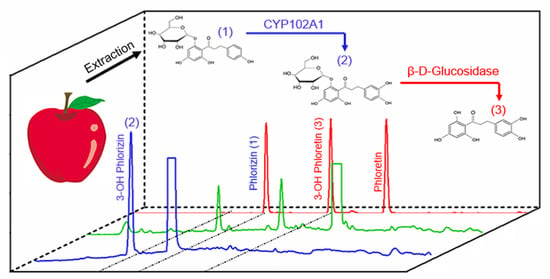
Enzymatic Production of 3-OH Phlorizin, a Possible Bioactive Polyphenol from Apples, by Bacillus megaterium CYP102A1 via Regioselective Hydroxylation
Abstract
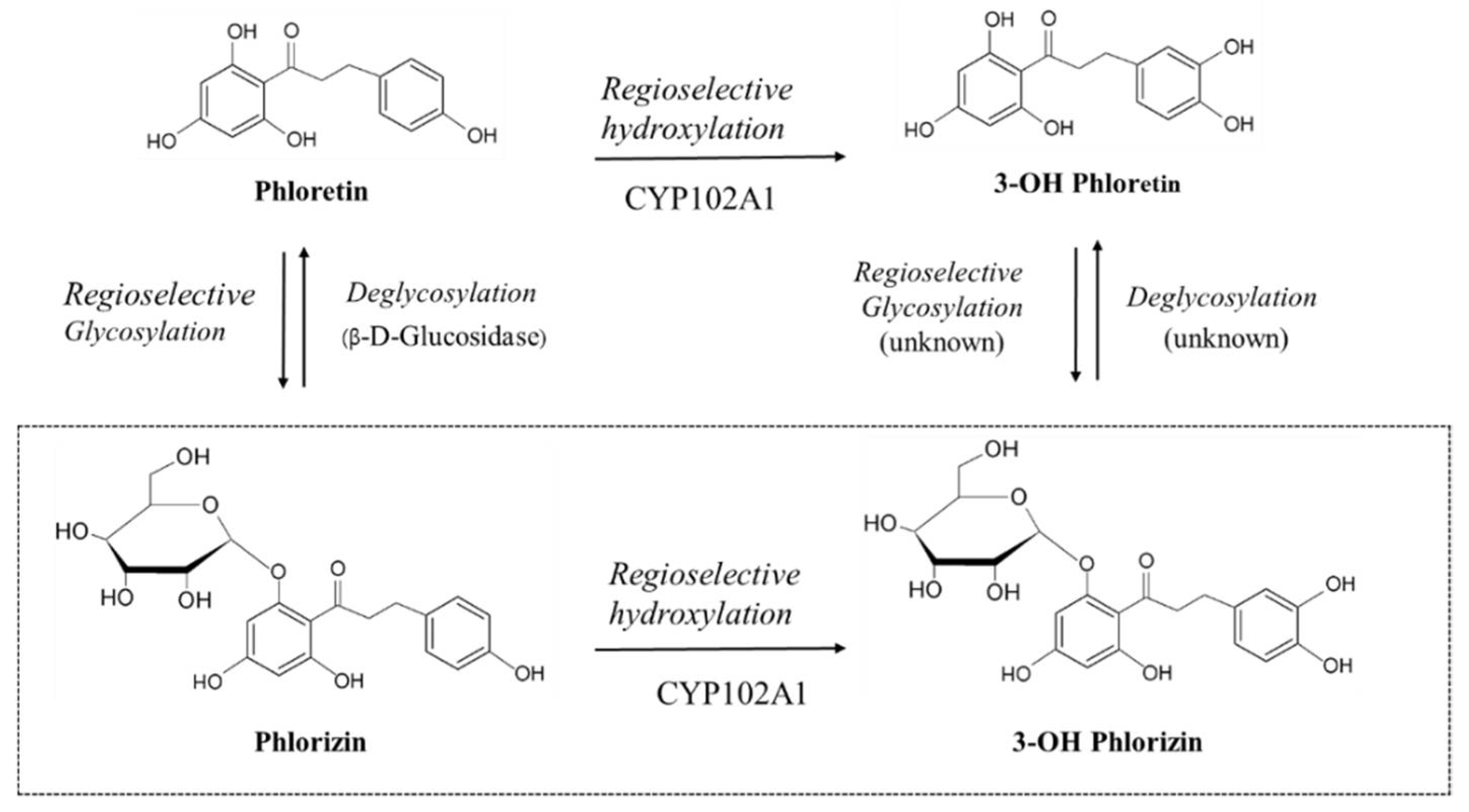
:1. Introduction
2. Materials and Methods
2.1. Materials
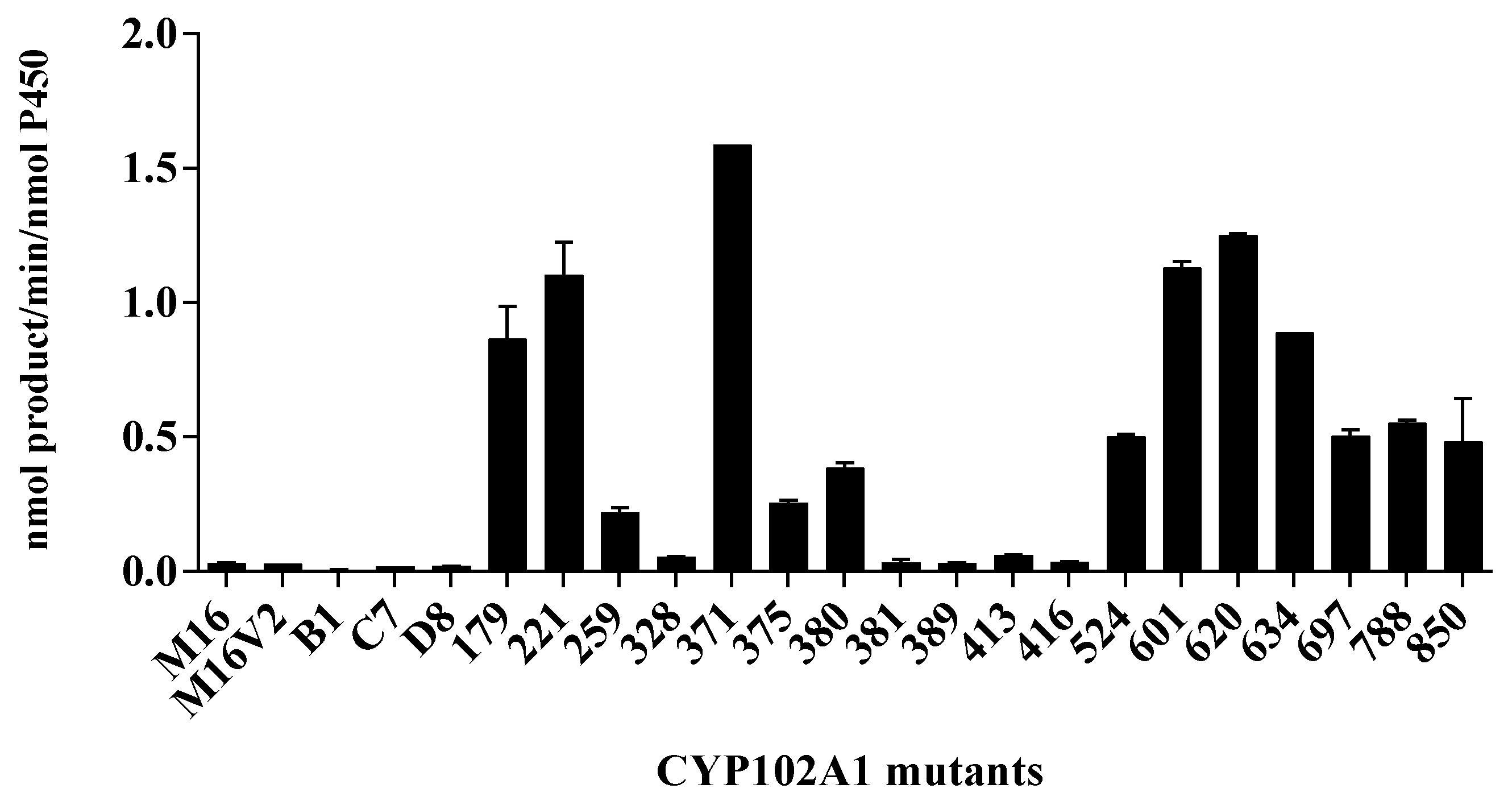
2.2. CYP102A1 Mutants to Screen Highly Active Phlorizin Hydroxylases
2.3. Construction of an Expression Plasmid for the CYP102A1 Gene
2.4. Expression of CYP102A1 Mutants
2.5. Hydroxylation of Phlorizin Catalyzed by CYP102A1 Mutants
2.6. LC–MS Analysis
2.7. Spectral Binding Titration
3. Results
3.1. Hydroxylation of Phlorizin Catalyzed by CYP102A1 Mutants
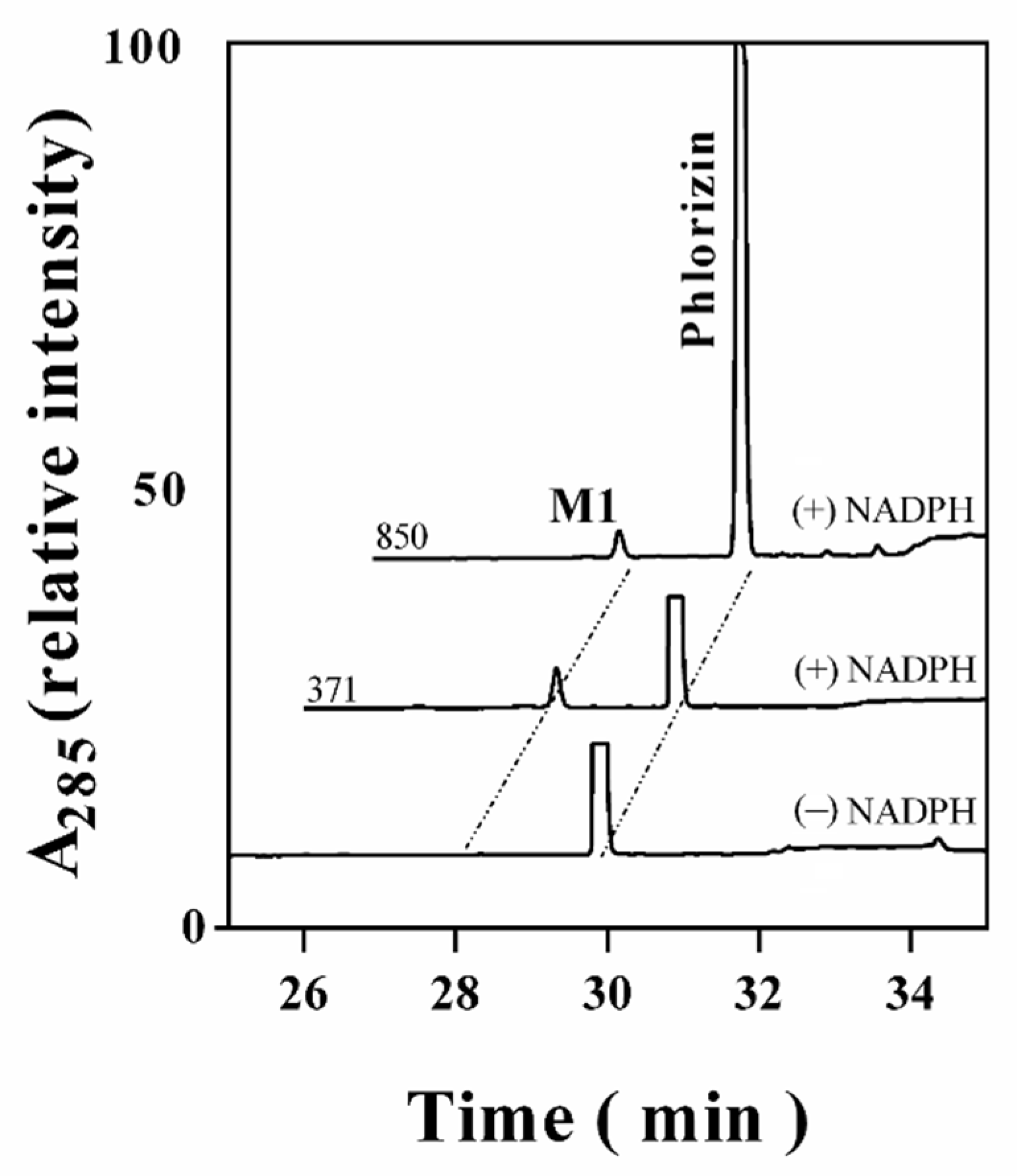
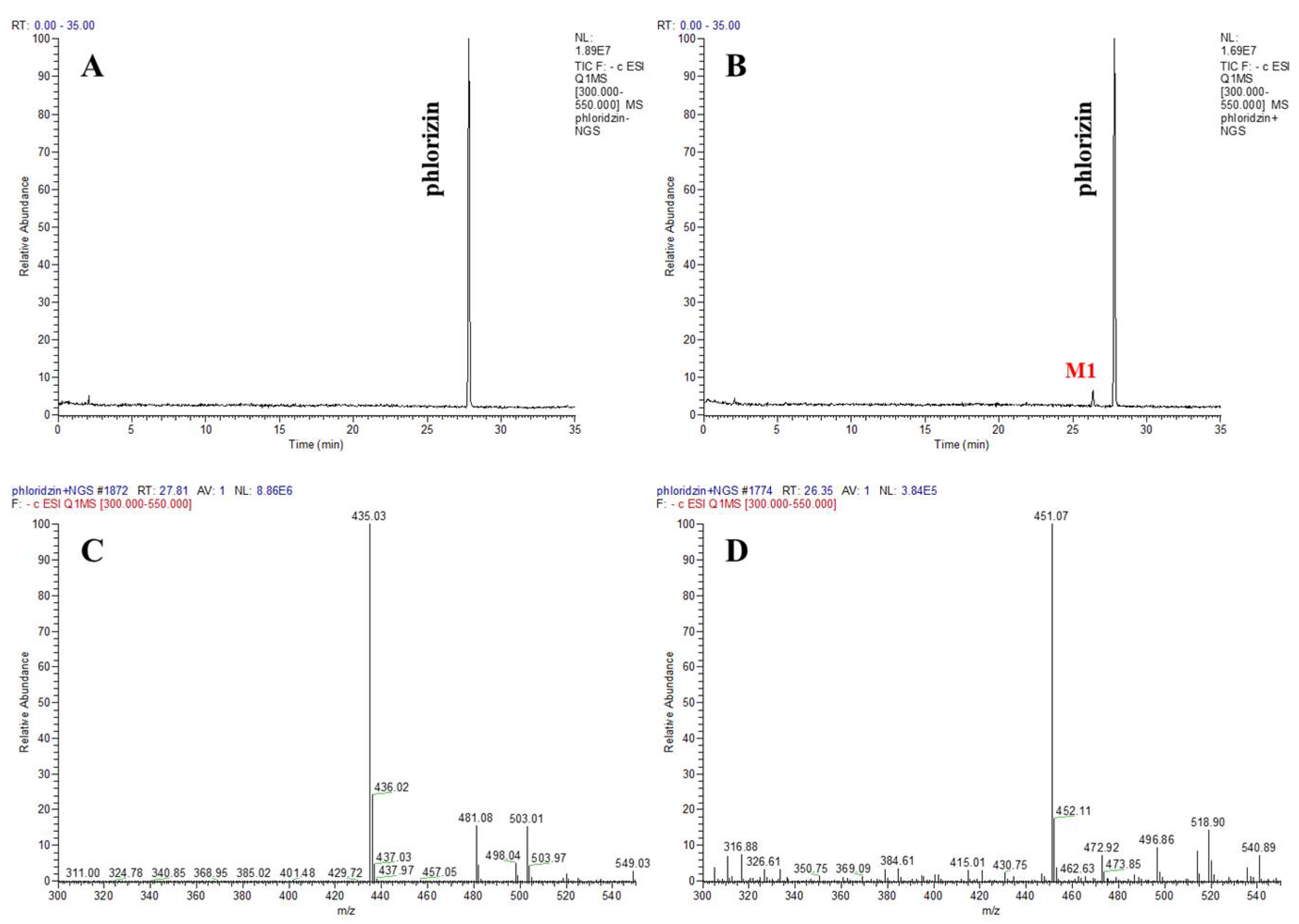
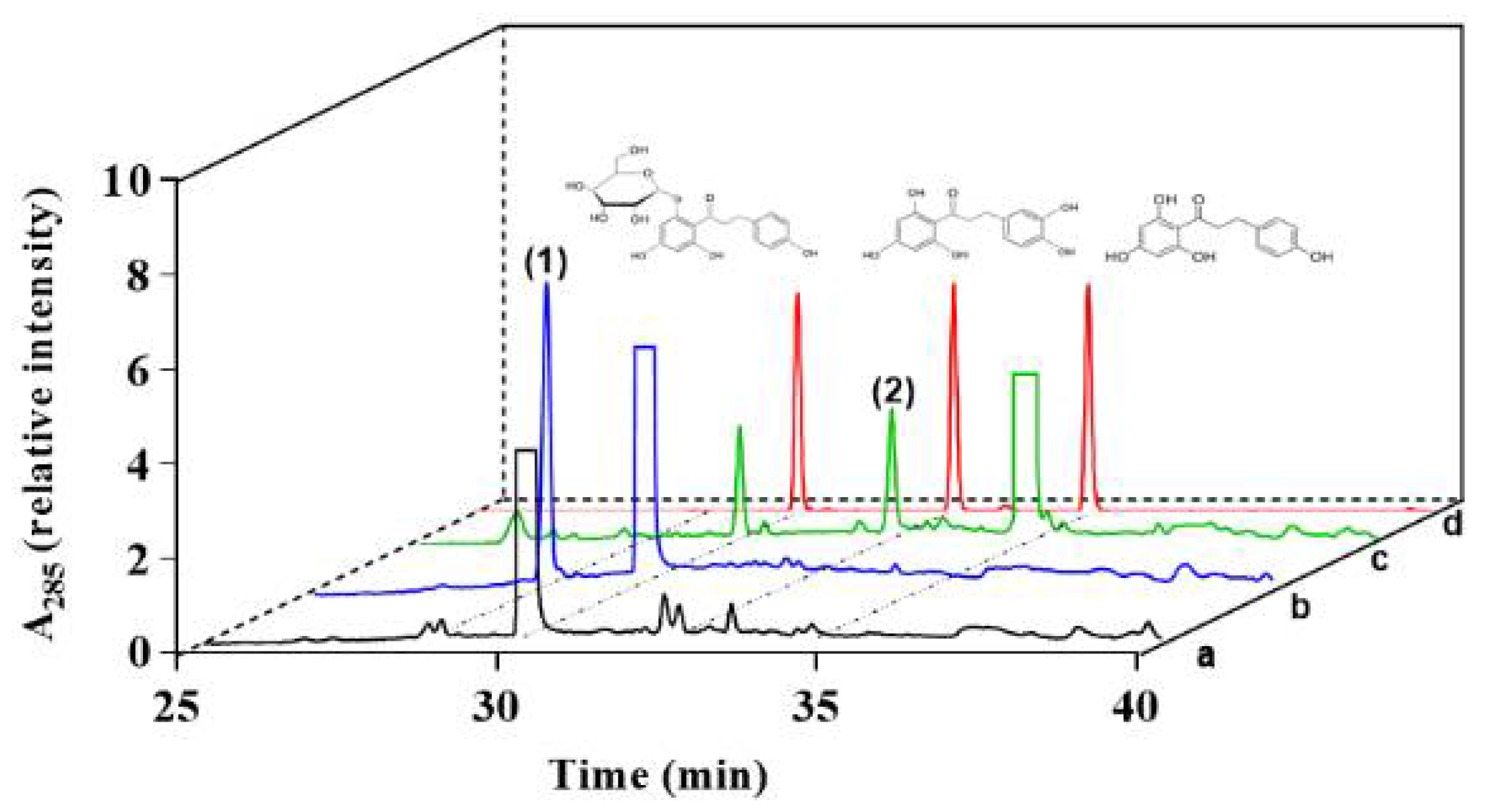
3.2. Characterizing a Major Product of Phlorizin by CYP102A1 and Subsequent β-Glucosidase
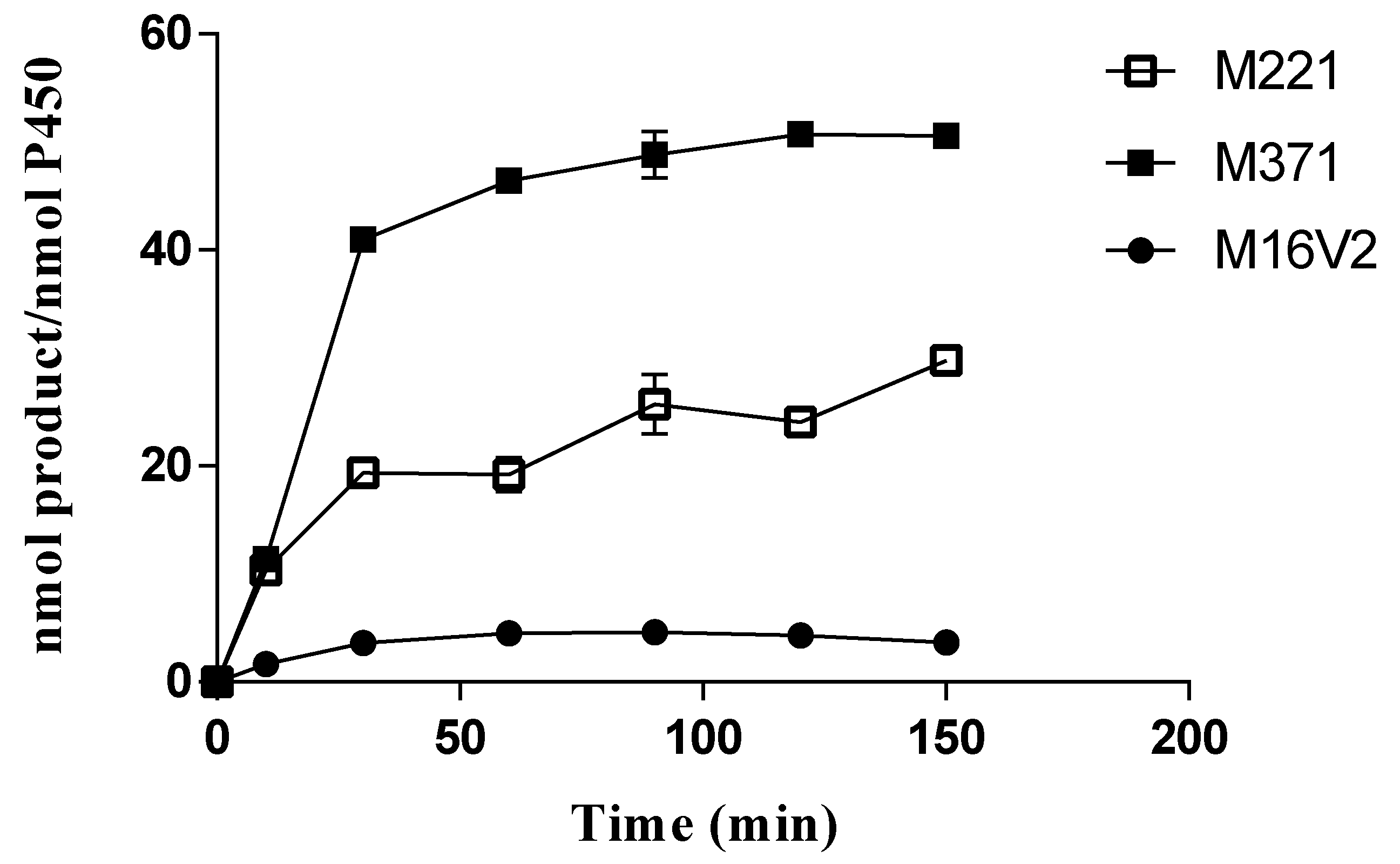
3.3. Kinetic Parameters and TTNs of Phlorizin Hydroxylation Catalyzed by CYP102A1 Mutants
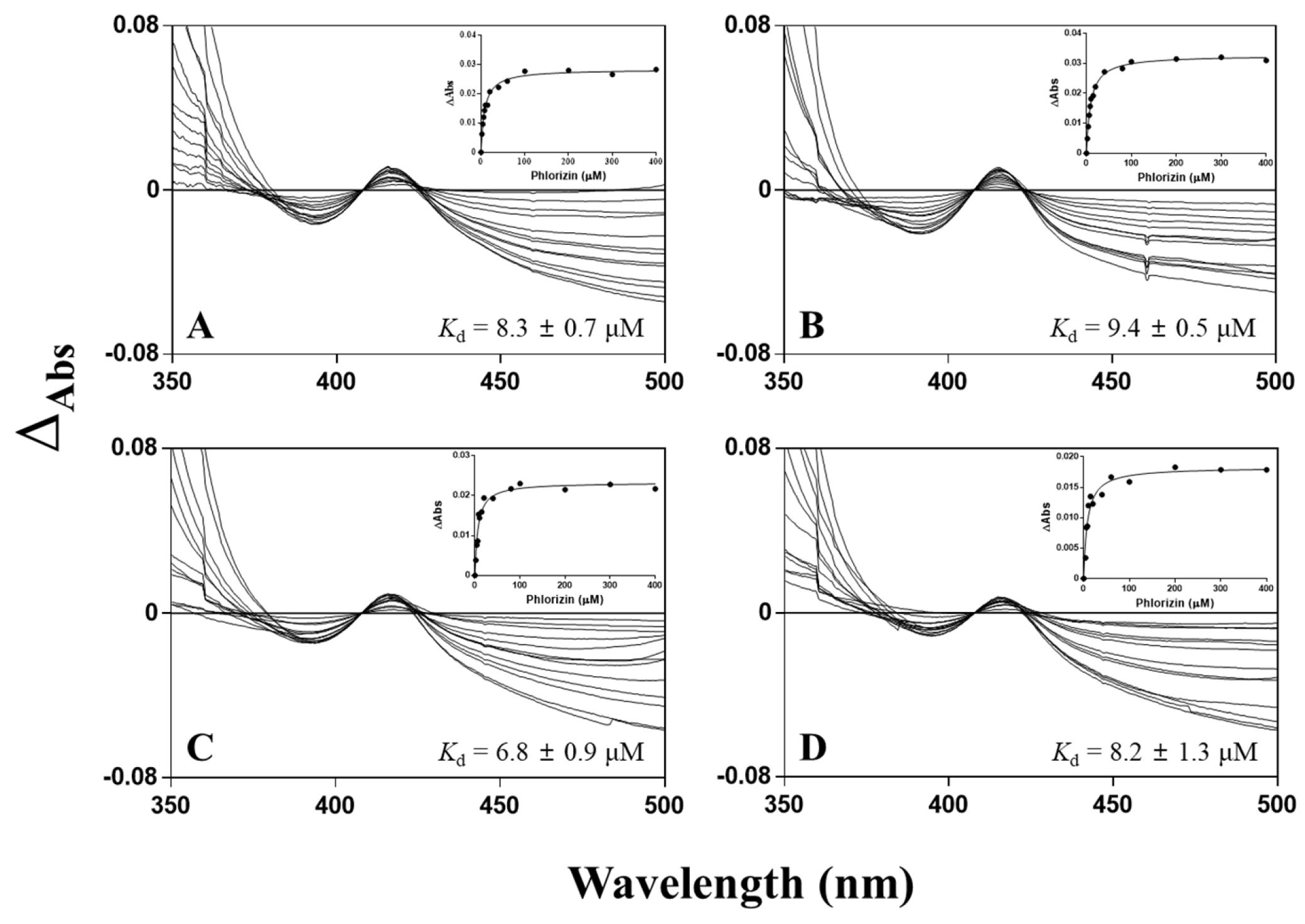
3.4. Spectral Titration of Phlorizin toward CYP102A1 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bars-Cortina, D.; Macià, A.; Iglesias, I.; Romero, M.P.; Motilva, M.J. Phytochemical profiles of new red-fleshed apple varieties compared with traditional and new white-fleshed varieties. J. Agric. Food Chem. 2017, 65, 1684–1696. [Google Scholar] [CrossRef]
- Skinner, R.C.; Gigliotti, J.C.; Ku, K.-M.; Tou, J.C. A comprehensive analysis of the composition, health benefits, and safety of apple pomace. Nutr. Rev. 2018, 76, 893–909. [Google Scholar] [CrossRef]
- Xie, L.; Cao, Y.; Zhao, Z.; Ren, C.; Xing, M.; Wu, B.; Zhang, B.; Xu, C.; Chen, K.; Li, X. Involvement of MdUGT75B1 and MdUGT71B1 in flavonol galactoside/glucoside biosynthesis in apple fruit. Food Chem. 2020, 312, 126124. [Google Scholar] [CrossRef]
