Live Cell Assays for the Assessment of Antioxidant Activities of Plant Extracts
Abstract
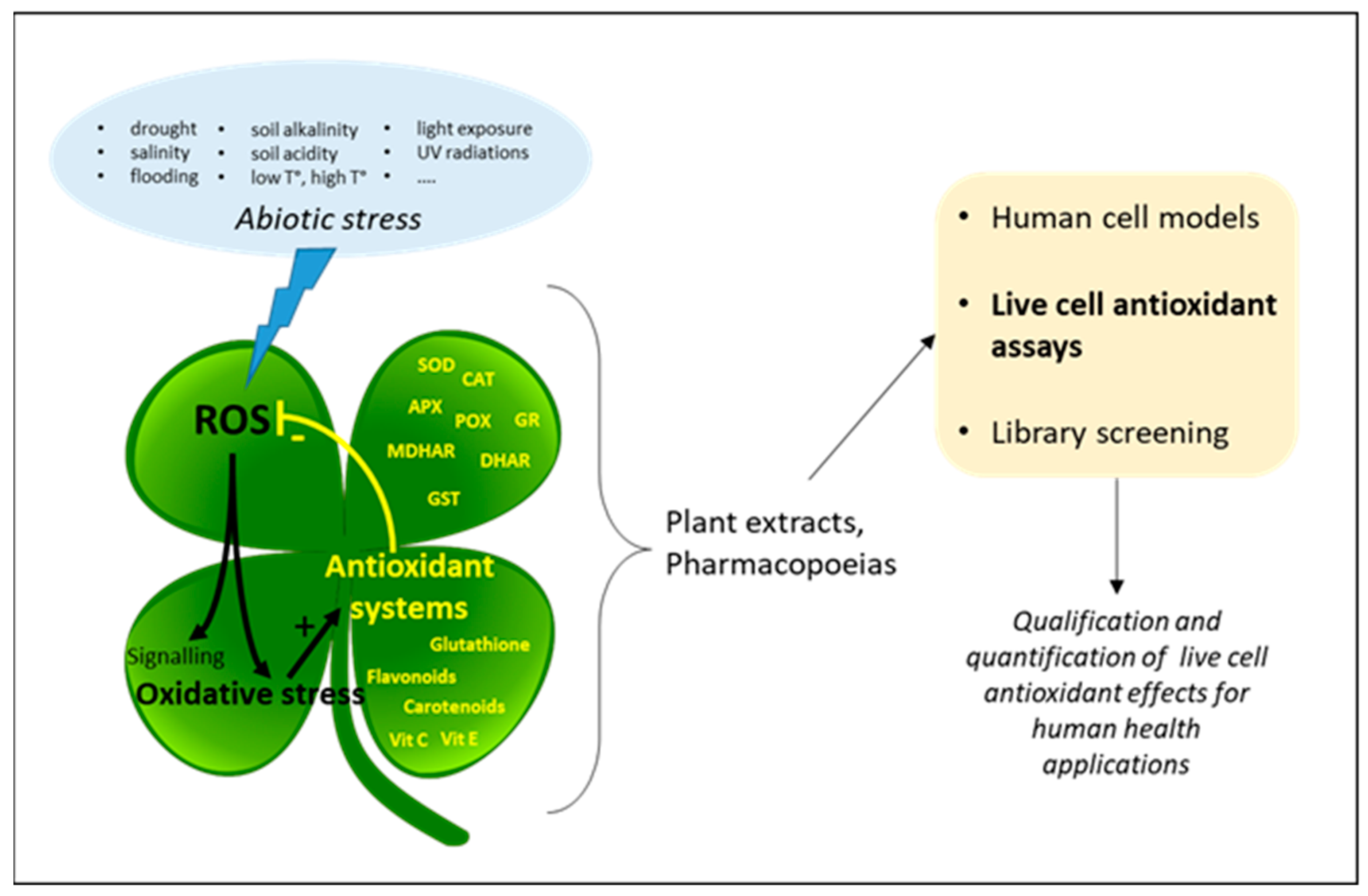
:1. Introduction
2. Cell Models
3. Antioxidant Effects in Live Cells
4. Live Cell Antioxidant Assays
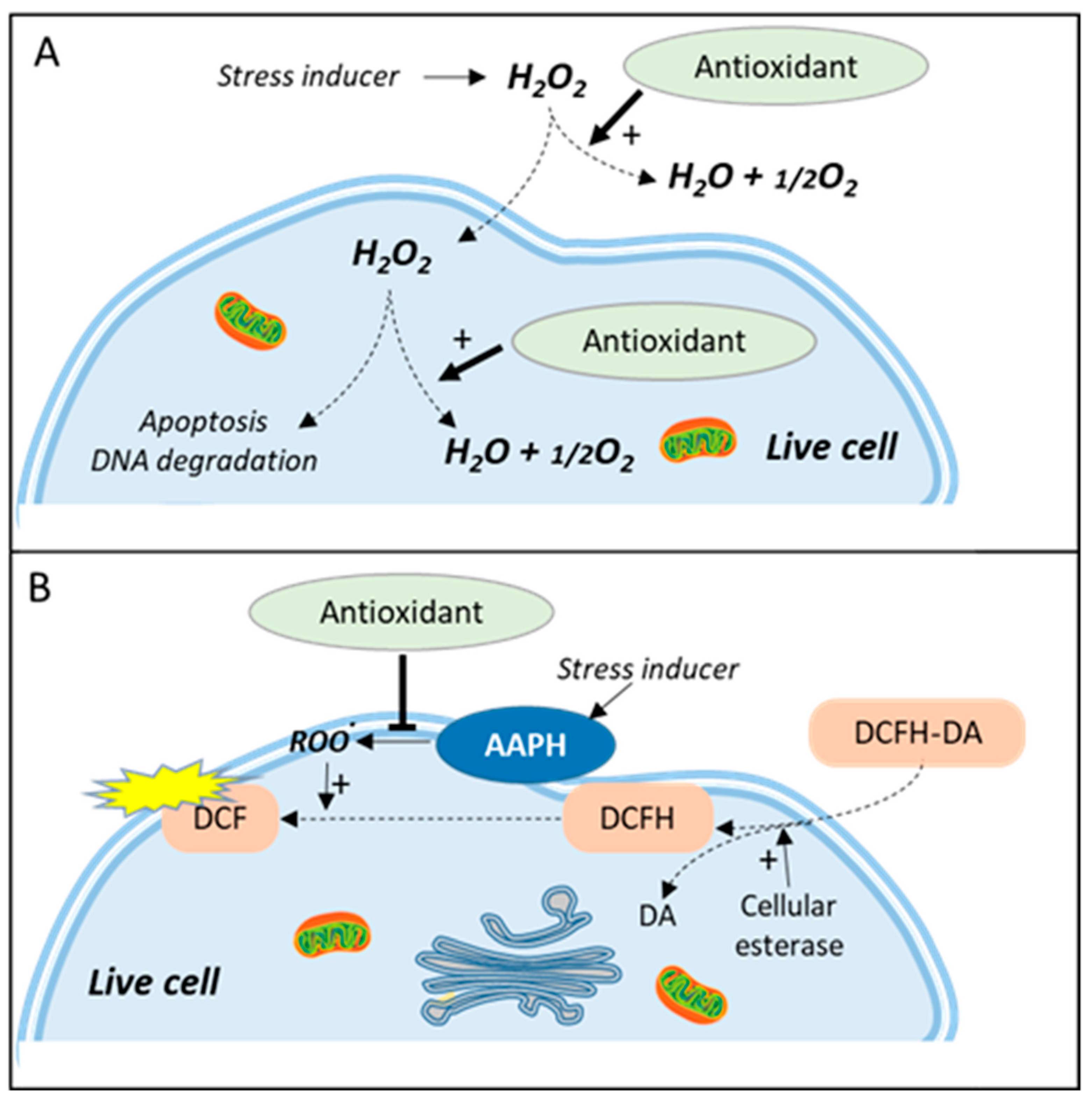
5. Catalase-Like Assay
6. Cell Antioxidant Assay (CAA)
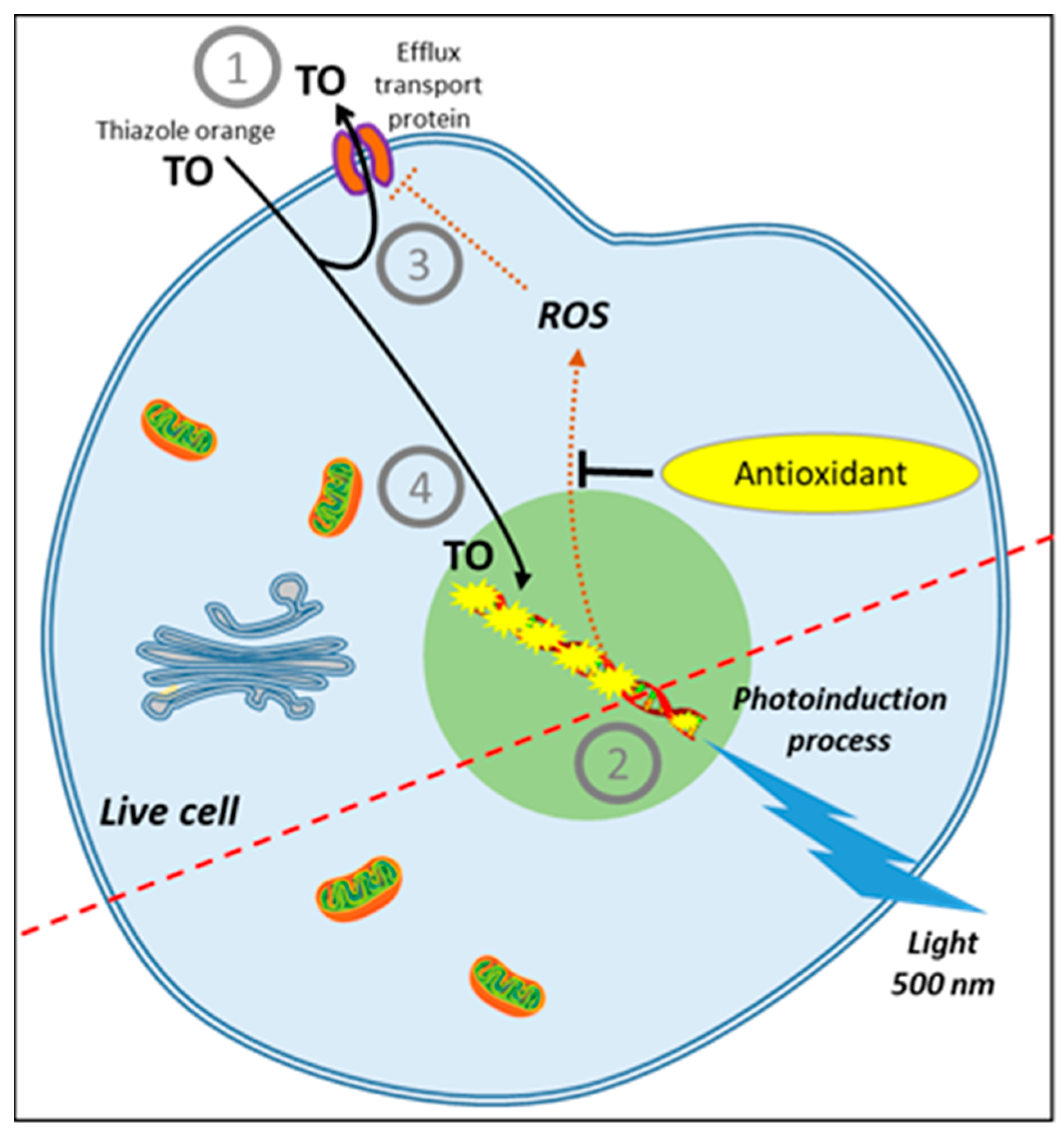
7. Toward a Better Monitoring of ROS Production in the Cell
8. AOP1, a New Antioxidant Live Cell Approach Based on Photoinduction
9. Measure of Indirect Antioxidant Effects by KEAP1/Nrf2/ARE Activation
10. Outlook—Toward an Industrial Standard of Cell-Based Assays
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Anee, T.I.; Nahar, K.; Hossen, M.S.; Zulfiqar, F.; Alam, M.M.; Fujita, M. Regulation of ROS Metabolism in Plants under Environmental Stress: A Review of Recent Experimental Evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- Soares, C.; Carvalho, M.E.A.; Azevedo, R.A.; Fidalgo, F. Plants Facing Oxidative Challenges—A Little Help from the Antioxidant Networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Singh, R.; Singh, S.; Parihar, P.; Mishra, R.K.; Tripathi, D.K.; Singh, V.P.; Chauhan, D.K.; Prasad, S.M. Reactive Oxygen Species (ROS): Beneficial Companions of Plants’ Developmental Processes. Front. Plant. Sci. 2016, 7, 1299. [Google Scholar] [CrossRef] [Green Version]
- Del Río, L.A. ROS and RNS in Plant Physiology: An Overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Lu, Y.; Saredy, J.; Wang, X.; Drummer Iv, C.; Shao, Y.; Saaoud, F.; Xu, K.; Liu, M.; Yang, W.Y.; et al. ROS Systems Are a New Integrated Network for Sensing Homeostasis and Alarming Stresses in Organelle Metabolic Processes. Redox Biol. 2020, 37, 101696. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant. Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramel, F.; Mialoundama, A.S.; Havaux, M. Nonenzymic Carotenoid Oxidation and Photooxidative Stress Signalling in Plants. J. Exp. Bot. 2013, 64, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I.; Alegre, L.; Van Breusegem, F.; Munné-Bosch, S. How Relevant Are Flavonoids as Antioxidants in Plants? Trends Plant. Sci. 2009, 14, 125–132. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The Significance of Reactive Oxygen Species and Antioxidant Defense System in Plants: A Concise Overview. Front. Plant. Sci. 2020, 11, 552969. [Google Scholar] [CrossRef]
- Ivanova, A.; Gerasimova, E.; Gazizullina, E. Study of Antioxidant Properties of Agents from the Perspective of Their Action Mechanisms. Molecules 2020, 25, 4251. [Google Scholar] [CrossRef]
- Gulcin, İ. Antioxidants and Antioxidant Methods: An Updated Overview. Arch. Toxicol. 2020, 94, 651–715. [Google Scholar] [CrossRef] [Green Version]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef]
- Pompella, A.; Sies, H.; Wacker, R.; Brouns, F.; Grune, T.; Biesalski, H.K.; Frank, J. The Use of Total Antioxidant Capacity as Surrogate Marker for Food Quality and its Effect on Health is to be Discouraged. Nutrition 2014, 30, 791–793. [Google Scholar] [CrossRef] [PubMed]
