tentler, D.; Wexler, E.M.; Pandolfo, M.; Geschwind, D.H. Gene expression profiling in frataxin deficient mice: Microarray evidence for significant expression changes without detectable neurodegeneration. Neurobiol. Dis. 2006, 22, 302–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaassen, C.D.; Reisman, S.A. Nrf2 the rescue: Effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010, 244, 57–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vauzour, D.; Buonfiglio, M.; Corona, G.; Chirafisi, J.; Vafeiadou, K.; Angeloni, C.; Hrelia, S.; Hrelia, P.; Spencer, J.P.E. Sulforaphane protects cortical neurons against 5-S-cysteinyl-dopamine-induced toxicity through the activation of ERK1/2, NrF-2 and the upregulation of detoxification enzymes. Mol. Nutr. Food Res. 2010, 54, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Walker, A.J.; Walder, K.; Berk, M.; Marx, W.; Carvalho, A.F.; Maes, M.; Puri, B.K. Increasing Nrf2 Activity as a Treatment Approach in Neuropsychiatry. Mol. Neurobiol. 2021. [Google Scholar] [CrossRef]
- Li, A.L.; Li, G.H.; Li, Y.R.; Wu, X.Y.; Ren, D.M.; Lou, H.X.; Wang, X.N.; Shen, T. Lignan and flavonoid support the prevention of cinnamon against oxidative stress related diseases. Phytomedicine 2019, 53, 143–153. [Google Scholar] [CrossRef]
- Bendini, A.; Cerretani, L.; Carrasco-Pancorbo, A.; Gómez-Caravaca, A.M.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Lercker, G. Phenolic molecules in virgin olive oils: A survey of their sensory properties, health effects, antioxidant activity and analytical methods. An overview of the last decade. Molecules 2007, 12, 1679–1719. [Google Scholar] [CrossRef]
- Dai, W.; Wang, H.; Fang, J.; Zhu, Y.; Zhou, J.; Wang, X.; Zhou, Y.; Zhou, M. Curcumin provides neuroprotection in model of traumatic brain injury via the Nrf2-ARE signaling pathway. Brain Res. Bull. 2018, 140, 65–71. [Google Scholar] [CrossRef]
- Tarozzi, A.; Angeloni, C.; Malaguti, M.; Morroni, F.; Hrelia, S.; Hrelia, P. Sulforaphane as a Potential protective phytochemical against neurodegenerative diseases. Oxid. Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef]
- Rungratanawanich, W.; Cenini, G.; Mastinu, A.; Sylvester, M.; Wilkening, A.; Abate, G.; Bonini, S.A.; Aria, F.; Marziano, M.; Maccarinelli, G.; et al. γ-oryzanol improves cognitive function and modulates hippocampal proteome in mice. Nutrients 2019, 11, 753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandes, M.S.; Gray, N.E. NRF2 as a Therapeutic Target in Neurodegenerative Diseases. ASN Neuro 2020, 12. [Google Scholar] [CrossRef]
