Extensive Thiol Profiling for Assessment of Intracellular Redox Status in Cultured Cells by HPLC-MS/MS
Abstract
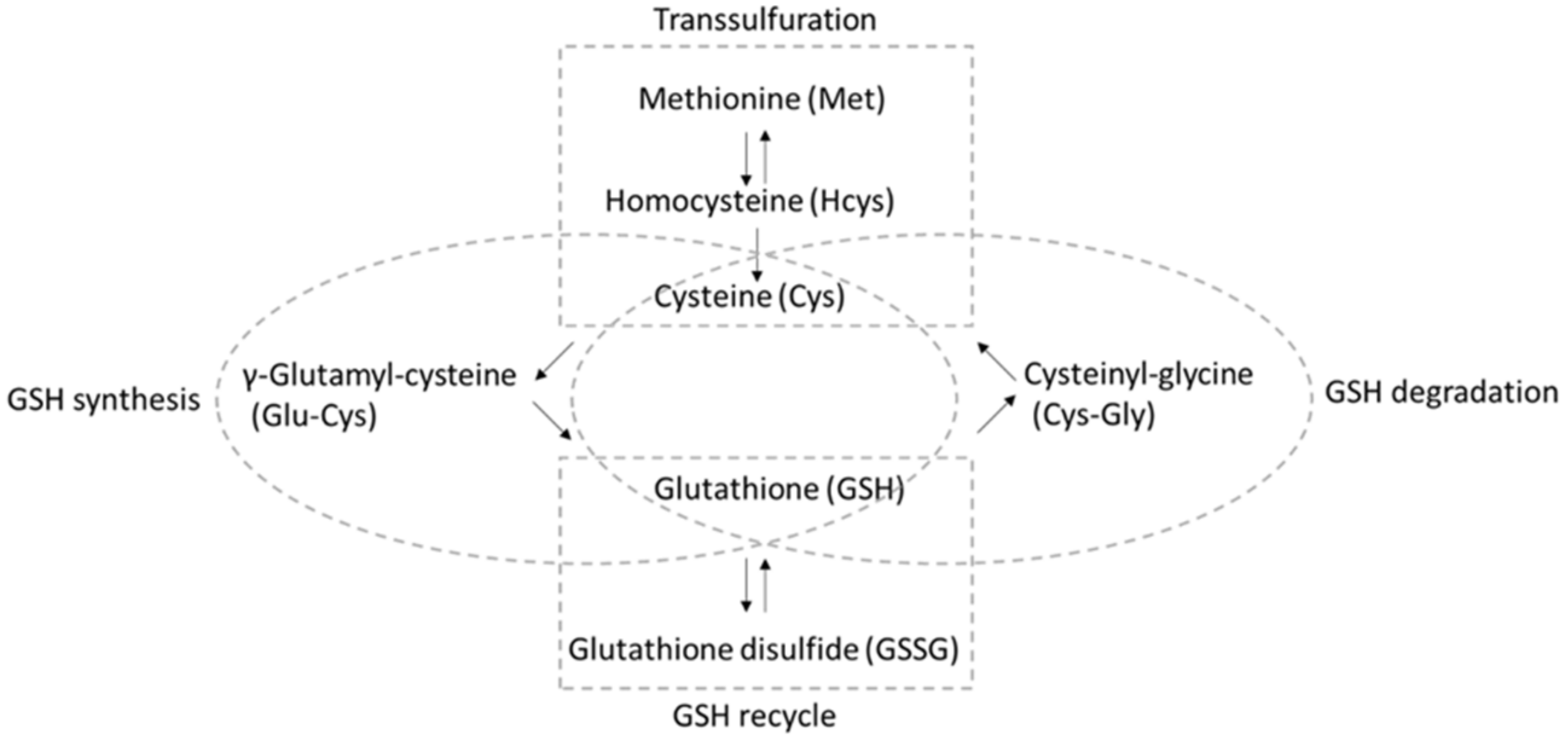
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Standard Preparation
2.3. Cell Culture
2.4. Cell Sample Preparation
2.5. Treatment with tBHP
2.6. HPLC-MS/MS Method
2.7. Statistical Analysis
3. Results and Discussion
3.1. HPLC-MS/MS Method Performance and Validation
3.2. Comprehensive Thiol Profiling of Different Cell Lines
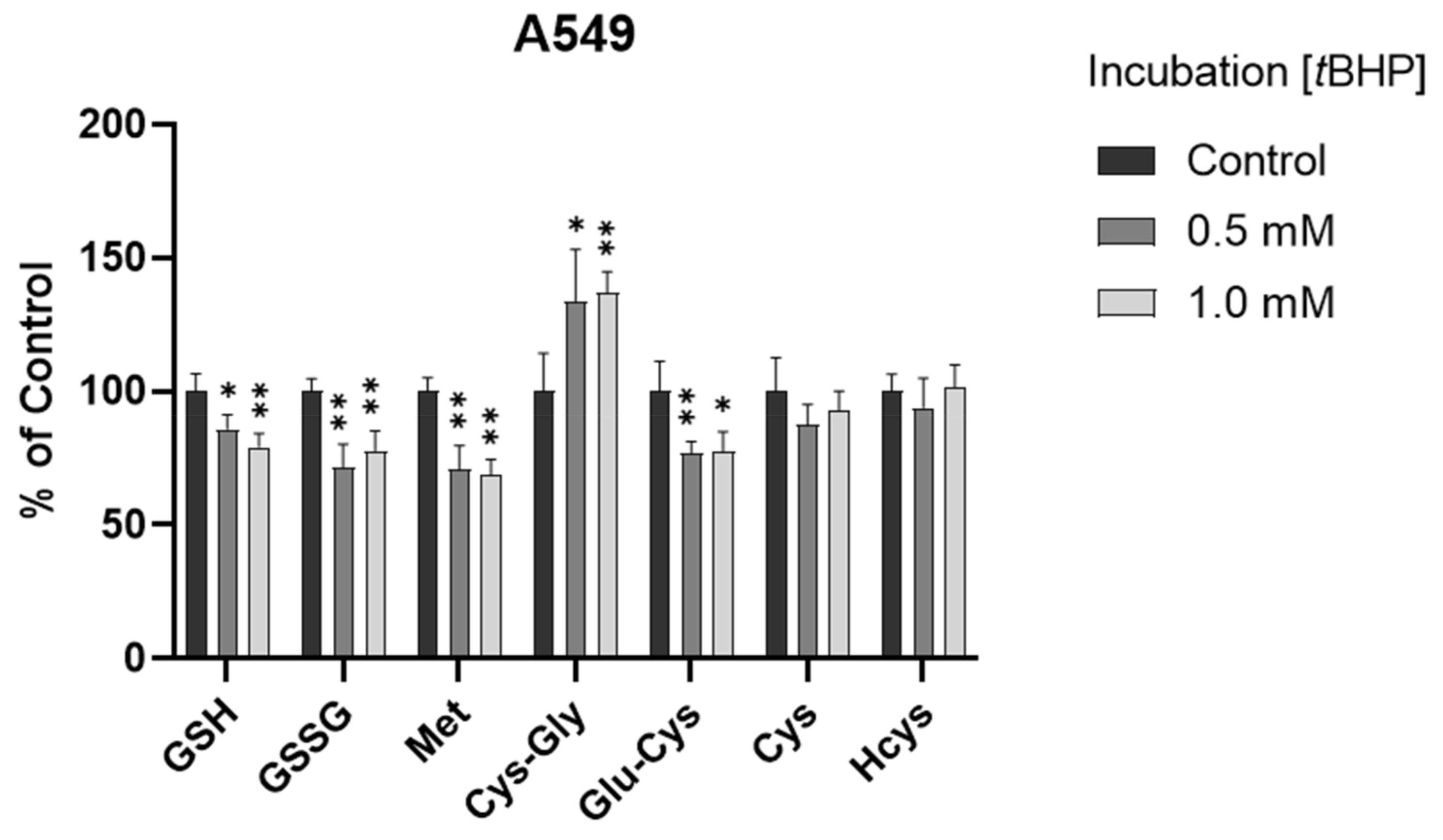
3.3. Assessment of Intracellular Redox Status in A549 Cells with tBHP Treatment
3.4. Assessment of Intracellular Redox Status in B3 Cells with tBHP Treatment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Halliwell, B. Oxidative stress and cancer: Have we moved forward? Biochem. J. 2007, 401, 1–11. [Google Scholar] [CrossRef]
- Patel, R.S.; Ghasemzadeh, N.; Eapen, D.J.; Sher, S.; Arshad, S.; Ko, Y.A.; Veledar, E.; Samady, H.; Zafari, A.M.; Sperling, L.; et al. Novel Biomarker of Oxidative Stress Is Associated With Risk of Death in Patients With Coronary Artery Disease. Circulation 2016, 133, 361–369. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.C.; Grey, A.C.; Zahraei, A.; Donaldson, P.J. Age-dependent changes in glutathione metabolism pathways in the lens: New insights into therapeutic strategies to prevent cataract formation-A review. Clin. Exp. Ophthalmol. 2020, 48, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Rathbun, W.B. Activity of glutathione synthesis enzymes in the rhesus monkey lens related to age: A model for the human lens. Curr. Eye Res. 1986, 5, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.R.; Minnion, M.; Barbarino, F.; Koster, G.; Fernandez, B.O.; Cumpstey, A.F.; Wischmann, P.; Madhani, M.; Frenneaux, M.P.; Postle, A.D.; et al. A robust and versatile mass spectrometry platform for comprehensive assessment of the thiol redox metabolome. Redox Biol. 2018, 16, 359–380. [Google Scholar] [CrossRef]
- Wu, J.; Sigler, A.; Pfaff, A.; Cen, N.; Ercal, N.; Shi, H. Development of a HPLC-MS/MS method for assessment of thiol redox status in human tear fluids. Anal. Biochem. 2021, 629, 114295. [Google Scholar] [CrossRef]