- Emerson, S.D.; Carbert, N.S. An apple a day: Protective associations between nutrition and the mental health of immigrants in canada. Soc. Psychiatry. Psychiatr. Epidemiol. 2019, 54, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, C.K.B.; Percário, S.; Silva, J.C.C.B.; da Silva Torres, E.A.F. An apple plus a brazil nut a day keeps the doctors away: Antioxidant capacity of foods and their health benefits. Curr. Pharm. Des. 2016, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.H.; Woo, S.-M.; Nguyen, N.A.; Cha, G.-S.; Yeom, S.-J.; Kang, H.-S.; Yun, C.-H. Regioselective hydroxylation of naringin dihydrochalcone to produce neoeriocitrin dihydrochalcone by CYP102A1 (BM3) mutants. Catalysts 2020, 10, 823. [Google Scholar] [CrossRef]
- Le, T.-K.; Jang, H.-H.; Nguyen, H.T.H.; Doan, T.T.M.; Lee, G.-Y.; Park, K.D.; Ahn, T.; Joung, Y.H.; Kang, H.-S.; Yun, C.-H. Highly regioselective hydroxylation of polydatin, a resveratrol glucoside, for one-step synthesis of astringin, a piceatannol glucoside, by P450 BM3. Enzym. Microb. Technol. 2017, 97, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Márquez, K.; Contreras, D.; Salgado, P.; Mardones, C. Production of hydroxyl radicals and their relationship with phenolic compounds in white wines. Food Chem. 2019, 271, 80–86. [Google Scholar] [CrossRef]
- Perussello, C.A.; Zhang, Z.; Marzocchella, A.; Tiwari, B.K. Valorization of apple pomace by extraction of valuable compounds. Compr. Rev. Food Sci. Food Saf. 2017, 16, 776–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojdyło, A.; Oszmiański, J.; Laskowski, P. Polyphenolic compounds and antioxidant activity of new and old apple varieties. J. Agric. Food Chem. 2008, 56, 6520–6530. [Google Scholar] [CrossRef]
- Kschonsek, J.; Wolfram, T.; Stöckl, A.; Böhm, V. Polyphenolic Compounds analysis of old and new apple cultivars and contribution of polyphenolic profile to the in vitro antioxidant capacity. Antioxidants 2018, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Łata, B.; Trampczynska, A.; Paczesna, J. Cultivar variation in apple peel and whole fruit phenolic composition. Sci. Hortic. 2009, 121, 176–181. [Google Scholar] [CrossRef]
- Iwashina, T. The Structure and distribution of the flavonoids in plants. J. Plant. Res. 2000, 113, 287–299. [Google Scholar] [CrossRef]