- Patapoutian, A.; Reichardt, L.F. Trk receptors: Mediators of neurotrophin action. Curr. Opin. Neurobiol. 2001, 11, 272–280. [Google Scholar] [CrossRef]
- Poo, M. Neurotrophins as synaptic modulators. Nat. Rev. Neurosci. 2001, 2, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Colucci-D’amato, L.; Speranza, L.; Volpicelli, F. Neurotrophic factor bdnf, physiological functions and therapeutic potential in depression, neurodegeneration and brain cancer. Int. J. Mol. Sci. 2020, 21, 7777. [Google Scholar] [CrossRef]
- Fahnestock, M.; Garzon, D.; Holsinger, R.M.D.; Michalski, B. Neurotrophic factors and Alzheimer’s disease: Are we focusing on the wrong molecule? In Proceedings of the Journal of Neural Transmission, Supplement; Springer: Wien, Austria, 2002; pp. 241–252. [Google Scholar]
- Siegel, G.J.; Chauhan, N.B. Neurotrophic factors in Alzheimer’s and Parkinson’s disease brain. Brain Res. Rev. 2000, 33, 199–227. [Google Scholar] [CrossRef]
- Arancibia, S.; Silhol, M.; Moulière, F.; Meffre, J.; Höllinger, I.; Maurice, T.; Tapia-Arancibia, L. Protective effect of BDNF against beta-amyloid induced neurotoxicity in vitro and in vivo in rats. Neurobiol. Dis. 2008, 31, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.; Atlas, R.; Lange, A.; Ginzburg, I. Brain-derived neurotrophic factor induces a rapid dephosphorylation of tau protein through a PI-3Kinase signalling mechanism. Eur. J. Neurosci. 2005, 22, 1081–1089. [Google Scholar] [CrossRef]
- Casamenti, F.; Stefani, M. Olive polyphenols: New promising agents to combat aging-associated neurodegeneration. Expert Rev. Neurother. 2017, 17, 345–358. [Google Scholar] [CrossRef]
- Hung, C.Y.; Hsu, T.I.; Chuang, J.Y.; Su, T.P.; Chang, W.C.; Hung, J.J. Sp1 in Astrocyte is Important for Neurite Outgrowth and Synaptogenesis. Mol. Neurobiol. 2020, 57, 261–277. [Google Scholar] [CrossRef]
- Moutinho, D.; Veiga, S.; Requena, J.R. Human VGF-Derived Antidepressant Neuropeptide TLQP62 Promotes SH-SY5Y Neurite Outgrowth. J. Mol. Neurosci. 2020, 70, 1293–1302. [Google Scholar] [CrossRef]
- Xiong, L.L.; Qiu, D.L.; Xiu, G.H.; Al-Hawwas, M.; Jiang, Y.; Wang, Y.C.; Hu, Y.; Chen, L.; Xia, Q.J.; Wang, T.H. DPYSL2 is a novel regulator for neural stem cell differentiation in rats: Revealed by Panax notoginseng saponin administration. Stem Cell Res. Ther. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Kakumoto, K.; Yoshida, T.; Kuwako, K.I.; Miyazaki, T.; Yamaguchi, J.; Konno, A.; Hata, J.; Uchiyama, Y.; Hirai, H.; et al. Elavl3 is essential for the maintenance of Purkinje neuron axons. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Singh, M.; Kakkar, A.; Sharma, R.; Kharbanda, O.P.; Monga, N.; Kumar, M.; Chowdhary, S.; Airan, B.; Mohanty, S. Synergistic Effect of BDNF and FGF2 in Efficient Generation of Functional Dopaminergic Neurons from human Mesenchymal Stem Cells. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palasz, E.; Wysocka, A.; Gasiorowska, A.; Chalimoniuk, M.; Niewiadomski, W.; Niewiadomska, G. BDNF as a promising therapeutic agent in parkinson’s disease. Int. J. Mol. Sci. 2020, 21, 1170. [Google Scholar] [CrossRef] [Green Version]
- Sarchielli, E.; Marini, M.; Ambrosini, S.; Peri, A.; Mazzanti, B.; Pinzani, P.; Barletta, E.; Ballerini, L.; Paternostro, F.; Paganini, M.; et al. Multifaceted roles of BDNF and FGF2 in human striatal primordium development: An in vitro study. Exp. Neurol. 2014, 257, 130–147. [Google Scholar] [CrossRef]
- Rached, M.B.; Galaverna, G.; Cirlini, M.; Boujneh, D.; Zarrouk, M.; Guerfel, M. Pedologic factors affecting virgin olive oil quality of “Chemlali” olive trees (Olea europaea L.). J. Oleo Sci. 2017, 66, 907–915. [Google Scholar] [CrossRef] [Green Version]
- Caruso, G.; Gucci, R.; Urbani, S.; Esposto, S.; Taticchi, A.; Di Maio, I.; Selvaggini, R.; Servili, M. Effect of different irrigation volumes during fruit development on quality of virgin olive oil of cv. Frantoio. Agric. Water Manag. 2014, 134, 94–103. [Google Scholar] [CrossRef]
- Caruso, G.; Gucci, R.; Sifola, M.I.; Selvaggini, R.; Urbani, S.; Esposto, S.; Taticchi, A.; Servili, M. Irrigation and fruit canopy position modify oil quality of olive trees (cv. Frantoio). J. Sci. Food Agric. 2017, 97, 3530–3539. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Kwon, H.; Cho, E.; jeon, J.; Kang, R.H.; Youn, K.; Jun, M.; Lee, Y.C.; Ryu, J.H.; Kim, D.H. The effects of pinoresinol on cholinergic dysfunction-induced memory impairments and synaptic plasticity in mice. Food Chem. Toxicol. 2019, 125, 376–382. [Google Scholar] [CrossRef]
- In, S.J.; Seo, K.H.; Song, N.Y.; Lee, D.S.; Kim, Y.C.; Baek, N.I. Lignans and neolignans from the stems of Vibrunum erosum and their neuroprotective and anti-inflammatory activity. Arch. Pharm. Res. 2015, 38, 26–34. [Google Scholar] [CrossRef]