- Soccio, M.; Laus, M.N.; Alfarano, M.; Pastore, D. The Soybean Lipoxygenase-Fluorescein Reaction may be used to Assess Antioxidant Capacity of Phytochemicals and Serum. Anal. Methods 2016, 8, 4354–4362. [Google Scholar] [CrossRef] [Green Version]
- Soccio, M.; Laus, M.N.; Alfarano, M.; Dalfino, G.; Panunzio, M.F.; Pastore, D. Antioxidant/Oxidant Balance as a novel approach to evaluate the effect on serum of long-term intake of plant antioxidant-rich foods. J. Funct. Foods 2018, 40, 778–784. [Google Scholar] [CrossRef]
- Franco, R.; Navarro, G.; Martínez-Pinilla, E. Antioxidants versus Food Antioxidant Additives and Food Preservatives. Antioxidants 2019, 8, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Oxidative Stress in Cell Culture: An under-Appreciated Problem? FEBS Lett. 2003, 540, 3–6. [Google Scholar] [CrossRef] [Green Version]
- Halliwell, B. Cell Culture, Oxidative Stress, and Antioxidants: Avoiding Pitfalls. Biomed. J. 2014, 37, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Woo, H.A.; Kil, I.S.; Bae, S.H. Peroxiredoxin Functions as a Peroxidase and a Regulator and Sensor of Local Peroxides. J. Biol. Chem. 2012, 287, 4403–4410. [Google Scholar] [CrossRef] [Green Version]
- Long, L.H.; Halliwell, B. The Effects of Oxaloacetate on Hydrogen Peroxide Generation from Ascorbate and Epigallocatechin Gallate in Cell Culture Media: Potential for Altering Cell Metabolism. Biochem. Biophys. Res. Commun. 2012, 417, 446–450. [Google Scholar] [CrossRef]
- Liu, R.H.; Finley, J. Potential Cell Culture Models for Antioxidant Research. J. Agric. Food Chem. 2005, 53, 4311–4314. [Google Scholar] [CrossRef] [PubMed]
- Cheli, F.; Baldi, A. Nutrition-Based Health: Cell-Based Bioassays for Food Antioxidant Activity Evaluation. J. Food Sci. 2011, 76, R197–R205. [Google Scholar] [CrossRef] [PubMed]
- Furger, C. Live Cell Assays. From Research to Health and Regulatory Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; ISBN 978-1-84821-858-1. [Google Scholar]
- Allen, D.D.; Caviedes, R.; Cárdenas, A.M.; Shimahara, T.; Segura-Aguilar, J.; Caviedes, P.A. Cell Lines as in Vitro Models for Drug Screening and Toxicity Studies. Drug Dev. Ind. Pharm. 2005, 31, 757–768. [Google Scholar] [CrossRef]
- Goya, L.; Martin, M.; Ramos, S.; Mateos, R.; Bravo, L. A Cell Culture Model for the Assessment of the Chemopreventive Potential of Dietary Compounds. Curr. Nutr. Food Sci. 2009, 5, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Scott, C.W.; Peters, M.F.; Dragan, Y.P. Human Induced Pluripotent Stem Cells and Their Use in Drug Discovery for Toxicity Testing. Toxicol. Lett. 2013, 219, 49–58. [Google Scholar] [CrossRef]
- Fraga, C.G.; Oteiza, P.I.; Galleano, M. In Vitro Measurements and Interpretation of Total Antioxidant Capacity. Biochim. Biophys. Acta 2014, 1840, 931–934. [Google Scholar] [CrossRef]
- López-Alarcón, C.; Denicola, A. Evaluating the Antioxidant Capacity of Natural Products: A Review on Chemical and Cellular-Based Assays. Anal. Chim. Acta 2013, 763, 1–10. [Google Scholar] [CrossRef]