- Dong, H.; Ning, Z.; Yu, L.; Li, L.; Lin, L.; Huang, J. Preparative separation and identification of the flavonoid phlorhizin from the crude extract of lithocarpus polystachyus rehd. Molecules 2007, 12, 552–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.; Chen, S.-N.; Joike, M.K.; Pendland, S.L.; Pauli, G.F.; Farnsworth, N.R. Inhibition of uropathogenic escherichia coli by cranberry juice: A new antiadherence assay. J. Agric. Food Chem. 2005, 53, 8940–8947. [Google Scholar] [CrossRef] [PubMed]
- Hilt, P.; Schieber, A.; Yildirim, C.; Arnold, G.; Klaiber, I.; Conrad, J.; Beifuss, U.; Carle, R. Detection of phloridzin in strawberries (Fragaria x Ananassa Duch.) by HPLC−PDA−MS/MS and NMR spectroscopy. J. Agric. Food Chem. 2003, 51, 2896–2899. [Google Scholar] [CrossRef]
- Hvattum, E. Determination of phenolic compounds in rose hip (Rosa Canina) using liquid chromatography coupled to electrospray ionisation tandem mass spectrometry and diode-array detection. Rapid Commun. Mass Spectrom. 2002, 16, 655–662. [Google Scholar] [CrossRef]
- Gosch, C.; Halbwirth, H.; Stich, K. Phloridzin: Biosynthesis, distribution and physiological relevance in plants. Phytochemistry 2010, 71, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Najafian, M.; Jahromi, M.Z.; Nowroznejhad, M.J.; Khajeaian, P.; Kargar, M.M.; Sadeghi, M.; Arasteh, A. Phloridzin reduces blood glucose levels and improves lipids metabolism in streptozotocin-induced diabetic rats. Mol. Biol. Rep. 2012, 39, 5299–5306. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Zhang, X.; Wang, Z.; Gao, Z.; Liu, G.; Hu, H.; Zou, L.; Li, X. Insulin sensitivity-enhancing activity of phlorizin is associated with lipopolysaccharide decrease and gut microbiota changes in obese and type 2 diabetes (Db/Db) mice. J. Agric. Food Chem. 2016, 64, 7502–7511. [Google Scholar] [CrossRef]
- Mendes, R.A.; Silva, E.B.L.S.; Takeara, R.; Freitas, R.G.; Brown, A.; de Souza, G.L.C. Probing the antioxidant potential of phloretin and phlorizin through a computational investigation. J. Mol. Model. 2018, 24, 101. [Google Scholar] [CrossRef] [PubMed]