- Dinda, B.; Dinda, M.; Kulsi, G.; Chakraborty, A.; Dinda, S. Therapeutic potentials of plant iridoids in Alzheimer’s and Parkinson’s diseases: A review. Eur. J. Med. Chem. 2019, 169, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Borah, P.; Sanjeev, A.; Mattaparthi, V.S.K. Computational investigation on the effect of Oleuropein aglycone on the α-synuclein aggregation. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Casamenti, F.; Grossi, C.; Rigacci, S.; Pantano, D.; Luccarini, I.; Stefani, M. Oleuropein Aglycone: A Possible Drug against Degenerative Conditions. In Vivo Evidence of its Effectiveness against Alzheimer’s Disease. J. Alzheimer’s Dis. 2015, 45, 679–688. [Google Scholar] [CrossRef] [PubMed]


| Gene | 5′-Forward-3′ | 5′-Reverse-3′ | RefSeq Accession No. |
|---|---|---|---|
| HMOX1 | CAACAAAGTGCAAGATTCTG | TGCATTCACATGGCATAAAG | NM_002133 |
| BDNF | CAAAAGTGGAGAACATTTGC | AACTCCAGTCAATAGGTCAG | NM_001143811 |
| NQO1 | AGTATCCACAATAGCTGACG | TTTGTGGGTCTGTAGAAATG | NM_000903 |
| GSR | GACCTATTCAACGAGCTTTAC | CAACCACCTTTTCTTCCTTG | NM_000637 |
| TXNRD1 | AGACAGTTAAGCATGATTGG | AATTGCCCATAAGCATTCTC | NM_001093771 |
| 18S rRNA | CAGAAGGATGTAAAGGATGG | TATTTCTTCTTGGACACACC | NM_022551 |
| Soil Categories | Hill | Plain |
|---|---|---|
| % ± SEM | % ± SEM | |
| Clay | 8.7 ± 0.08 * | 8.4 ± 0.08 |
| Silt | 38 ± 0.46 *** | 46.1 ± 0.50 |
| Sandy | 53.3 ± 0.68 ** | 45.6 ± 0.94 |
| pH | 5.4 ± 0.04 *** | 6.9 ± 0.05 |
| Ppm ± | Ppm ± | |
| K | 110 ± 0.83 *** | 54.7 ± 0.44 |
| Mg | 73.7 ± 0.47 *** | 63.6 ± 0.37 |
| Ca | 1188 ± 10.29 *** | 1428 ± 15.46 |
| P | 48 ± 0.36 *** | 34 ± 0.22 |
| Total N | 2600 ± 24.02 *** | 3000 ± 27.71 |
| Phenolic Compound | Hill | Plain |
|---|---|---|
| (μg/g ± SD) | ||
| Hydroxytyrosol ** | 678.21 ± 24.27 | 2167.85 ± 93.35 |
| Tyrosol ** | 771.82 ± 38.75 | 1697.46 ± 123.03 |
| Vanillic acid ** | 6.83 ± 0.34 | 4.05 ± 0.15 |
| p-Coumaric acid ** | 3.22 ± 0.19 | 10.30 ± 0.67 |
| ferulic acid * | 0.56 ± 0.04 | 0.89 ± 0.05 |
| 3,4-DHPEA-EDA ** | 803.33 ± 46.12 | 2610.92 ± 169.48 |
| p-HPEA-EDA | 1102.60 ± 71.26 | 1316.25 ± 95.03 |
| Pinoresinol ** | 1819.41 ± 51.98 | 664.64 ± 23.78 |
| Acetoxypinoresinol ** | 1854.62 ± 133.24 | 1018.40 ± 80.94 |
| Luteolin | 64.27 ± 4.15 | 85.78 ± 6.19 |
| 3,4-DHPEA-EA * | 330.78 ± 9.45 | 244.55 ± 8.75 |
| p-HPEA-EA ** | 756.07 ± 54.32 | 251.74 ± 20.01 |
| Apigenin * | 25.08 ± 1.08 | 40.08 ± 2.02 |
| Spot n. | Protein Name | ID | Gene | Coverage (%) | Peptides | Unic Peptide | MW (kDa) | pI | Ratio (Hill/Ctrl) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| 2417 | Heterogeneous nuclear ribonucleoprotein A3 | P51991 | HNRNPA3 | 29 | 10 | 10 | 39 | 9.1 | 7.84 | 0.017 |
| 2514 | ELAV-like protein 3, Iso 1,2 | Q14576 | ELAVL3 | 11, 11 | 3 | 1 | 39/38 | 9.3 | 7.3 | 0.012 |
