Typical 2-Cys Peroxiredoxins as a Defense Mechanism against Metal-Induced Oxidative Stress in the Solitary Ascidian Ciona robusta
Abstract
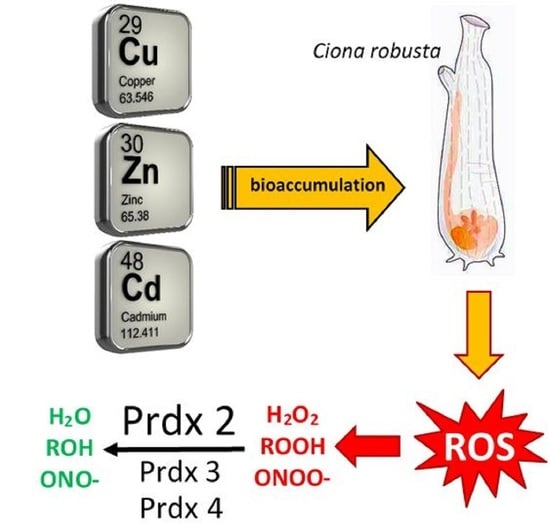
:1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Primers Design, Total RNA Extraction, cDNA Synthesis, Amplification, and Sequencing
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Tissue Activity of 2-Cys Prdxs
