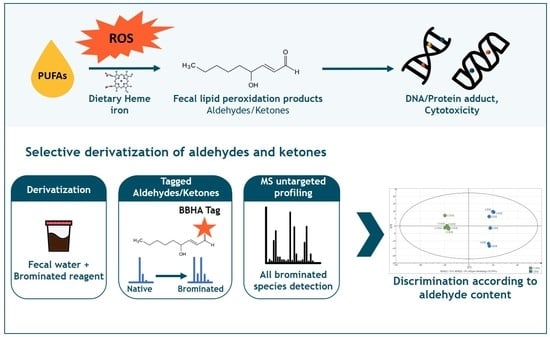
Towards Aldehydomics: Untargeted Trapping and Analysis of Reactive Diet-Related Carbonyl Compounds Formed in the Intestinal Lumen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Animal Experiment and Fecal Water Preparation
2.3. Sample Treatment
2.4. Liquid Chromatography–Mass Spectrometry
2.5. Data Processing and Statistical Treatment
3. Results and Discussion
3.1. Set-Up of the Derivatization Method
3.2. MS/MS Fragmentation of Aldehyde BBHA Derivatives
3.3. Profiling of Diet-Related Peroxidation Carbonyl Compounds Formed in the Intestinal Lumen
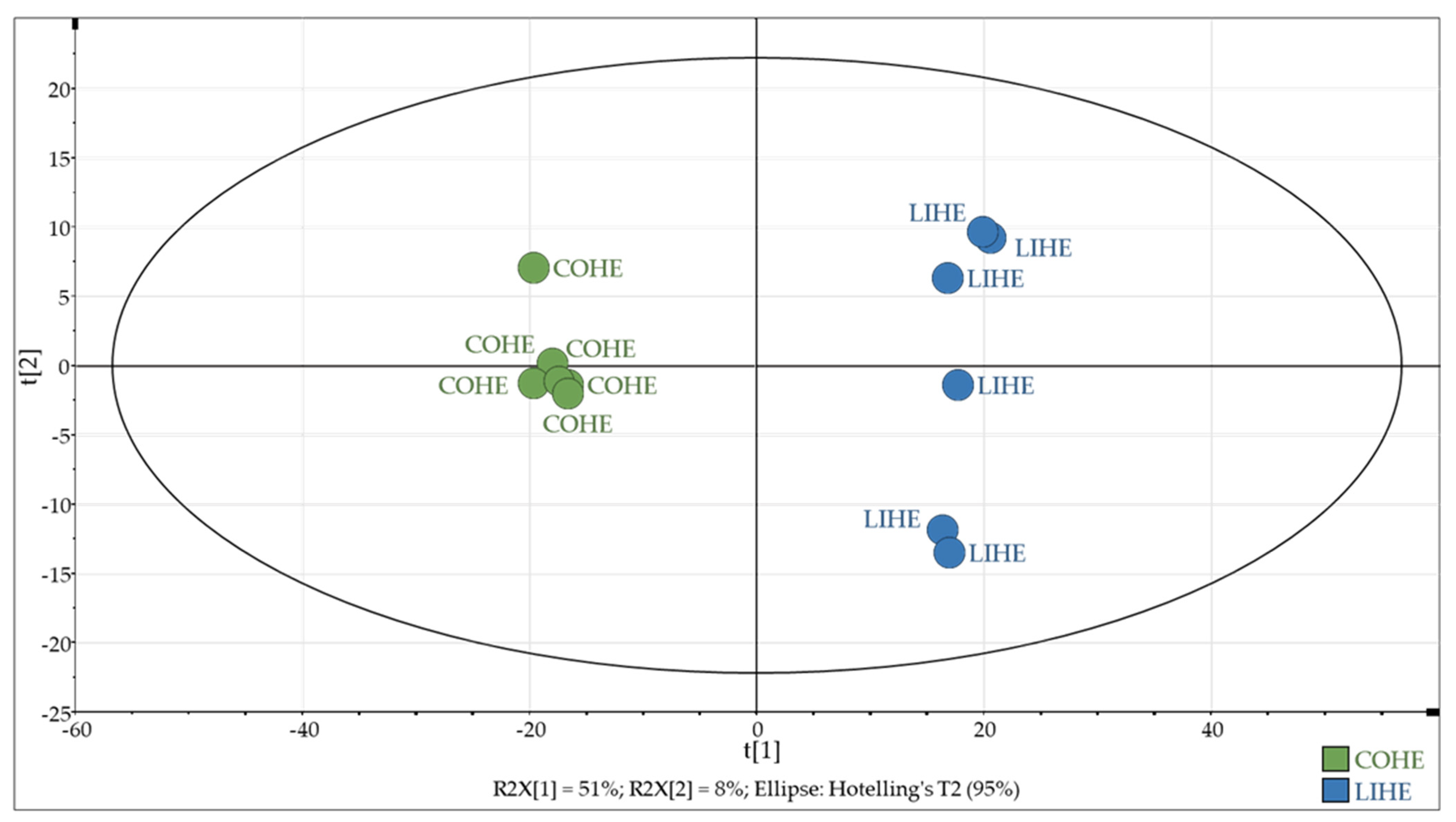
3.4. Untargeted “Aldehydomics” Approach: Statistical Data Analysis and Identification of Aldehydes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bastide, N.M.; Pierre, F.H.F.; Corpet, D.E. Heme iron from meat and risk of colorectal cancer: A meta-analysis and a review of the mechanisms involved. Cancer Prev. Res. 2011, 4, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Vieira, A.R.; Abar, L.; Chan, D.S.M.; Vingeliene, S.; Polemiti, E.; Stevens, C.; Greenwood, D.; Norat, T. Foods and beverages and colorectal cancer risk: A systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR Continuous Update Project. Ann. Oncol. 2017, 28, 1788–1802. [Google Scholar] [CrossRef]
- Lippi, G.; Mattiuzzi, C.; Cervellin, G. Meat consumption and cancer risk: A critical review of published meta-analyses. Crit. Rev. Oncol. Hematol. 2016, 97, 1–14. [Google Scholar] [CrossRef]
- WCRF World Cancer Research Fund/American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of Cancer: A Global Perspective; AICR: Washington, DC, USA, 2007. [Google Scholar]
- WCRF World Cancer Research Fund/American Institute for Cancer Research. Diet, Nutrition, Physical Activity and Cancer: A Global Perspective Continuous Update Project Expert Report; AICR: Washington, DC, USA, 2018. [Google Scholar]