- Li, B.; Li, L.; Donaldson, P.J.; Lim, J.C. Dynamic regulation of GSH synthesis and uptake pathways in the rat lens epithelium. Exp. Eye Res. 2010, 90, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Sbodio, J.I.; Snyder, S.H.; Paul, B.D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 2019, 176, 583–593. [Google Scholar] [CrossRef]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef]
- Tyagi, N.; Sedoris, K.C.; Steed, M.; Ovechkin, A.V.; Moshal, K.S.; Tyagi, S.C. Mechanisms of homocysteine-induced oxidative stress. Am. J. Physiol. Heart Circ. Physiol. 2005, 289, H2649–H2656. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, P.; Kok, F.J.; Kruyssen, D.A.; Schouten, E.G.; Witteman, J.C.; Grobbee, D.E.; Ueland, P.M.; Refsum, H. Plasma total homocysteine, B vitamins, and risk of coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 989–995. [Google Scholar] [CrossRef]
- Mayer, E.L.; Jacobsen, D.W.; Robinson, K. Homocysteine and coronary atherosclerosis. J. Am. Coll. Cardiol. 1996, 27, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Martinez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Mas, D.; Valdivie, M.; Hu, C.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef]
- Catanesi, M.; Brandolini, L.; d’Angelo, M.; Benedetti, E.; Tupone, M.G.; Alfonsetti, M.; Cabri, E.; Iaconis, D.; Fratelli, M.; Cimini, A.; et al. l-Methionine Protects against Oxidative Stress and Mitochondrial Dysfunction in an In Vitro Model of Parkinson’s Disease. Antioxidants 2021, 10, 1467. [Google Scholar] [CrossRef]
- Babizhayev, M.A.; Yegorov, Y.E. Biomarkers of oxidative stress and cataract. Novel drug delivery therapeutic strategies targeting telomere reduction and the expression of telomerase activity in the lens epithelial cells with N-acetylcarnosine lubricant eye drops: Anti-cataract which helps to prevent and treat cataracts in the eyes of dogs and other animals. Curr. Drug Deliv. 2014, 11, 24–61. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, V.M.; Beyer, E.C. Oxidative stress, lens gap junctions, and cataracts. Antioxid. Redox Signal. 2009, 11, 339–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbay, A.; Ozer, O.F.; Altinisik, M.; Elbay, A.E.; Sezer, T.; Bayraktar, H.; Ozdemir, H. A novel tool reflecting the role of oxidative stress in the cataracts: Thiol/disulfide homeostasis. Scand. J. Clin. Lab. Investig. 2017, 77, 223–227. [Google Scholar] [CrossRef]
- Vinson, J.A. Oxidative stress in cataracts. Pathophysiology 2006, 13, 151–162. [Google Scholar] [CrossRef]