- Amorati, R.; Valgimigli, L. Advantages and Limitations of Common Testing Methods for Antioxidants. Free Radic. Res. 2015, 49, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Honzel, D.; Carter, S.G.; Redman, K.A.; Schauss, A.G.; Endres, J.R.; Jensen, G.S. Comparison of Chemical and Cell-Based Antioxidant Methods for Evaluation of Foods and Natural Products: Generating Multifaceted Data by Parallel Testing Using Erythrocytes and Polymorphonuclear Cells. J. Agric. Food Chem. 2008, 56, 8319–8325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Duanghathaipornsuk, S.; Farrell, E.J.; Alba-Rubio, A.C.; Zelenay, P.; Kim, D.-S. Detection Technologies for Reactive Oxygen Species: Fluorescence and Electrochemical Methods and Their Applications. Biosensors 2021, 11, 30. [Google Scholar] [CrossRef]
- Mello, L.D.; Kisner, A.; Goulart, M.O.F.; Kubota, L.T. Biosensors for Antioxidant Evaluation in Biological Systems. Comb. Chem. High. Throughput Screen. 2013, 16, 109–120. [Google Scholar] [CrossRef]
- Mason, R.P. Imaging Free Radicals in Organelles, Cells, Tissue, and in Vivo with Immuno-Spin Trapping. Redox Biol. 2016, 8, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Andina, D.; Leroux, J.-C.; Luciani, P. Ratiometric Fluorescent Probes for the Detection of Reactive Oxygen Species. Chemistry 2017, 23, 13549–13573. [Google Scholar] [CrossRef]
- Winterbourn, C.C. The Challenges of Using Fluorescent Probes to Detect and Quantify Specific Reactive Oxygen Species in Living Cells. Biochim. Biophys. Acta 2014, 1840, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Debowska, K.; Debski, D.; Hardy, M.; Jakubowska, M.; Kalyanaraman, B.; Marcinek, A.; Michalski, R.; Michalowski, B.; Ouari, O.; Sikora, A.; et al. Toward Selective Detection of Reactive Oxygen and Nitrogen Species with the Use of Fluorogenic Probes--Limitations, Progress, and Perspectives. Pharm. Rep. 2015, 67, 756–764. [Google Scholar] [CrossRef] [Green Version]
- Gough, D.R.; Cotter, T.G. Hydrogen Peroxide: A Jekyll and Hyde Signalling Molecule. Cell Death Dis. 2011, 2, e213. [Google Scholar] [CrossRef] [Green Version]
- Rincheval, V.; Bergeaud, M.; Mathieu, L.; Leroy, J.; Guillaume, A.; Mignotte, B.; Le Floch, N.; Vayssière, J.-L. Differential Effects of Bcl-2 and Caspases on Mitochondrial Permeabilization during Endogenous or Exogenous Reactive Oxygen Species-Induced Cell Death: A Comparative Study of H2O2, Paraquat, t-BHP, Etoposide and TNF-α-Induced Cell Death. Cell Biol. Toxicol. 2012, 28, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Nascimento da Silva, L.C.; Bezerra Filho, C.M.; de Paula, R.A.; Silva e Silva, C.S.; Oliveira de Souza, L.I.; da Silva, M.V.; Correia, M.T.D.S.; de Figueiredo, R.C.B.Q. In Vitro Cell-Based Assays for Evaluation of Antioxidant Potential of Plant-Derived Products. Free Radic. Res. 2016, 50, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; Wang, R.; Luo, X.; Li, Y.; Chen, Z. Protective Effects of Rice Dreg Protein Hydrolysates against Hydrogen Peroxide-Induced Oxidative Stress in HepG-2 Cells. Food Funct. 2016, 7, 1429–1437. [Google Scholar] [CrossRef]
- Fang, S.; Lin, F.; Qu, D.; Liang, X.; Wang, L. Characterization of Purified Red Cabbage Anthocyanins: Improvement in HPLC Separation and Protective Effect against H2O2-Induced Oxidative Stress in HepG2 Cells. Molecules 2018, 24, 124. [Google Scholar] [CrossRef] [Green Version]