- Bastide, N.M.; Chenni, F.; Audebert, M.; Santarelli, R.L.; Tache, S.; Naud, N.; Baradat, M.; Jouanin, I.; Surya, R.; Hobbs, D.A.; et al. A central role for heme iron in colon carcinogenesis associated with red meat intake. Cancer Res. 2015, 75, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Sesink, A.L.A.; Termont, D.S.M.L.; Kleibeuker, J.H.; Van der Meer, R. Red meat and colon cancer: The cytotoxic and hyperproliferative effects of dietary heme meat and colon cancer. Cancer Res 1999, 59, 5704–5709. [Google Scholar]
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef]
- Guéraud, F.; Atalay, M.; Bresgen, N.; Cipak, A.; Eckl, P.M.; Huc, L.; Jouanin, I.; Siems, W.; Uchida, K. Chemistry and biochemistry of lipid peroxidation products. Free Radic. Res. 2010, 44, 1098–1124. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.; Chevolleau, S.; Noguer-Meireles, M.-H.; Pujos-Guillot, E.; Delosière, M.; Chantelauze, C.; Joly, C.; Blas-y-Estrada, F.; Jouanin, I.; Durand, D.; et al. Heme-iron-induced production of 4-hydroxynonenal in intestinal lumen may have extra-intestinal consequences through protein-adduct formation. Antioxidants 2020, 9, 1293. [Google Scholar] [CrossRef]
- Baradat, M.; Jouanin, I.; Dalleau, S.; Taché, S.; Gieules, M.; Debrauwer, L.; Canlet, C.; Huc, L.; Dupuy, J.; Pierre, F.H.F.; et al. 4-Hydroxy-2(E)-nonenal metabolism differs in apc+/+ cells and in apcmin/+ cells: It may explain colon cancer promotion by heme iron. Chem. Res. Toxicol. 2011, 24, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Takeda, S.; Terao, J. Lipidomic analysis for lipid peroxidation-derived aldehydes using gas chromatography-mass spectrometry. Chem. Res. Toxicol. 2007, 20, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, S.; Inaba, Y.; Kunugita, N. Derivatization of carbonyl compounds with 2,4-dinitrophenylhydrazine and their subsequent determination by high-performance liquid chromatography. J. Chromatogr. B 2011, 879, 1282–1289. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Carini, M.; Orioli, M.; Vistoli, G.; Regazzoni, L.; Colombo, G.; Rossi, R.; Milzani, A.; Aldini, G. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic. Biol. Med. 2009, 46, 1411–1419. [Google Scholar] [CrossRef]