- Herzog, K.; IJlst, L.; van Cruchten, A.G.; van Roermund, C.W.T.; Kulik, W.; Wanders, R.J.A.; Waterham, H.R. An UPLC-MS/MS Assay to Measure Glutathione as Marker for Oxidative Stress in Cultured Cells. Metabolites 2019, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Berger, R.S.; Heinrich, P.; Marchiq, I.; Pouyssegur, J.; Renner, K.; Oefner, P.J.; Dettmer, K. Optimized Protocol for the In Situ Derivatization of Glutathione with N-Ethylmaleimide in Cultured Cells and the Simultaneous Determination of Glutathione/Glutathione Disulfide Ratio by HPLC-UV-QTOF-MS. Metabolites 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Giustarini, D.; Tsikas, D.; Colombo, G.; Milzani, A.; Dalle-Donne, I.; Fanti, P.; Rossi, R. Pitfalls in the analysis of the physiological antioxidant glutathione (GSH) and its disulfide (GSSG) in biological samples: An elephant in the room. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1019, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Giustarini, D.; Dalle-Donne, I.; Milzani, A.; Fanti, P.; Rossi, R. Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat. Protoc. 2013, 8, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.S.T.; Napylov, A.; Paquet, A.; Vuckovic, D. Comparison of N-ethyl maleimide and N-(1-phenylethyl) maleimide for derivatization of biological thiols using liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2020, 412, 1639–1652. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Beltz, J.; Chernatynskaya, A.; Pfaff, A.; Ercal, N. Protective effects of tiopronin on oxidatively challenged human lung carcinoma cells (A549). Free Radic. Res. 2020, 54, 319–329. [Google Scholar] [CrossRef]
- Sigler, A.; He, X.; Bose, M.; Cristea, A.; Liu, W.; Nam, P.K.; James, D.; Burton, C.; Shi, H. Simultaneous Determination of Eight Urinary Metabolites by HPLC-MS/MS for Noninvasive Assessment of Traumatic Brain Injury. J. Am. Soc. Mass Spectrom. 2020, 31, 1910–1917. [Google Scholar] [CrossRef]
- Beltz, J.; Pfaff, A.; Ercal, N. Simultaneous determination of tiopronin and its primary metabolite in plasma and ocular tissues by HPLC. Biomed. Chromatogr. 2019, 33, e4375. [Google Scholar] [CrossRef]