- Somanah, J.; Bourdon, E.; Bahorun, T. Extracts of Mauritian Carica Papaya (Var. Solo) Protect SW872 and HepG2 Cells against Hydrogen Peroxide Induced Oxidative Stress. J. Food Sci. Technol. 2017, 54, 1917–1927. [Google Scholar] [CrossRef]
- Singh, S. Nanomaterials Exhibiting Enzyme-Like Properties (Nanozymes): Current Advances and Future Perspectives. Front. Chem. 2019, 7, 46. [Google Scholar] [CrossRef]
- Yarjanli, Z.; Ghaedi, K.; Esmaeili, A.; Zarrabi, A.; Rahgozar, S. The Antitoxic Effects of Quercetin and Quercetin-Conjugated Iron Oxide Nanoparticles (QNPs) against H2O2-Induced Toxicity in PC12 Cells. Int. J. Nanomed. 2019, 14, 6813–6830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, M.O.; Branco, C.S.; Sene, J.; DallAgnol, R.; Agostini, F.; Moura, S.; Salvador, M. Antioxidant and Antigenotoxic Activities of the Brazilian Pine Araucaria Angustifolia (Bert.) O. Kuntze. Antioxidants 2014, 3, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.D.; Acosta, D. Failure of Gentamicin to Elevate Cellular Malondialdehyde Content or Increase Generation of Intracellular Reactive Oxygen Species in Primary Cultures of Renal Cortical Epithelial Cells. Biochem. Pharm. 1990, 40, 1523–1526. [Google Scholar] [CrossRef]
- Rosenkranz, A.R.; Schmaldienst, S.; Stuhlmeier, K.M.; Chen, W.; Knapp, W.; Zlabinger, G.J. A Microplate Assay for the Detection of Oxidative Products Using 2′,7′-Dichlorofluorescin-Diacetate. J. Immunol. Methods 1992, 156, 39–45. [Google Scholar] [CrossRef]
- Wang, H.; Joseph, J.A. Quantifying Cellular Oxidative Stress by Dichlorofluorescein Assay Using Microplate Reader. Free Radic. Biol. Med. 1999, 27, 612–616. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Liu, R.H. Cellular Antioxidant Activity (CAA) Assay for Assessing Antioxidants, Foods, and Dietary Supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.L.; Kang, X.; He, X.; Dong, M.; Zhang, Q.; Liu, R.H. Cellular Antioxidant Activity of Common Fruits. J. Agric. Food Chem. 2008, 56, 8418–8426. [Google Scholar] [CrossRef] [PubMed]
- De la Haba, C.; Palacio, J.R.; Martínez, P.; Morros, A. Effect of Oxidative Stress on Plasma Membrane Fluidity of THP-1 Induced Macrophages. Biochim. Biophys. Acta BBA Biomembr. 2013, 1828, 357–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Yang, J.-H.; Yoon, S.J.; Lee, J.-H.; Yang, E.S.; Park, J.-W. Lipid Peroxidation-Mediated Cytotoxicity and DNA Damage in U937 Cells. Biochimie 2002, 84, 1199–1205. [Google Scholar] [CrossRef]
- Sunitha, D. A Review on Antioxidant Methods. Asian J. Pharm. Clin. Res. 2016, 9, 14–32. [Google Scholar] [CrossRef]
- Kellett, M.E.; Greenspan, P.; Pegg, R.B. Modification of the Cellular Antioxidant Activity (CAA) Assay to Study Phenolic Antioxidants in a Caco-2 Cell Line. Food Chem. 2018, 244, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Swift, L.M.; Sarvazyan, N. Localization of Dichlorofluorescin in Cardiac Myocytes: Implications for Assessment of Oxidative Stress. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H982–H990. [Google Scholar] [CrossRef]