- Douny, C.; Bayram, P.; Brose, F.; Degand, G.; Scippo, M.-L. Development of an LC-MS/MS Analytical Method for the simultaneous measurement of aldehydes from polyunsaturated fatty acids degradation in animal feed. Drug Test. Anal. 2016, 8, 458–464. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.-N.; Chen, D.; Zhang, T.-Y.; Ding, J.; Feng, Y.-Q. Use of ammonium sulfite as a post-column derivatization reagent for rapid detection and quantification of aldehydes by LC-MS. Talanta 2020, 206, 120172. [Google Scholar] [CrossRef]
- Cao, Y.; Guan, Q.; Sun, T.; Qi, W.; Guo, Y. Charged tag founded in n-(1-chloroalkyl)pyridinium quaternization for quantification of fatty aldehydes. Anal. Chim. Acta 2016, 937, 80–86. [Google Scholar] [CrossRef]
- Guo, N.; Peng, C.-Y.; Zhu, Q.-F.; Yuan, B.-F.; Feng, Y.-Q. Profiling of carbonyl compounds in serum by stable isotope labeling-double precursor ion scan-mass spectrometry analysis. Anal. Chim. Acta 2017, 967, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zou, X.-G.; Deng, L.; Fan, Y.-W.; Li, H.; Li, J.; Deng, Z.-Y. Analysis of nonpolar lipophilic aldehydes/ketones in oxidized edible oils using HPLC-QqQ-MS for the evaluation of their parent fatty acids. Food Res. Int. 2014, 64, 901–907. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Singer, H.P.; Slobodnik, J.; Ipolyi, I.M.; Oswald, P.; Krauss, M.; Schulze, T.; Haglund, P.; Letzel, T.; Grosse, S.; et al. Non-target screening with high-resolution mass spectrometry: Critical review using a collaborative trial on water analysis. Anal. Bioanal. Chem. 2015, 407, 6237–6255. [Google Scholar] [CrossRef] [PubMed]
- Jamin, E.L.; Costantino, R.; Mervant, L.; Martin, J.-F.; Jouanin, I.; Blas-Y-Estrada, F.; Guéraud, F.; Debrauwer, L. Global profiling of toxicologically relevant metabolites in urine: Case study of reactive aldehydes. Anal. Chem. 2020, 92, 1746–1754. [Google Scholar] [CrossRef]
- Pourchet, M.; Debrauwer, L.; Oberacher, M.; Krauss, M.; Vlaanderen, A.; Covaci, A.; Damont, A.; Klanova, J.; Lamoree, M.; Sarigiannis, D.; et al. Suspect and non-targeted screening of chemicals of emerging concern for human biomonitoring and environmental health studies: Current capabilities, promises, and challenges. Environ. Int. 2020, 139, 105545. [Google Scholar] [CrossRef] [PubMed]