- Antika, L.D.; Lee, E.-J.; Kim, Y.-H.; Kang, M.-K.; Park, S.-H.; Kim, D.Y.; Oh, H.; Choi, Y.-J.; Kang, Y.-H. Dietary phlorizin enhances osteoblastogenic bone formation through enhancing β-catenin activity via gsk-3β inhibition in a model of senile osteoporosis. J. Nutr. Biochem. 2017, 49, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Zhang, Y.; Zhang, J.; Kang, W. Analysis of chemical constituents changing in physical process and nutritional components of Malus halliana Koehne tea. J. Food Qual. 2017, 2017, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Leu, S.J.; Lin, Y.P.; Lin, R.D.; Wen, C.L.; Cheng, K.T.; Hsu, F.L.; Lee, M.H. Phenolic constituents of Malus Doumeri Var. Formosana in the field of skin care. Biol. Pharm. Bull. 2006, 29, 740–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondonno, N.P.; Bondonno, C.P.; Ward, N.C.; Hodgson, J.M.; Croft, K.D. The cardiovascular health benefits of apples: Whole fruit vs. isolated compounds. Trends Food Sci. Technol. 2017, 69, 243–256. [Google Scholar] [CrossRef]
- Waldbauer, K.; McKinnon, R.; Kopp, B. Apple pomace as potential source of natural active compounds. Planta Med. 2017, 83, 994–1010. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.A.; Jang, J.; Le, T.-K.; Nguyen, T.H.H.; Woo, S.-M.; Yoo, S.-K.; Lee, Y.J.; Park, K.D.; Yeom, S.-J.; Kim, G.-J.; et al. Biocatalytic production of a potent inhibitor of adipocyte differentiation from phloretin using engineered CYP102A1. J. Agric. Food Chem. 2020, 68, 6683–6691. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Cao, N.T.; Nguyen, T.H.H.; Le, T.-K.; Cha, G.S.; Choi, S.-K.; Pan, J.-G.; Yeom, S.-J.; Kang, H.-S.; Yun, C.-H. Regioselective hydroxylation of phloretin, a bioactive compound from apples, by human cytochrome P450 enzymes. Pharmaceuticals 2020, 13, 330. [Google Scholar] [CrossRef]
- Nelson, D.R. A world of cytochrome P450s. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, S.H.; Cha, G.S.; Choi, D.S.; Nam, D.H.; Lee, J.H.; Lee, J.-K.; Yun, C.-H.; Jeong, K.J.; Park, C.B. Cofactor-free light-driven whole-cell cytochrome P450 catalysis. Angew. Chem. Int. Ed. 2015, 54, 969–973. [Google Scholar] [CrossRef]