| 1601 | Non-POU domain-containing octamer-binding protein | Q15233 | NONO | 14 | 4 | 4 | 54 | 9.0 | 4.59 | 0.03 |
| 1030 | Pyruvate dehydrogenase phosphatase regulatory subunit, mitochondrial, Iso 1 | Q8NCN5 | PDPR | 4 | 3 | 3 | 99 | 5.7 | 2.48 | 0.017 |
| 1278 | Heat shock 70 kDa protein 1A, Iso 1,2 | P0DMV8 | HSPA1A | 13 | 6 | 1 | 66/70 | 5.4/5.5 | 2.3 | 0.011 |
| 1278 | Heat shock 70 kDa protein 1B | P0DMV9 | HSPA1B | 13 | 6 | 1 | 70 | 5.4 | 2.3 | 0.011 |
| 1278 | Heat shock cognate 71 kDa protein | P11142 | HSPA8 | 34 | 18 | 14 | 71 | 5.3 | 2.29 | 0.011 |
| 1367 | Dihydropyrimidinase-related protein 3, Iso LCRMP-4 | Q14195 | DPYSL3 | 28 | 14 | 14 | 74 | 5.9 | 2.20 | 0.022 |
| 1606 | Pyruvate kinase PKM, Iso M2 | P14618 | PKM | 38 | 18 | 18 | 58 | 7.9 | 1.8 | 0.034 |
| 1085 | Membrane primary amine oxidase, Iso 1,2 | Q16853 | AOC3 | 3 | 2 | 2 | 84/70 | 6.0/7.1 | 1.8 | 0.023 |
| 1085 | Vitamin D-binding protein, Iso 1,3 | P02774 | GC | 8 | 2 | 2 | 53/55 | 5.1/5.4 | 1.77 | 0.023 |
| 894 | Filamin-A, Iso 1,2 | P21333 | FLNA | 4 | 7 | 7 | 280 | 5.7 | 1.69 | 0.016 |
| 1136 | Ezrin | P15311 | EZR | 10 | 5 | 2 | 69 | 5.9 | 1.66 | 0.005 |
| 895 | Filamin-A, Iso 1,2 | P21333 | FLNA | 3 | 6 | 6 | 280 | 5.7 | 1.54 | 0.01 |
| 1093 | ATP-dependent 6-phosphofructokinase, muscle type, Iso 1, 3 | P08237 | PFKM | 15 | 9 | 9 | 85/93 | 8.2 | 1.50 | 0.005 |
| 945 | Prolyl 3-hydroxylase 3 | Q8IVL6 | P3H3 | 6 | 4 | 4 | 82 | 5.8 | 1.50 | 0.008 |
| 1427 | Dihydropyrimidinase-related protein 2, Iso 1,2 | Q16555 | DPYSL2 | 15/16 | 5 | 5 | 62/58 | 5.9/5.7 | 1.49 | 0.033 |
| 2475 | Eukaryotic translation initiation factor 2 subunit 1 | P05198 | EIF2S1 | 30 | 7 | 7 | 36 | 5.0 | 1.46 | 0.029 |
| 569 | Hypoxia up-regulated protein 1 | Q9Y4L1 | HYOU1 | 45 | 32 | 32 | 111 | 5.1 | 1.45 | 0.014 |
| 781 | Heat shock 70 kDa protein 4 | P34932 | HSPA4 | 32 | 18 | 18 | 94 | 5.1 | 1.39 | 0.044 |
| 1485 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial, Iso 1,2,3 | P49748 | ACADVL | 4 | 2 | 2 | 70/68 | 7.7/8.7 | 1.38 | 0.001 |
| 1280 | Heterogeneous nuclear ribonucleoprotein M, Iso 1,2 | P52272 | HNRNPM | 18 | 8 | 8 | 77/74 | 8.8/ 8.9 | 1.36 | 0.019 |
| 1371 | Probable ATP-dependent RNA helicase DDX17, Iso 1,2,3,4 | Q92841 | DDX17 | 8 | 4 | 4 | 80/72 | 8.5/ 8.8 | 1.36 | 0.019 |
| 1371 | Calcium-binding mitochondrial carrier protein Aralar2, Iso 1,2 | Q9UJS0 | SLC25A13 | 17 | 8 | 8 | 74 | 8.7 | 1.36 | 0.019 |
| 2074 | Vimentin | P08670 | VIM | 27 | 10 | 10 | 53 | 5.0 | 1.34 | 0.004 |
| 952 | 26S proteasome non-ATPase regulatory subunit 2 | Q13200 | PSMD2 | 19 | 10 | 10 | 100 | 5.1 | 1.34 | 0.017 |