- Eggink, M.; Wijtmans, M.; Ekkebus, R.; Lingeman, H.; de Esch, I.J.P.; Kool, J.; Niessen, W.M.A.; Irth, H. Development of a selective esi-ms derivatization reagent: Synthesis and optimization for the analysis of aldehydes in biological mixtures. Anal. Chem. 2008, 80, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Eggink, M.; Charret, S.; Wijtmans, M.; Lingeman, H.; Kool, J.; Niessen, W.M.A.; Irth, H. Development of an on-line weak-cation exchange liquid chromatography–tandem mass spectrometric method for screening aldehyde products in biological matrices. J. Chromatogr. B 2009, 877, 3937–3945. [Google Scholar] [CrossRef] [PubMed]
- Eggink, M.; Wijtmans, M.; Kretschmer, A.; Kool, J.; Lingeman, H.; de Esch, I.J.P.; Niessen, W.M.A.; Irth, H. Targeted LC–MS derivatization for aldehydes and carboxylic acids with a new derivatization agent 4-APEBA. Anal. Bioanal. Chem. 2010, 397, 665–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Liu, P.; Wang, Y.-L.; Yu, Q.-W.; Yuan, B.-F.; Feng, Y.-Q. Profiling of aldehyde-containing compounds by stable isotope labelling-assisted mass spectrometry analysis. Analyst 2015, 140, 5276–5286. [Google Scholar] [CrossRef]
- Tie, C.; Hu, T.; Jia, Z.-X.; Zhang, J.-L. Derivatization strategy for the comprehensive characterization of endogenous fatty aldehydes using hplc-multiple reaction monitoring. Anal. Chem. 2016, 88, 7762–7768. [Google Scholar] [CrossRef]
- El-Maghrabey, M.; Kishikawa, N.; Kuroda, N. Novel isotope-coded derivatization method for aldehydes using 14N/15N-ammonium acetate and 9,10-phenanthrenequinone. Anal. Chem. 2018, 90, 13867–13875. [Google Scholar] [CrossRef]
- Cuyckens, F.; Balcaen, L.I.L.; De Wolf, K.; De Samber, B.; Van Looveren, C.; Hurkmans, R.; Vanhaecke, F. Use of the bromine isotope ratio in HPLC-ICP-MS and HPLC-ESI-MS analysis of a new drug in development. Anal. Bioanal. Chem. 2008, 390, 1717–1729. [Google Scholar] [CrossRef] [PubMed]
- Bierła, K.; Riu, A.; Debrauwer, L.; Zalko, D.; Bouyssiere, B.; Szpunar, J. Screening for polybrominated diphenyl ethers in biological samples by reversed-phase fast HPLC-ICP MS. J. Anal. At. Spectrom. 2010, 25, 889. [Google Scholar] [CrossRef]