- Afri, M.; Frimer, A.A.; Cohen, Y. Active Oxygen Chemistry within the Liposomal Bilayer. Part IV: Locating 2′,7′-Dichlorofluorescein (DCF), 2′,7′-Dichlorodihydrofluorescein (DCFH) and 2′,7′-Dichlorodihydrofluorescein Diacetate (DCFH-DA) in the Lipid Bilayer. Chem. Phys. Lipids 2004, 131, 123–133. [Google Scholar] [CrossRef]
- González, E.; Vaillant, F.; Rojas, G.; Pérez, A. Novel Semiautomated Method for Assessing in Vitro Cellular Antioxidant Activity Using the Light-Scattering Properties of Human Erythrocytes. J. Agric. Food Chem. 2010, 58, 1455–1461. [Google Scholar] [CrossRef]
- González, E.; Vaillant, F.; Pérez, A. In Vitro Cell-Mediated Antioxidant Protection of Human Erythrocytes by Some Common Tropical Fruits. J. Nutr. Food Sci. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Dobiasová, S.; Řehořová, K.; Kučerová, D.; Biedermann, D.; Káňová, K.; Petrásková, L.; Koucká, K.; Václavíková, R.; Valentová, K.; Ruml, T.; et al. Multidrug Resistance Modulation Activity of Silybin Derivatives and Their Anti-Inflammatory Potential. Antioxidants 2020, 9, 455. [Google Scholar] [CrossRef]
- Gong, E.S.; Liu, C.; Li, B.; Zhou, W.; Chen, H.; Li, T.; Wu, J.; Zeng, Z.; Wang, Y.; Si, X.; et al. Phytochemical Profiles of Rice and Their Cellular Antioxidant Activity against ABAP Induced Oxidative Stress in Human Hepatocellular Carcinoma HepG2 Cells. Food Chem. 2020, 318, 126484. [Google Scholar] [CrossRef]
- Nizioł-Łukaszewska, Z.; Furman-Toczek, D.; Zagórska-Dziok, M. Antioxidant Activity and Cytotoxicity of Jerusalem Artichoke Tubers and Leaves Extract on HaCaT and BJ Fibroblast Cells. Lipids Health Dis. 2018, 17, 280. [Google Scholar] [CrossRef] [Green Version]
- Royall, J.A.; Ischiropoulos, H. Evaluation of 2′,7′-Dichlorofluorescin and Dihydrorhodamine 123 as Fluorescent Probes for Intracellular H2O2 in Cultured Endothelial Cells. Arch. Biochem. Biophys. 1993, 302, 348–355. [Google Scholar] [CrossRef]
- Yazdani, M. Concerns in the Application of Fluorescent Probes DCDHF-DA, DHR 123 and DHE to Measure Reactive Oxygen Species in Vitro. Toxicol. Vitr. 2015, 30, 578–582. [Google Scholar] [CrossRef]
- Zhang, D.; Xie, L.; Wei, Y.; Liu, Y.; Jia, G.; Zhou, F.; Ji, B. Development of a Cell-Based Antioxidant Activity Assay Using Dietary Fatty Acid as Oxidative Stressor. Food Chem. 2013, 141, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, Z.; Xu, Z.; Chen, L.; Wang, Y. 2′,7′-Dichlorodihydrofluorescein as a Fluorescent Probe for Reactive Oxygen Species Measurement: Forty Years of Application and Controversy. Free Radic. Res. 2010, 44, 587–604. [Google Scholar] [CrossRef] [PubMed]
- Quintanar-Escorza, M.A.; González-Martínez, M.T.; del Pilar, I.-O.M.; Calderón-Salinas, J.V. Oxidative Damage Increases Intracellular Free Calcium [Ca2+]i Concentration in Human Erythrocytes Incubated with Lead. Toxicol. In Vitro 2010, 24, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gao, G.; Zhang, S.; Wang, H.; Ke, L.; Zhou, J.; Rao, P.; Wang, Q.; Li, J. Influences of Calcium and Magnesium Ions on Cellular Antioxidant Activity (CAA) Determination. Food Chem. 2020, 320, 126625. [Google Scholar] [CrossRef]