| 808 | Insulin-degrading enzyme | P14735 | IDE | 3 | 3 | 3 | 117 | 6.2 | 1.33 | 0.043 |
| 1216 | 78 kDa glucose-regulated protein | P11021 | HSPA5 | 46 | 30 | 30 | 72 | 5.0 | 1.29 | 0.003 |
| 2374 | Alcohol dehydrogenase class-3 | P11766 | ADH5 | 18 | 4 | 4 | 39 | 7.6 | 1.29 | 0.037 |
| 2417 | Heterogeneous nuclear ribonucleoproteins A2/B1, Iso B1 | P22626 | HNRNPA2B1 | 12 | 3 | 3 | 37 | 8.9 | 1.29 | 0.037 |
| 1413 | Prelamin-A/C, Iso A,C | P02545 | LMNA | 9 | 5 | 5 | 74/65 | 6.5/6.4 | 1.28 | 0.023 |
| 1650 | Non-POU domain-containing octamer-binding protein | Q15233 | NONO | 25 | 9 | 9 | 54 | 9.0 | 1.28 | 0.041 |
| 901 | Programmed cell death 6-interacting protein, Iso 1,2 | Q8WUM4 | PDCD6IP | 21 | 11 | 11 | 96 | 1.28 | 0.006 | |
| 1371 | Heterogeneous nuclear ribonucleoprotein M, Iso 1,2 | P52272 | HNRNPM | 16/17 | 8 | 8 | 77/75 | 8.8/8.9 | 1.27 | 0.023 |
| 1461 | Asparagine--tRNA ligase, cytoplasmic | O43776 | NARS | 31 | 13 | 13 | 63 | 5.9 | 1.26 | 0.037 |
| 1467 | Prelamin-A/C, Iso A,C | P02545 | LMNA | 16/19 | 9 | 9 | 74/65 | 6.5/6.4 | 1.26 | 0.043 |
| 811 | Neutral alpha-glucosidase AB, Iso 1,2 | Q14697 | GANAB | 10 | 8 | 8 | 106/109 | 1.26 | 0.001 | |
| 1012 | Gelsolin, Iso 1,2,3,4 | P06396 | GSN | 6 | 3 | 3 | 85/80 | 5.7/5.5 | 1.24 | 0.041 |
| 1363 | Lamin-B1 | P20700 | LMNB1 | 33 | 20 | 20 | 66 | 5.1 | 1.24 | 0.03 |
| 1330 | Beta-catenin-like protein 1, Iso 1,4 | Q8WYA6 | CTNNBL1 | 7 | 2 | 2 | 65/61 | 4.9/5.0 | 1.24 | 0.03 |
| 1330 | Ubiquilin-2 | Q9UHD9 | UBQLN2 | 5 | 2 | 2 | 65 | 5.1 | 1.24 | 0.03 |
| 1084 | Neurosecretory protein VGF | O15240 | VGF | 14 | 6 | 6 | 67 | 4.7 | 1.22 | 0.013 |
| 1084 | Secretogranin-2 | P13521 | SCG2 | 14 | 7 | 7 | 71 | 4.6 | 1.22 | 0.013 |
| 2593 | Elongation factor 1-delta, Iso 1,2 | P29692 | EEF1D | 26 | 6 | 6 | 31 | 4.9/6.0 | 1.16 | 0.015 |
| 1427 | Phosphoacetylglucosamine mutase, Iso 1,2,3 | O95394 | PGM3 | 6 | 3 | 3 | 59/62 | 5.8/5.6 | 1.07 | 0.023 |
| 3410 | Proteasome subunit alpha type-2 | P25787 | PSMA2 | 39 | 7 | 7 | 26 | 7.1 | 0.81 | 0.002 |
| 2315 | Actin, cytoplasmic 1 | P60709 | ACTB | 11 | 4 | 4 | 42 | 5.2 | 0.75 | 0.014 |
| 2315 | Actin, cytoplasmic 2 | P63261 | ACTG1 | 11 | 4 | 4 | 42 | 5.3 | 0.75 | 0.014 |
| 1979 | Ribonuclease inhibitor | P13489 | RNH1 | 19 | 6 | 6 | 50 | 4.7 | 0.73 | 0.017 |
| 2230 | DNA-directed RNA polymerases I and III subunit RPAC1, Iso 1,2 | O15160 | POLR1C | 17/16 | 4 | 4 | 39/38 | 5.3/5.6 | 0.67 | 0.007 |
| 2830 | Voltage-dependent anion-selective channel protein 2, Iso 1,2,3 | P45880 | VDAC2 | 15/16 | 3 | 3 | 33/30 | 7.5/6.8 | 0.66 | 0.023 |