- Chevolleau, S.; Noguer-Meireles, M.-H.; Jouanin, I.; Naud, N.; Pierre, F.; Gueraud, F.; Debrauwer, L. Development and validation of an ultra high performance liquid chromatography-electrospray tandem mass spectrometry method using selective derivatisation, for the quantification of two reactive aldehydes produced by lipid peroxidation, HNE (4-Hydroxy-2( E )-Nonenal) and HHE (4-Hydroxy-2( E )-Hexenal) in faecal water. J. Chromatogr. B 2018, 1083, 171–179. [Google Scholar] [CrossRef]
- Chandra, A.; Srivastava, S.K. A Synthesis of 4-hydroxy-2-trans-nonenal and 4-(3h) 4-hydroxy-2-trans-nonenal. Lipids 1997, 32, 779–782. [Google Scholar] [CrossRef]
- Jouanin, I.; Chevolleau, S.; Canlet, C.; Lorber, C.; Pierre, F.; Guéraud, F.; Debrauwer, L. facile oxime ether synthesis: Free carbonyl compound derivatization by a brominated o -benzylhydroxylamine. Synth. Commun. 2015, 45, 1585–1591. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Böttcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, A.K.; Matthews, S.B.; Vassel, N.; Cox, C.D.; Naseem, R.; Chaichi, J.; Holland, I.B.; Green, J.; Wann, K.T. Bacterial metabolic ‘toxins’: A new mechanism for lactose and food intolerance, and irritable bowel syndrome. Toxicology 2010, 278, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Spickett, C.M. The lipid peroxidation product 4-hydroxy-2-nonenal: Advances in chemistry and analysis. Redox Biol. 2013, 1, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Guéraud, F.; Taché, S.; Steghens, J.-P.; Milkovic, L.; Borovic-Sunjic, S.; Zarkovic, N.; Gaultier, E.; Naud, N.; Héliès-Toussaint, C.; Pierre, F.; et al. Dietary polyunsaturated fatty acids and heme iron induce oxidative stress biomarkers and a cancer promoting environment in the colon of rats. Free Radic. Biol. Med. 2015, 83, 192–200. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical analysis working group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Gabbs, M.; Leng, S.; Devassy, J.G.; Monirujjaman, M.; Aukema, H.M. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv. Nutr. 2015, 6, 513–540. [Google Scholar] [CrossRef] [Green Version]