- Whitehouse, C.J.C.; Bell, S.G.; Wong, L.-L. P450 BM3 (CYP102A1): Connecting the dots. Chem. Soc. Rev. 2012, 41, 1218–1260. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012, 30, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.-H.; Kim, K.-H.; Kim, D.-H.; Jung, H.-C.; Pan, J.-G. The bacterial P450 BM3: A prototype for a biocatalyst with human P450 activities. Trends Biotechnol. 2007, 25, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Otey, C.R.; Bandara, G.; Lalonde, J.; Takahashi, K.; Arnold, F.H. Preparation of Human metabolites of propranolol using laboratory-evolved bacterial cytochromes P450. Biotechnol. Bioeng. 2006, 93, 494–499. [Google Scholar] [CrossRef]
- Kim, D.-H.; Kim, K.-H.; Kim, D.; Jung, H.-C.; Pan, J.-G.; Chi, Y.-T.; Ahn, T.; Yun, C.-H. Oxidation of human cytochrome P450 1A2 substrates by bacillus megaterium cytochrome P450 BM3. J. Mol. Catal. B Enzym. 2010, 63, 179–187. [Google Scholar] [CrossRef]
- Jang, H.-H.; Ryu, S.-H.; Le, T.-K.; Doan, T.T.M.; Nguyen, T.H.H.; Park, K.D.; Yim, D.-E.; Kim, D.-H.; Kang, C.-K.; Ahn, T.; et al. Regioselective C-H hydroxylation of omeprazole sulfide by bacillus megaterium CYP102A1 to produce a human metabolite. Biotechnol. Lett. 2017, 39, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-Y.; Kim, S.-Y.; Kim, D.; Kim, D.-H.; Shin, S.-M.; Park, S.-H.; Kim, K.-H.; Jung, H.-C.; Pan, J.-G.; Joung, Y.H.; et al. Characterization of diverse natural variants of CYP102A1 found within a species of Bacillus Megaterium. AMB Express 2011, 1, 1. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Kim, K.-H.; Kim, D.-H.; Liu, K.-H.; Jung, H.-C.; Pan, J.-G.; Yun, C.-H. Generation of human metabolites of 7-ethoxycoumarin by bacterial cytochrome P450 BM3. Drug Metab. Dispos. 2008, 36, 2166–2170. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-Y.; Ryu, S.H.; Park, S.-H.; Cha, G.S.; Kim, D.-H.; Kim, K.-H.; Hong, A.W.; Ahn, T.; Pan, J.-G.; Joung, Y.H.; et al. Chimeric cytochromes P450 engineered by domain swapping and random mutagenesis for producing human metabolites of drugs. Biotechnol. Bioeng. 2014, 111, 1313–1322. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [CrossRef]