2.5. Analyses of Tissue Metal Content
2.6. Statistical Analyses
2.7. Gene and Protein Organization Analyses
2.8. Sequence Alignments and Phylogenetic Analyses
2.9. In Situ Hybridization (ISH)
3. Results
3.1. Gene and Transcript Organizations
3.2. Protein Organizations
3.3. Molecular Phylogeny
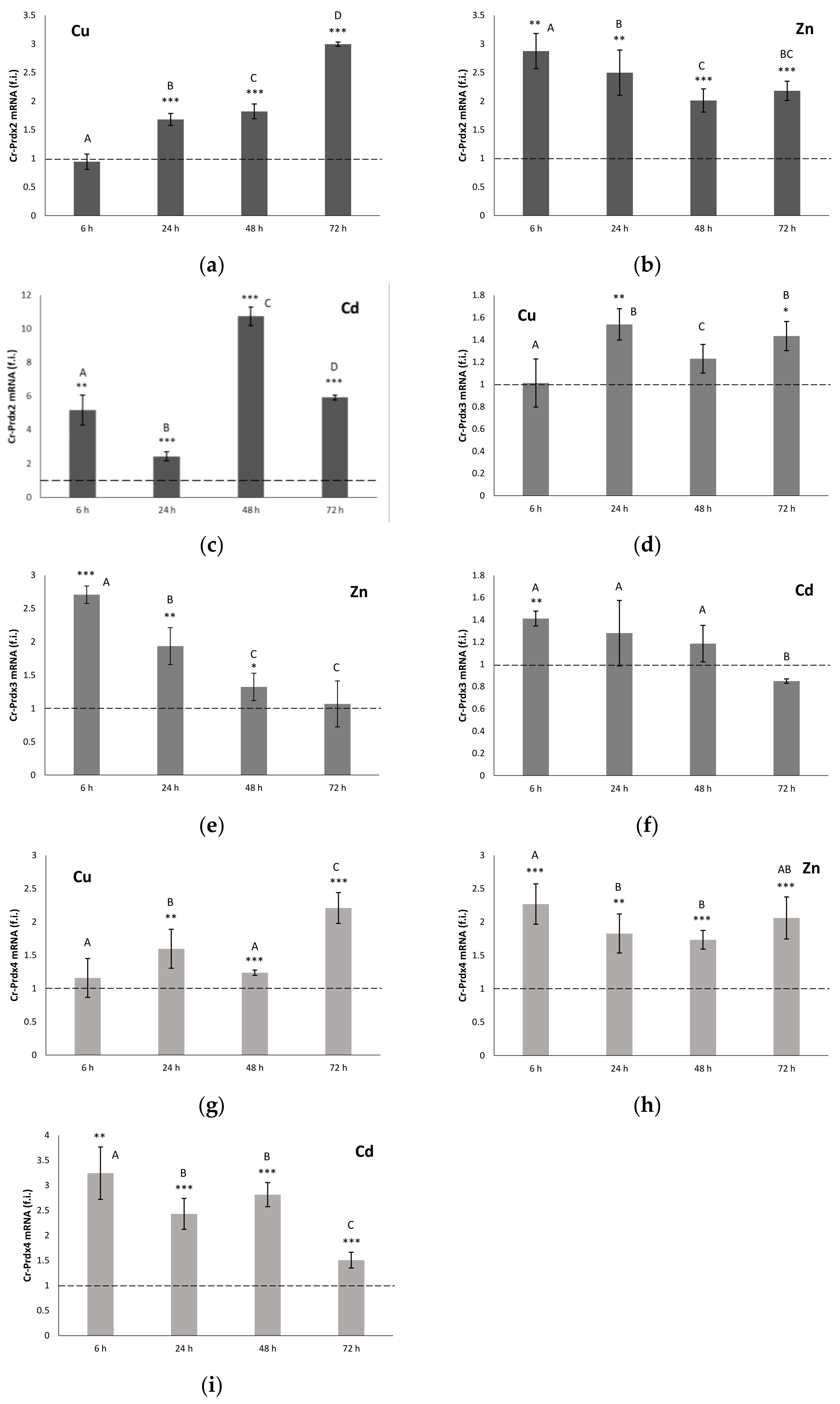