- Wojtovich, A.P.; Foster, T.H. Optogenetic Control of ROS Production. Redox Biol. 2014, 2, 368–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulina, M.E.; Chudakov, D.M.; Britanova, O.V.; Yanushevich, Y.G.; Staroverov, D.B.; Chepurnykh, T.V.; Merzlyak, E.M.; Shkrob, M.A.; Lukyanov, S.; Lukyanov, K.A. A Genetically Encoded Photosensitizer. Nat. Biotechnol. 2006, 24, 95–99. [Google Scholar] [CrossRef]
- Paardekooper, L.M.; van Vroonhoven, E.; Ter Beest, M.; van den Bogaart, G. Radical Stress Is More Cytotoxic in the Nucleus than in Other Organelles. Int. J. Mol. Sci. 2019, 20, 4147. [Google Scholar] [CrossRef] [Green Version]
- Vitriol, E.A.; Uetrecht, A.C.; Shen, F.; Jacobson, K.; Bear, J.E. Enhanced EGFP-Chromophore-Assisted Laser Inactivation Using Deficient Cells Rescued with Functional EGFP-Fusion Proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 6702–6707. [Google Scholar] [CrossRef] [Green Version]
- Trewin, A.J.; Berry, B.J.; Wei, A.Y.; Bahr, L.L.; Foster, T.H.; Wojtovich, A.P. Light-Induced Oxidant Production by Fluorescent Proteins. Free Radic. Biol. Med. 2018, 128, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, K.; Matsuda, T.; Sakai, N.; Fu, D.; Noda, M.; Uchiyama, S.; Kotera, I.; Arai, Y.; Horiuchi, M.; Fukui, K.; et al. SuperNova, a Monomeric Photosensitizing Fluorescent Protein for Chromophore-Assisted Light Inactivation. Sci. Rep. 2013, 3, 2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, A.E.; Gallagher, W.M.; Byrne, A.T. Porphyrin and Nonporphyrin Photosensitizers in Oncology: Preclinical and Clinical Advances in Photodynamic Therapy. Photochem. Photobiol. 2009, 85, 1053–1074. [Google Scholar] [CrossRef] [PubMed]
- Agostinis, P.; Berg, K.; Cengel, K.A.; Foster, T.H.; Girotti, A.W.; Gollnick, S.O.; Hahn, S.M.; Hamblin, M.R.; Juzeniene, A.; Kessel, D.; et al. Photodynamic Therapy of Cancer: An Update. CA Cancer J. Clin. 2011, 61, 250–281. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.K. Photodynamic Therapy: Current Role in the Treatment of Chorioretinal Conditions. Eye 2016, 30, 202–210. [Google Scholar] [CrossRef] [Green Version]
- Kessel, D. Photodynamic Therapy: A Brief History. J. Clin. Med. 2019, 8, 1581. [Google Scholar] [CrossRef] [Green Version]
- Lubitz, I.; Zikich, D.; Kotlyar, A. Specific High-Affinity Binding of Thiazole Orange to Triplex and G-Quadruplex DNA. Biochemistry 2010, 49, 3567–3574. [Google Scholar] [CrossRef]
- Karunakaran, V.; Pérez Lustres, J.L.; Zhao, L.; Ernsting, N.P.; Seitz, O. Large Dynamic Stokes Shift of DNA Intercalation Dye Thiazole Orange Has Contribution from a High-Frequency Mode. J. Am. Chem. Soc. 2006, 128, 2954–2962. [Google Scholar] [CrossRef] [PubMed]
- Nygren, J.; Svanvik, N.; Kubista, M. The Interactions between the Fluorescent Dye Thiazole Orange and DNA. Biopolymers 1998, 46, 39–51. [Google Scholar] [CrossRef]
- Armitage, B.A. Cyanine Dye–Nucleic Acid Interactions. In Heterocyclic Polymethine Dyes; Strekowski, L., Ed.; Topics in Heterocyclic Chemistry; Springer: Berlin/Heidelberg, Germany, 2008; Volume 14, pp. 11–29. ISBN 978-3-540-79063-1. [Google Scholar]
- Derick, S.; Gironde, C.; Perio, P.; Reybier, K.; Nepveu, F.; Jauneau, A.; Furger, C. LUCS (Light-Up Cell System), a Universal High Throughput Assay for Homeostasis Evaluation in Live Cells. Sci. Rep. 2017, 7, 18069. [Google Scholar] [CrossRef] [PubMed]