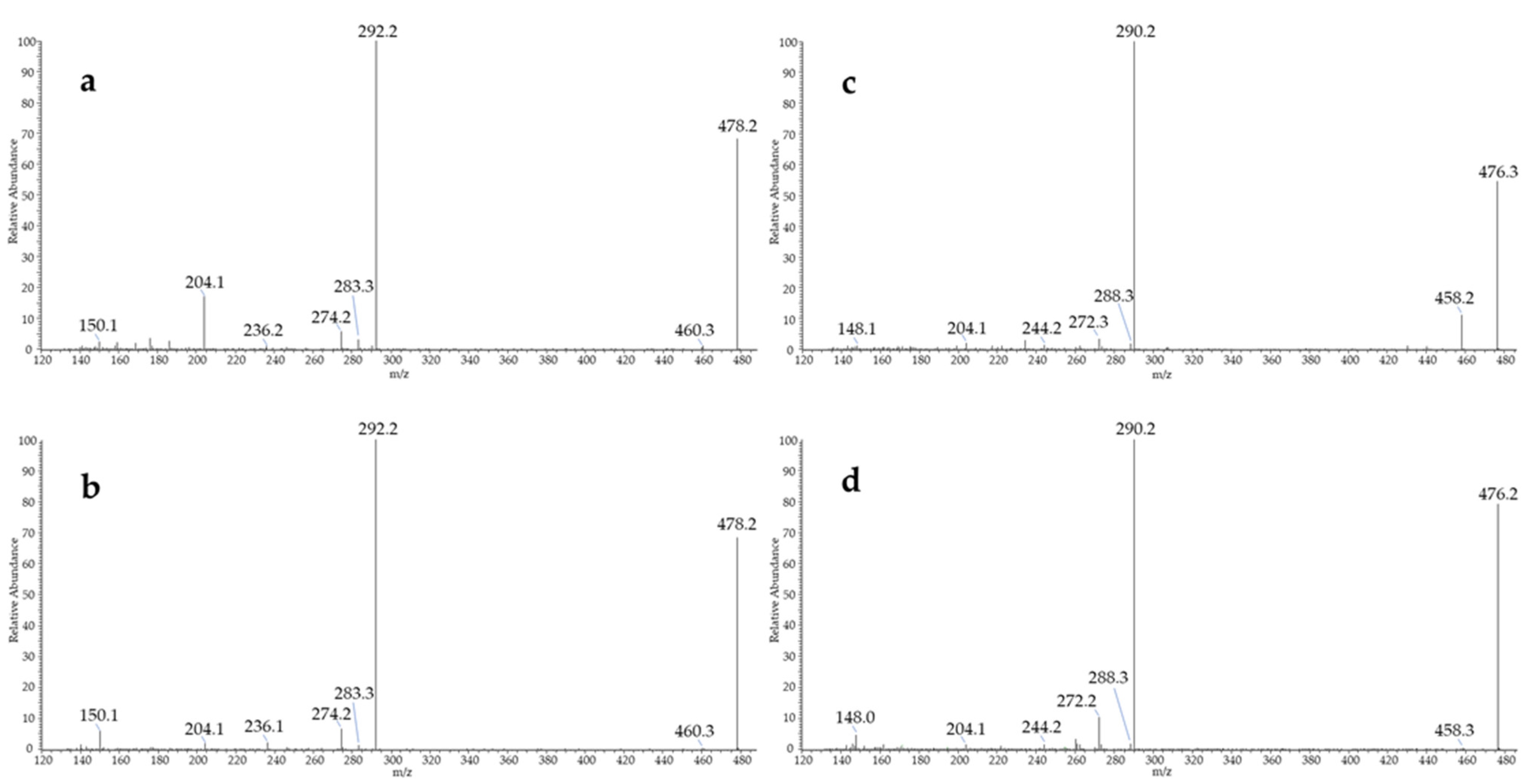
| Compound | Abbreviation | Chemical Formula BBHA Derivative | Molecular Mass | Calculated Exact Mass (79Br/81Br) or (79Br79Br/79Br81Br/81Br81Br) [M+H]+ | Main Observed Fragment Ions (m/z) from MS/MS Carried Out on the 79Br Isotopomer | Retention Time (min.) |
|---|---|---|---|---|---|---|
| 4-OH-hexenal | HHE | C13H16O2NBr | 297/299 | 298.04372/300.04167 | 168.9645; 224.0069 | 15.6; 15.7 |
| 4-Oxo-hexenal | OHE | C13H14O2NBr | 295/297 | 296.02807/298.02602 | 168.9645; 240.0020; 268.0329; 221.9912 | 16.8 |
| 4-OH-nonenal | HNE | C16H22O2NBr | 339/341 | 340.09067/342.08862 | 168.9645; 224.0069 | 17.6; 17.8 |
| 4-Oxo-nonenal | ONE | C16H20O2NBr | 337/339 | 338.07502/340.07297 | 168.9646; 240.0021; 221.9914; 310.0803 | 18.5 |
| 4,5-Epoxy-2-decenal | EDE | C17H22O2NBr | 351/353 | 352.09067/354.08862 | 168.9645; 254.0174; 201.9860 | 17.8 |
| 2,4-Decadienal | DDE | C17H22ONBr | 335/337 | 336.09575/338.09371 | 168.9649; 150.1279; 248.0072; 256,1698 | 20.3 |
| 4-Hydroperoxynonenal | HPNE | C16H22O3NBr | 355/357 | 356.08558/358.08354 | 240.0022; 168.9648; 170.1176; 322.0807 | 17.7; 17.9 |
| 4-OH-nonanal | OHN | C16H24O2NBr | 341/343 | 342.10632/344.10427 | 324.0960; 168.9642; 141.1273; 254,0175 | 17.6 |
| Malondialdehyde | MDA | C17H16O2N2Br2 1 | 438/440/442 | 438.96513/440.96308/442.96104 | 168.9645; 254.0048; 237.0021 | 18.7 |
| Pyruvaldehyde | PRA | C17H16O2N2Br2 1 | 438/440/442 | 438.96513/440.96308/442.96104 | 168.9650; 235.9948; 336.9224 | 19.8 |
| 5-Oxo-pentanoic acid | 5-OPA | C12H14O3NBr | 299/301 | 300.02298/302.02094 | 168.9649; 282.0128; 96.0044 | 14.5 |
| 6-Oxo-hexanoic acid | 6-OHA | C13H16O3NBr | 313/315 | 314.03863/316.03659 | 168.9649; 296.0286; 183.9758; 110.0601 | 15.0 |
| 7-Oxo-heptanoic acid | 7-OHA | C14H18O3NBr | 327/329 | 328.05428/330.05224 | 168.9648; 310.0441; 183.9761; 124.0755 | 15.5 |
| 8-Oxo-octanoic acid | 8-OOA | C15H20O3NBr | 341/343 | 342.06993/344.06789 | 168.9649; 324.0599; 183.9758; 138.0914 | 16.1 |
| 9-Oxo-nonanoic acid | 9-ONA | C16H22O3NBr | 355/357 | 356.08558/358.08354 | 168.9646; 338.0751; 183.9756; 152.1070 | 16.7 |