- Tomin, T.; Schittmayer, M.; Birner-Gruenberger, R. Addressing Glutathione Redox Status in Clinical Samples by Two-Step Alkylation with N-ethylmaleimide Isotopologues. Metabolites 2020, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Giustarini, D.; Colombo, G.; Garavaglia, M.L.; Astori, E.; Portinaro, N.M.; Reggiani, F.; Badalamenti, S.; Aloisi, A.M.; Santucci, A.; Rossi, R.; et al. Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, and cultured cells. Free Radic. Biol. Med. 2017, 112, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.C.; Schott, K.; Charao, M.; Moro, A.; Bulcao, R.; Grotto, D.; Valentini, J.; Bohrer, D.; Cardoso, S.; Pomblum, V. Quantification of reduced glutathione by HPLC-UV in erythrocytes of hemodialysis patients. Biomed. Chromatogr. 2008, 22, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chi, D.; Song, D.; Su, G.; Li, L.; Shao, L. Quantification of glutathione in plasma samples by HPLC using 4-fluoro-7-nitrobenzofurazan as a fluorescent labeling reagent. J. Chromatogr. Sci. 2012, 50, 119–122. [Google Scholar] [CrossRef]
- Moore, T.; Le, A.; Niemi, A.K.; Kwan, T.; Cusmano-Ozog, K.; Enns, G.M.; Cowan, T.M. A new LC-MS/MS method for the clinical determination of reduced and oxidized glutathione from whole blood. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 929, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Arismendi-Morillo, G.; Mukherjee, P.; Chinopoulos, C. On the Origin of ATP Synthesis in Cancer. iScience 2020, 23, 101761. [Google Scholar] [CrossRef]
- Spector, A.; Wang, R.R.; Ma, W.; Kleiman, N.J. Development and characterization of an H2O2-resistant immortal lens epithelial cell line. Investig. Ophthalmol. Vis. Sci. 2000, 41, 832–843. [Google Scholar]
- Griffith, O.W. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 1999, 27, 922–935. [Google Scholar] [CrossRef]
- Rahman, I.; MacNee, W. Oxidative stress and regulation of glutathione in lung inflammation. Eur. Respir. J. 2000, 16, 534–554. [Google Scholar] [CrossRef]
- Wild, A.C.; Mulcahy, R.T. Regulation of gamma-glutamylcysteine synthetase subunit gene expression: Insights into transcriptional control of antioxidant defenses. Free Radic. Res. 2000, 32, 281–301. [Google Scholar] [CrossRef]
- Franklin, C.C.; Backos, D.S.; Mohar, I.; White, C.C.; Forman, H.J.; Kavanagh, T.J. Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase. Mol. Asp. Med. 2009, 30, 86–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krejsa, C.M.; Franklin, C.C.; White, C.C.; Ledbetter, J.A.; Schieven, G.L.; Kavanagh, T.J. Rapid activation of glutamate cysteine ligase following oxidative stress. J. Biol. Chem. 2010, 285, 16116–16124. [Google Scholar] [CrossRef] [Green Version]