- Gironde, C.; Rigal, M.; Dufour, C.; Furger, C. AOP1, a New Live Cell Assay for the Direct and Quantitative Measure of Intracellular Antioxidant Effects. Antioxidants 2020, 9, 471. [Google Scholar] [CrossRef] [PubMed]
- Vigliante, I.; Mannino, G.; Maffei, M.E. OxiCyan®, a Phytocomplex of Bilberry (Vaccinium Myrtillus) and Spirulina (Spirulina Platensis), Exerts Both Direct Antioxidant Activity and Modulation of ARE/Nrf2 Pathway in HepG2 Cells. J. Funct. Foods 2019, 61, 103508. [Google Scholar] [CrossRef]
- Bugaj, L.J.; Lim, W.A. High-Throughput Multicolor Optogenetics in Microwell Plates. Nat. Protoc. 2019, 14, 2205–2228. [Google Scholar] [CrossRef]
- Thomas, O.S.; Hörner, M.; Weber, W. A Graphical User Interface to Design High-Throughput Optogenetic Experiments with the OptoPlate-96. Nat. Protoc. 2020, 15, 2785–2787. [Google Scholar] [CrossRef]
- Niture, S.K.; Khatri, R.; Jaiswal, A.K. Regulation of Nrf2-an Update. Free Radic. Biol. Med. 2014, 66, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Zhang, D.D.; Lo, S.-C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [Green Version]
- Levonen, A.-L.; Landar, A.; Ramachandran, A.; Ceaser, E.K.; Dickinson, D.A.; Zanoni, G.; Morrow, J.D.; Darley-Usmar, V.M. Cellular Mechanisms of Redox Cell Signalling: Role of Cysteine Modification in Controlling Antioxidant Defences in Response to Electrophilic Lipid Oxidation Products. Biochem. J. 2004, 378, 373–382. [Google Scholar] [CrossRef] [Green Version]
- Unoki, T.; Akiyama, M.; Kumagai, Y. Nrf2 Activation and Its Coordination with the Protective Defense Systems in Response to Electrophilic Stress. Int. J. Mol. Sci. 2020, 21, 545. [Google Scholar] [CrossRef] [Green Version]
- Baird, L.; Llères, D.; Swift, S.; Dinkova-Kostova, A.T. Regulatory Flexibility in the Nrf2-Mediated Stress Response Is Conferred by Conformational Cycling of the Keap1-Nrf2 Protein Complex. Proc. Natl. Acad. Sci. USA 2013, 110, 15259–15264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. P62 Links Autophagy and Nrf2 Signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Lv, Y.-F.; Zhao, J.-L.; You, Q.-D.; Jiang, Z.-Y. Regulation of Nrf2 by Phosphorylation: Consequences for Biological Function and Therapeutic Implications. Free Radic. Biol. Med. 2021, 168, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Chattopadhyay, A. Nrf2–ARE Signaling in Cellular Protection: Mechanism of Action and the Regulatory Mechanisms. J. Cell. Physiol. 2020, 235, 3119–3130. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant Response Elements: Discovery, Classes, Regulation and Potential Applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Motahari, P.; Sadeghizadeh, M.; Behmanesh, M.; Sabri, S.; Zolghadr, F. Generation of Stable ARE-Driven Reporter System for Monitoring Oxidative Stress. J. Pharm. Sci. 2015, 23, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, S.; Hou, D.-X. Multiple Regulations of Keap1/Nrf2 System by Dietary Phytochemicals. Mol. Nutr. Food Res. 2016, 60, 1731–1755. [Google Scholar] [CrossRef]