- Miller, G.P.; Guengerich, F.P. Binding and oxidation of alkyl 4-nitrophenyl ethers by rabbit cytochrome P450 1A2: Evidence for two binding sites. Biochemistry 2001, 40, 7262–7272. [Google Scholar] [CrossRef] [PubMed]
- Slámová, K.; Kapešová, J.; Valentová, K. “Sweet Flavonoids”: Glycosidase-catalyzed modifications. Int. J. Mol. Sci. 2018, 19, 2126. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.-C.; Hinkelmann, J.; Schwab, W. Glucosylation of aroma chemicals and hydroxy fatty acids. J. Biotechnol. 2015, 216, 100–109. [Google Scholar] [CrossRef]
- Sato, M.F.; Vieira, R.G.; Zardo, D.M.; Falcão, L.D.; Nogueira, A.; Wosiacki, G. Apple pomace from eleven cultivars: An approach to identify sources of bioactive compounds. Acta Sci. Agron. 2010, 32, 29–35. [Google Scholar] [CrossRef]
- Rana, S.; Gupta, S.; Rana, A.; Bhushan, S. Functional properties, phenolic constituents and antioxidant potential of industrial apple pomace for utilization as active food ingredient. Food Sci. Hum. Wellness 2015, 4, 180–187. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Ying, D.; Guo, B.; Cheng, L.J.; May, B.; Bird, T.; Sanguansri, L.; Cao, Y.; Augustin, M. Extrusion of apple pomace increases antioxidant activity upon in vitro digestion. Food Funct. 2019, 10, 951–963. [Google Scholar] [CrossRef]
- Barreira, J.C.M.; Arraibi, A.A.; Ferreira, I.C.F.R. Bioactive and functional compounds in apple pomace from juice and cider manufacturing: Potential use in dermal formulations. Trends Food Sci. Technol. 2019, 90, 76–87. [Google Scholar] [CrossRef]
- Choi, B.Y. Biochemical basis of anti-cancer-effects of phloretin—A natural dihydrochalcone. Molecules 2019, 24, 278. [Google Scholar] [CrossRef] [Green Version]
- Oleszek, M.; Pecio, Ł.; Kozachok, S.; Lachowska-Filipiuk, Ż.; Oszust, K.; Frąc, M. Phytochemicals of apple pomace as prospect bio-fungicide agents against mycotoxigenic fungal species—In vitro experiments. Toxins 2019, 11, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.-C.; Chen, M.-C.; Liu, S.; Callahan, V.M.; Bracci, N.R.; Lehman, C.W.; Dahal, B.; de la Fuente, C.L.; Lin, C.-C.; Wang, T.T.; et al. Phloretin inhibits zika virus infection by interfering with cellular glucose utilisation. Int. J. Antimicrob. Agents 2019, 54, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Barlas, N.; Özer, S.; Karabulut, G. The estrogenic effects of apigenin, phloretin and myricetin based on uterotrophic assay in immature Wistar albino rats. Toxicol. Lett. 2014, 226, 35–42. [Google Scholar] [CrossRef]