| 10-Oxo-decanoic acid | 10-ODA | C17H24O3NBr | 369/371 | 370.10123/372.09919 | 168.9648; 352.0910; 183.9758; 166.1228 | 17.2 |
| Compound | m/z [M+H]+ | Rt (min) | Identification Level | FoldLIHE/COHE |
|---|---|---|---|---|
| Malondialdehyde (MDA) | 438.96519 | 18.60 | 1 | 202.27 |
| 4-Hydroxy-hex-2-enal (HHE) | 298.04369 | 15.68 | 1 | 11.46 |
| 9-Oxo-octadecadienoic acid (9-oxoODE) | 478.19490 | 20.51 | 1 | 18.49 |
| 9-Oxo-octadecatrienoic acid (9-oxoOTrE) | 476.17916 | 19.93 | 1 | 78.39 |
| 4-Hydroxy-non-2-enal (HNE) | 340.09047 | 17.65 | 1 | 3.27 |
| α-Linolenic acid | 462.20024 | 19.90 | 1 | 371.22 |
| Eicosapentaenoic acid | 486.19979 | 19.84 | 1 | 59.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chevolleau, S.; Noguer-Meireles, M.-H.; Mervant, L.; Martin, J.-F.; Jouanin, I.; Pierre, F.; Naud, N.; Guéraud, F.; Debrauwer, L. Towards Aldehydomics: Untargeted Trapping and Analysis of Reactive Diet-Related Carbonyl Compounds Formed in the Intestinal Lumen. Antioxidants 2021, 10, 1261. https://doi.org/10.3390/antiox10081261
Chevolleau S, Noguer-Meireles M-H, Mervant L, Martin J-F, Jouanin I, Pierre F, Naud N, Guéraud F, Debrauwer L. Towards Aldehydomics: Untargeted Trapping and Analysis of Reactive Diet-Related Carbonyl Compounds Formed in the Intestinal Lumen. Antioxidants. 2021; 10(8):1261. https://doi.org/10.3390/antiox10081261
Chicago/Turabian StyleChevolleau, Sylvie, Maria-Helena Noguer-Meireles, Loïc Mervant, Jean-François Martin, Isabelle Jouanin, Fabrice Pierre, Nathalie Naud, Françoise Guéraud, and Laurent Debrauwer. 2021. "Towards Aldehydomics: Untargeted Trapping and Analysis of Reactive Diet-Related Carbonyl Compounds Formed in the Intestinal Lumen" Antioxidants 10, no. 8: 1261. https://doi.org/10.3390/antiox10081261
APA StyleChevolleau, S., Noguer-Meireles, M.-H., Mervant, L., Martin, J.-F., Jouanin, I., Pierre, F., Naud, N., Guéraud, F., & Debrauwer, L. (2021). Towards Aldehydomics: Untargeted Trapping and Analysis of Reactive Diet-Related Carbonyl Compounds Formed in the Intestinal Lumen. Antioxidants, 10(8), 1261. https://doi.org/10.3390/antiox10081261