| Spot n. | Protein Name | ID | Gene | Coverage (%) | Peptides | Unic Peptides | MW (kDa) | pI | Ratio (Plain/Ctrl) | Ratio (Hill/Ctrl) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2164 | Heterogeneous nuclear ribonucleoprotein D0 Iso 1,3 | Q14103 | HNRNPD | 9/10 | 2 | 2 | 38/32 | 7.6/8.2 | 1.77 | 1.44 | 0.005 |
| 2164 | Citrate synthase mitochondrial | O75390 | CS | 8 | 3 | 3 | 52 | 7.4 | 1.77 | 1.44 | 0.005 |
| 2164 | Protein arginine methyltransferase NDUFAF7 mitochondrial | Q7L592 | NDUFAF7 | 8 | 3 | 3 | 49 | 7.3 | 1.77 | 1.44 | 0.005 |
| 2164 | Cytochrome b-c1 complex subunit 2. mitochondrial | P22695 | UQCRC2 | 14 | 4 | 4 | 48 | 7.7 | 1.77 | 1.44 | 0.005 |
| 1001 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 | O43143 | DHX15 | 14 | 9 | 9 | 91 | 7.1 | 1.72 | 1.49 | 0.009 |
| 5503 | MICOS complex subunit MIC60, Iso 1,2, 4 | Q16891 | IMMT | 31/31 | 17 | 1 | 83/ 82 | 5.7/6.1 | 1.66 | 1.75 | 0.019 |
| 1057 | X-ray repair cross-complementing protein 5 | P13010 | XRCC5 | 13 | 7 | 7 | 83 | 5.5 | 1.62 | 1.43 | 0.0006 |
| 1993 | 26S proteasome regulatory subunit 7 | P35998 | PSMC2 | 38 | 14 | 14 | 48 | 5.7 | 1.58 | 1.23 | 0.005 |
| 881 | Elongation factor 2 | P13639 | EEF2 | 9 | 6 | 6 | 95 | 6.4 | 1.58 | 1.29 | 0.033 |
| 726 | Vinculin, Iso 1,2 | P18206 | VCL | 16/15 | 9 | 9 | 116/124 | 5.8/5.5 | 1.38 | 1.63 | 0.028 |
| 2654 | Annexin A2, Iso 1,2 | P07355 | ANXA2 | 46/44 | 18 | 18 | 38/40 | 7.5/ 8.5 | 1.24 | 1.22 | 0.036 |
| 3761 | Adenine phosphoribosyltransferase, Iso 1,2 | P07741 | APRT | 23/31 | 3 | 3 | 19/14 | 5.7/6.7 | 0.87 | 0.81 | 0.036 |
| 3761 | DNA-directed RNA polymerase II subunit RPB7 | P62487 | POLR2G | 23 | 3 | 3 | 19 | 5.3 | 0.87 | 0.81 | 0.036 |
| 3549 | Eukaryotic translation initiation factor 3 subunit K, Iso 1,2 | Q9UBQ5 | EIF3K | 11 | 2 | 2 | 25/24 | 4.8. 4.7 | 0.7 | 0.73 | 0.013 |
| 3549 | Proteasome subunit beta type-6 | P28072 | PSMB6 | 18 | 4 | 4 | 25 | 4.9 | 0.7 | 0.73 | 0.013 |
| 3549 | Translationally-controlled tumor protein | P13693 | TPT1 | 34 | 5 | 5 | 19 | 4.8 | 0.7 | 0.73 | 0.013 |
| 3977 | Diablo homolog. Mitochondrial, Iso 1,2 | Q9NR28 | DIABLO | 27/34 | 6 | 6 | 27/21 | 4.7/4.8 | 0.58 | 0.52 | 0.003 |
| 3977 | Ras-related protein Rap-2a | P10114 | RAP2A | 10 | 2 | 2 | 20 | 4.73 | 0.58 | 0.52 | 0.003 |
| Upstream Regulator | Molecule Type | Activation z-Score | p-Value of Overlap | Target Molecules |
|---|---|---|---|---|