| Analyte | Ion Pairs | DP * | CE * | CXP * | LOQ * | Linear Range | R2 |
|---|---|---|---|---|---|---|---|
| (m/z) | (V) | (V) | (V) | (ng/mL) | (ng/mL) | ||
| GSH | 433.2 → 304.1 | 46 | 20 | 20 | 0.01 | 0.01–500 | 0.9924 |
| GSSG | 613.2 → 355.2 | 116 | 29 | 24 | 5 | 5–500 | 0.9945 |
| Cys-Gly | 304.1 → 201.0 | 66 | 21 | 12 | 0.1 | 0.1–500 | 0.9992 |
| Glu-Cys | 376.1 → 247.1 | 71 | 21 | 12 | 0.05 | 0.05–500 | 0.9979 |
| Met | 150.2 → 103.8 | 41 | 15 | 8 | 0.1 | 0.1–500 | 0.9973 |
| Hcys | 261.1 → 55.8 | 51 | 41 | 8 | 0.05 | 0.05–500 | 0.9956 |
| Cys | 247.1 → 229.8 | 51 | 19 | 14 | 0.05 | 0.05–500 | 0.9926 |
| Analyte | Recovery (%) | RSD (%) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Spike level of 100 ng/mL | Spike level of 500 ng/mL | |||
| GSH | 115 | 5 | 103 | 1 |
| Spike level of 4 ng/mL | Spike level of 100 ng/mL | |||
| Cys-Gly | 97 | 9 | 105 | 4 |
| Glu-Cys | 95 | 3 | 103 | 2 |
| Met | 88 | 3 | 100 | 4 |
| Cys | 92 | 12 | 105 | 2 |
| GSSG | 126 | 7 | 124 | 4 |
| Hcys | 105 | 1 | 113 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Chernatynskaya, A.; Pfaff, A.; Kou, H.; Cen, N.; Ercal, N.; Shi, H. Extensive Thiol Profiling for Assessment of Intracellular Redox Status in Cultured Cells by HPLC-MS/MS. Antioxidants 2022, 11, 24. https://doi.org/10.3390/antiox11010024
Wu J, Chernatynskaya A, Pfaff A, Kou H, Cen N, Ercal N, Shi H. Extensive Thiol Profiling for Assessment of Intracellular Redox Status in Cultured Cells by HPLC-MS/MS. Antioxidants. 2022; 11(1):24. https://doi.org/10.3390/antiox11010024
Chicago/Turabian StyleWu, Jiandong, Anna Chernatynskaya, Annalise Pfaff, Huari Kou, Nan Cen, Nuran Ercal, and Honglan Shi. 2022. "Extensive Thiol Profiling for Assessment of Intracellular Redox Status in Cultured Cells by HPLC-MS/MS" Antioxidants 11, no. 1: 24. https://doi.org/10.3390/antiox11010024
APA StyleWu, J., Chernatynskaya, A., Pfaff, A., Kou, H., Cen, N., Ercal, N., & Shi, H. (2022). Extensive Thiol Profiling for Assessment of Intracellular Redox Status in Cultured Cells by HPLC-MS/MS. Antioxidants, 11(1), 24. https://doi.org/10.3390/antiox11010024