- Furue, M.; Uchi, H.; Mitoma, C.; Hashimoto-Hachiya, A.; Chiba, T.; Ito, T.; Nakahara, T.; Tsuji, G. Antioxidants for Healthy Skin: The Emerging Role of Aryl Hydrocarbon Receptors and Nuclear Factor-Erythroid 2-Related Factor-2. Nutrients 2017, 9, 223. [Google Scholar] [CrossRef]
- Balstad, T.R.; Carlsen, H.; Myhrstad, M.C.W.; Kolberg, M.; Reiersen, H.; Gilen, L.; Ebihara, K.; Paur, I.; Blomhoff, R. Coffee, Broccoli and Spices Are Strong Inducers of Electrophile Response Element-Dependent Transcription in Vitro and in Vivo—Studies in Electrophile Response Element Transgenic Mice. Mol. Nutr. Food Res. 2011, 55, 185–197. [Google Scholar] [CrossRef]
- Kolb, H.; Kempf, K.; Martin, S. Health Effects of Coffee: Mechanism Unraveled? Nutrients 2020, 12, 1842. [Google Scholar] [CrossRef]
- Roy, A.; McDonald, P.; Timmermann, B.N.; Gupta, M.; Chaguturu, R. Bioactivity Profiling of Plant Biodiversity of Panama by High Throughput Screening. Nat. Prod. Commun. 2019, 14, 71–74. [Google Scholar] [CrossRef]
- Zielonka, J.; Zielonka, M.; VerPlank, L.; Cheng, G.; Hardy, M.; Ouari, O.; Ayhan, M.M.; Podsiadły, R.; Sikora, A.; Lambeth, J.D.; et al. Mitigation of NADPH Oxidase 2 Activity as a Strategy to Inhibit Peroxynitrite Formation. J. Biol. Chem. 2016, 291, 7029–7044. [Google Scholar] [CrossRef] [Green Version]

| CAA Assay | AOP1 Assay |
|---|---|
| Based on the production of AAPH-induced peroxyl radicals | Based on the controlled production of 1O2 and free radicals by photoinduction |
| Measures effects of plasma membrane-based antioxidants | Measures effects of intracellular-based antioxidants |
| No control of ROS production | Easy control of ROS production by light intensity; allows monitoring ROS production at a sublethal level (i.e., more physiological concentrations) |
| Interpretation limited to AAPH effects | |
| Does not differentiate between antioxidant and cytotoxic effects | Can easily discriminate between antioxidant and cytotoxic effects |
| Results need to be confirmed by performing a cytotoxicity assay (e.g., MTT) | No other assay needed |
| DCFH-DA subject to auto-oxidation | Sensor not directly involved in the oxidation process |
| Subject to cell leakage | No cell leakage |
| Fluorescence levels vary according to cell density | No effect of cell density (measure on a ratio mode) |
| Needs culture medium washes that disrupt cell culture | No washes required |
| Difficult to standardize | Easy to standardize |
| Detection by fluorescence readers | Detection by fluorescence readers + illuminator |
| Limited to adherent cells | Works for adherent and suspension cells, and organotypic models |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furger, C. Live Cell Assays for the Assessment of Antioxidant Activities of Plant Extracts. Antioxidants 2021, 10, 944. https://doi.org/10.3390/antiox10060944
Furger C. Live Cell Assays for the Assessment of Antioxidant Activities of Plant Extracts. Antioxidants. 2021; 10(6):944. https://doi.org/10.3390/antiox10060944
Chicago/Turabian StyleFurger, Christophe. 2021. "Live Cell Assays for the Assessment of Antioxidant Activities of Plant Extracts" Antioxidants 10, no. 6: 944. https://doi.org/10.3390/antiox10060944