| TP53 | Transcription regulator | 0.295 | 8.48 × 10−11 | ACADVL, ACTB, ADH5, ANXA2, CS, EZR, GC, GSN, HNRNPA2B1, HSPA1A/HSPA1B, HSPA8, NARS1, PDCD6IP, PFKM, PGM3, PKM, PSMA2, PSMD2, RAP2A, SLC25A13, UBQLN2, VCL, VIM, XRCC5 |
| MAPT | Other | 7.74 × 10−9 | ACTB, ACTG1, CS, DPYSL2, DPYSL3, EEF2, HSPA1A/HSPA1B, HSPA5, HSPA8, PKM, PSMD2 | |
| BDNF | Growth factor | 0.29 | 2.53 × 10−7 | ANXA2, DPYSL3, EEF1D, EEF2, FLNA, HSPA5, RNH1, VGF, VIM |
| IL5 | Cytokine | 2.236 | 1.25 × 10−3 | ANXA2, HSPA5, LMNB1, PKM, VIM |
| SP1 | Transcription regulator | 2.211 | 5.07 × 10−3 | ADH5, EZR, FLNA, HSPA5, PKM, VIM |
| FGF2 | Growth factor | 2.391 | 1.1 × 10−3 | FLNA, LMNA, SCG2, VGF, VIM, XRCC5 |
| RET | Kinase | 2.186 | 1.6 × 10−5 | DPYSL2, HSPA1A/HSPA1B, HSPA8, UBQLN2, VGF |
| MYC | Transcription regulator | 2.185 | 1.32 × 10−5 | ACTB, EEF2, EIF2S1, EZR, FLNA, HNRNPA2B1, HNRNPD, NARS1, PFKM, PKM, POLR2G, VDAC2, VIM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbalace, M.C.; Zallocco, L.; Beghelli, D.; Ronci, M.; Scortichini, S.; Digiacomo, M.; Macchia, M.; Mazzoni, M.R.; Fiorini, D.; Lucacchini, A.; et al. Antioxidant and Neuroprotective Activity of Extra Virgin Olive Oil Extracts Obtained from Quercetano Cultivar Trees Grown in Different Areas of the Tuscany Region (Italy). Antioxidants 2021, 10, 421. https://doi.org/10.3390/antiox10030421
Barbalace MC, Zallocco L, Beghelli D, Ronci M, Scortichini S, Digiacomo M, Macchia M, Mazzoni MR, Fiorini D, Lucacchini A, et al. Antioxidant and Neuroprotective Activity of Extra Virgin Olive Oil Extracts Obtained from Quercetano Cultivar Trees Grown in Different Areas of the Tuscany Region (Italy). Antioxidants. 2021; 10(3):421. https://doi.org/10.3390/antiox10030421
Chicago/Turabian StyleBarbalace, Maria Cristina, Lorenzo Zallocco, Daniela Beghelli, Maurizio Ronci, Serena Scortichini, Maria Digiacomo, Marco Macchia, Maria Rosa Mazzoni, Dennis Fiorini, Antonio Lucacchini, and et al. 2021. "Antioxidant and Neuroprotective Activity of Extra Virgin Olive Oil Extracts Obtained from Quercetano Cultivar Trees Grown in Different Areas of the Tuscany Region (Italy)" Antioxidants 10, no. 3: 421. https://doi.org/10.3390/antiox10030421
APA StyleBarbalace, M. C., Zallocco, L., Beghelli, D., Ronci, M., Scortichini, S., Digiacomo, M., Macchia, M., Mazzoni, M. R., Fiorini, D., Lucacchini, A., Hrelia, S., Giusti, L., & Angeloni, C. (2021). Antioxidant and Neuroprotective Activity of Extra Virgin Olive Oil Extracts Obtained from Quercetano Cultivar Trees Grown in Different Areas of the Tuscany Region (Italy). Antioxidants, 10(3), 421. https://doi.org/10.3390/antiox10030421