- Anunciato Casarini, T.P.; Frank, L.A.; Pohlmann, A.R.; Guterres, S.S. Dermatological applications of the flavonoid phloretin. Eur. J. Pharmacol. 2020, 889, 173593. [Google Scholar] [CrossRef] [PubMed]
- Dugé de Bernonville, T.; Guyot, S.; Paulin, J.-P.; Gaucher, M.; Loufrani, L.; Henrion, D.; Derbré, S.; Guilet, D.; Richomme, P.; Dat, J.F.; et al. Dihydrochalcones: Implication in resistance to oxidative stress and bioactivities against advanced glycation end-products and vasoconstriction. Phytochemistry 2010, 71, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.-L.; Mo, H.; Smith, B.J.; Chen, C.-H.; Chen, L.; Chyu, M.-C.; Kwun, I.-S. Chapter 52—Green Tea and other fruit polyphenols attenuate deterioration of bone microarchitecture. In Polyphenols in Human Health and Disease; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 681–693. ISBN 9780123984562. [Google Scholar]
- Jugdé, H.; Nguy, D.; Moller, I.; Cooney, J.M.; Atkinson, R.G. Isolation and characterization of a novel glycosyltransferase that converts phloretin to phlorizin, a potent antioxidant in apple. FEBS J. 2008, 275, 3804–3814. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, F.; Crane, R.K. Studies on the mechanism of intestinal absorption of sugars VII. Phenylglycoside transport and its possible relationship to phlorizin inhibition of the active transport of sugars by the small intestine. Biochim. Biophys. Acta 1964, 93, 116–135. [Google Scholar] [CrossRef]
- Ehrenkranz, J.R.L.; Lewis, N.G.; Kahn, C.R.; Roth, J. Phlorizin: A Review. Diabetes Metab. Res. Rev. 2005, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Ahn, T.; Jung, H.-C.; Pan, J.-G.; Yun, C.-H. Generation of the human metabolite piceatannol from the anticancer-preventive agent resveratrol by bacterial cytochrome P450 BM3. Drug Metab. Dispos. 2009, 37, 932–936. [Google Scholar] [CrossRef] [Green Version]
- Park, C.M.; Park, H.S.; Cha, G.S.; Park, K.D.; Yun, C.-H. Regioselective hydroxylation of rhododendrol by CYP102A1 and tyrosinase. Catalysts 2020, 10, 1114. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhang, Y.; Chen, X.; Wang, Y.; Chen, W.; Xu, Q.; Li, P.; Ma, F. Extraction, Identification, and antioxidant and anticancer tests of seven dihydrochalcones from malus “Red Splendor” fruit. Food Chem. 2017, 231, 324–331. [Google Scholar] [CrossRef] [PubMed]

| Enzymes | kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) |
|---|---|---|---|
| M221 | 1.1 ± 0.2 | 1540 ± 500 | 0.00071 ± 0.00027 |
| M371 | 0.90 ± 0.2 | 979 ± 315 | 0.00091 ± 0.00036 |
| M850 | 0.40 ± 0.02 | 747 ± 102 | 0.00053 ± 0.00008 |
| M16V2 | 0.16 ± 0.01 | 267 ± 34 | 0.00059 ± 0.00008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.A.; Cao, N.T.; Nguyen, T.H.H.; Ji, J.-H.; Cha, G.S.; Kang, H.-S.; Yun, C.-H. Enzymatic Production of 3-OH Phlorizin, a Possible Bioactive Polyphenol from Apples, by Bacillus megaterium CYP102A1 via Regioselective Hydroxylation. Antioxidants 2021, 10, 1327. https://doi.org/10.3390/antiox10081327
Nguyen NA, Cao NT, Nguyen THH, Ji J-H, Cha GS, Kang H-S, Yun C-H. Enzymatic Production of 3-OH Phlorizin, a Possible Bioactive Polyphenol from Apples, by Bacillus megaterium CYP102A1 via Regioselective Hydroxylation. Antioxidants. 2021; 10(8):1327. https://doi.org/10.3390/antiox10081327
Chicago/Turabian StyleNguyen, Ngoc Anh, Ngoc Tan Cao, Thi Huong Ha Nguyen, Jung-Hwan Ji, Gun Su Cha, Hyung-Sik Kang, and Chul-Ho Yun. 2021. "Enzymatic Production of 3-OH Phlorizin, a Possible Bioactive Polyphenol from Apples, by Bacillus megaterium CYP102A1 via Regioselective Hydroxylation" Antioxidants 10, no. 8: 1327. https://doi.org/10.3390/antiox10081327
APA StyleNguyen, N. A., Cao, N. T., Nguyen, T. H. H., Ji, J.-H., Cha, G. S., Kang, H.-S., & Yun, C.-H. (2021). Enzymatic Production of 3-OH Phlorizin, a Possible Bioactive Polyphenol from Apples, by Bacillus megaterium CYP102A1 via Regioselective Hydroxylation. Antioxidants, 10(8), 1327. https://doi.org/10.3390/antiox10081327