3.4. qRT-PCR
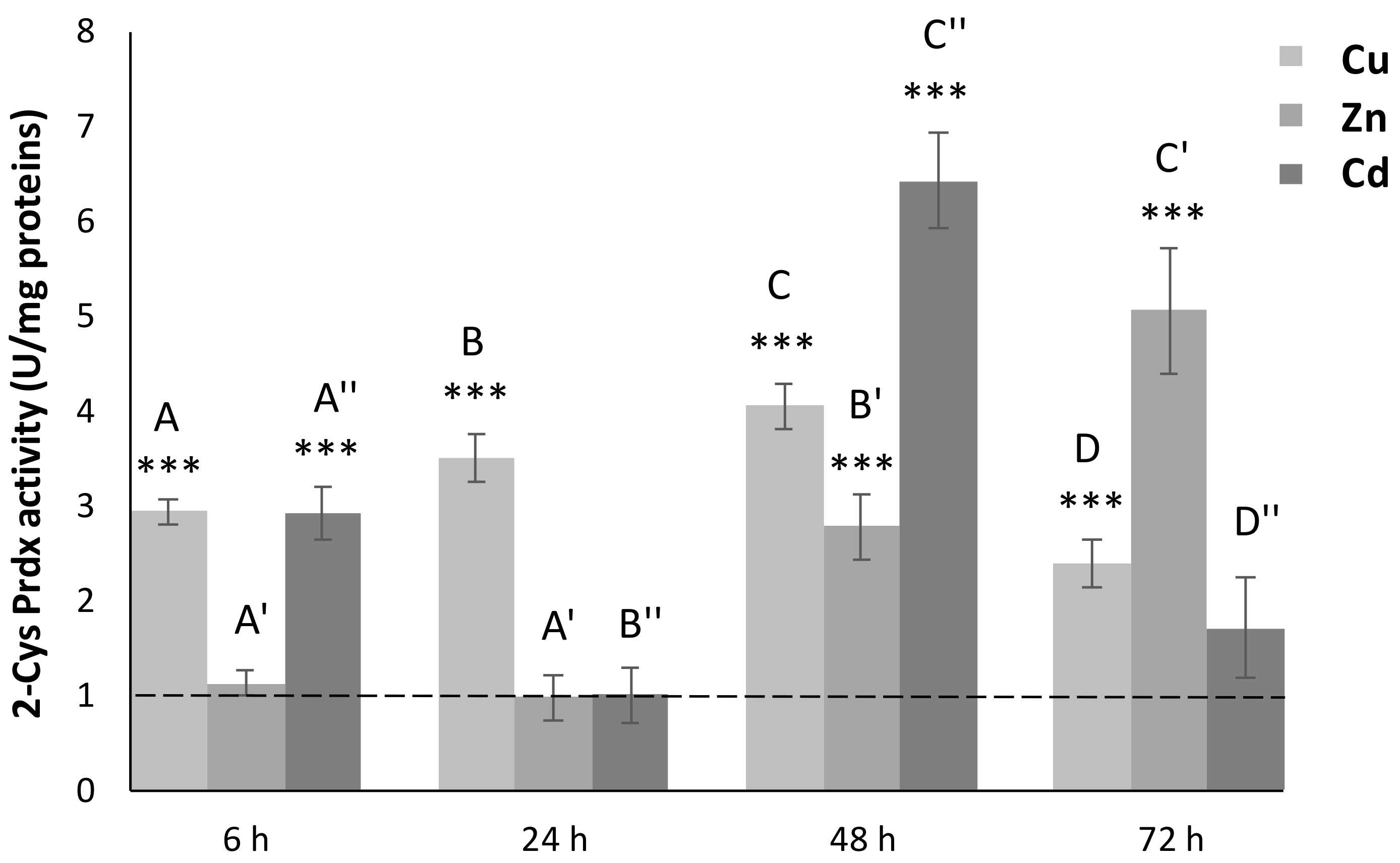
3.5. 2-Cys Prdx Activity
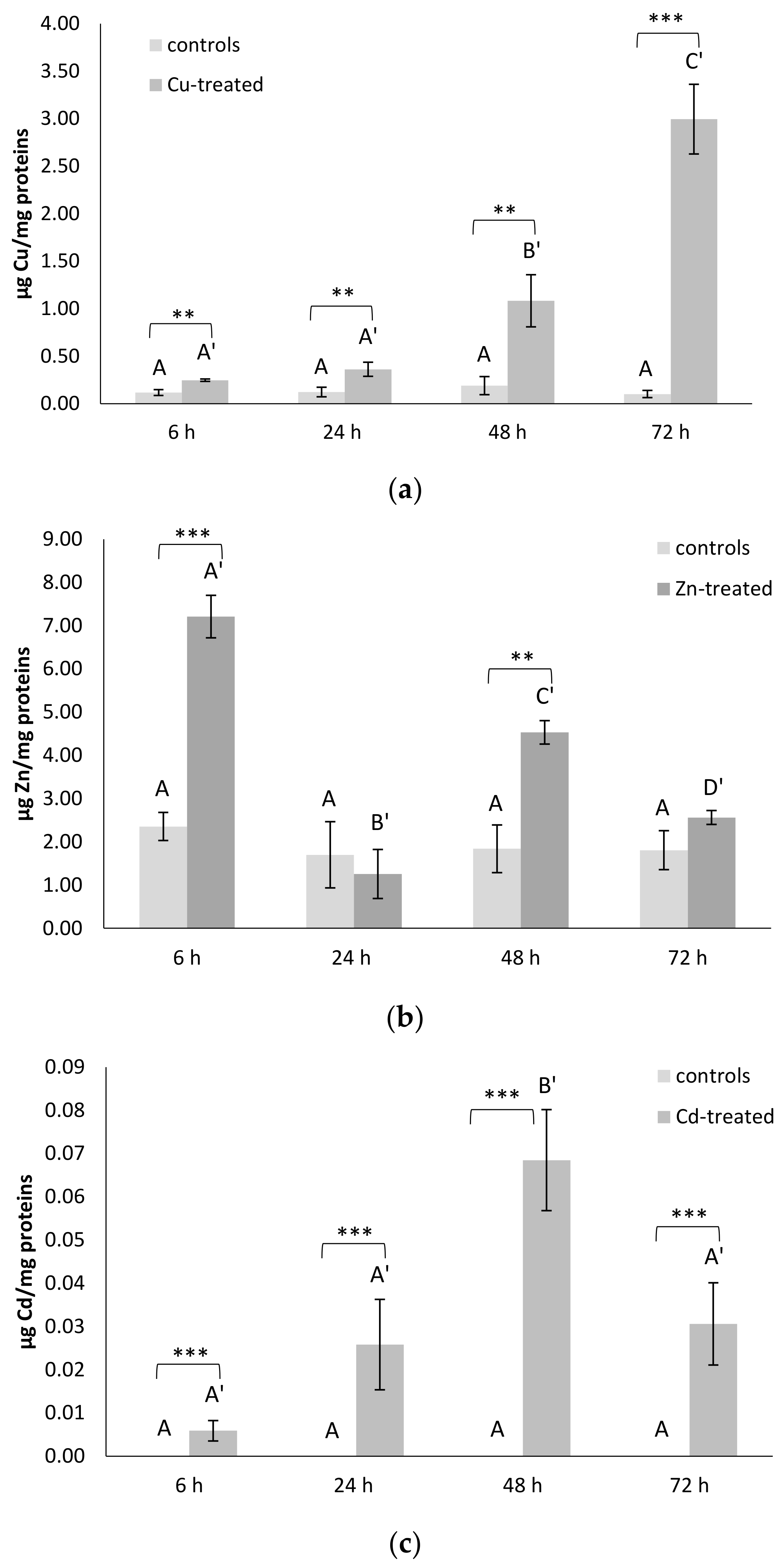
3.6. Metal Contents
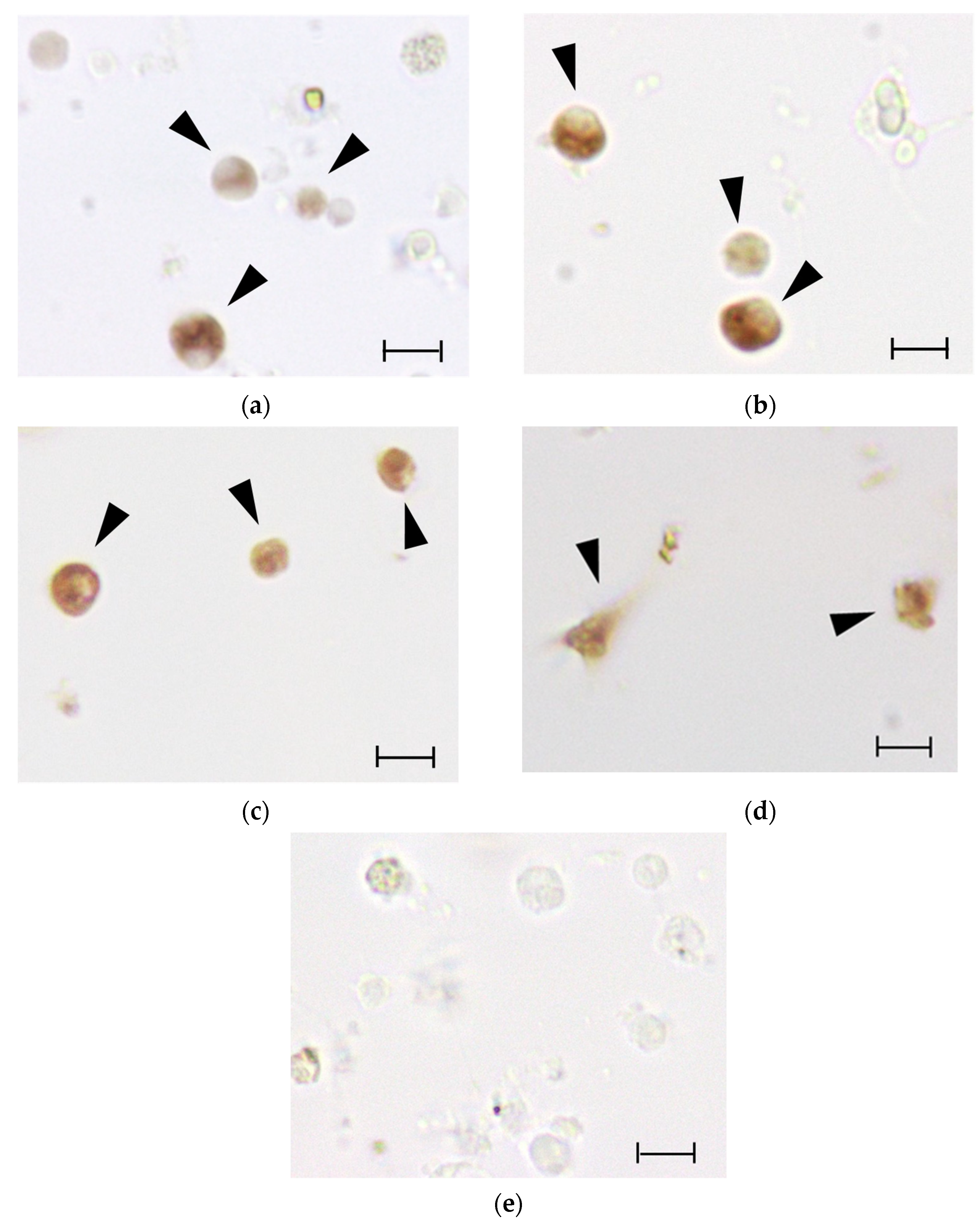
3.7. ISH
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lesser, M.P. Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 2006, 68, 253–278. [Google Scholar] [CrossRef] [Green Version]
- Boldrin, F.; Santovito, G.; Formigari, A.; Bisharyan, Y.; Cassidy-Hanley, D.; Clark, T.G.; Piccinni, E. MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp. Biochem. Physiol. C 2008, 147, 232–240. [Google Scholar] [CrossRef]
- Santovito, G.; Piccinni, E.; Irato, P. An improved method for rapid determination of the reduced and oxidized states of metallothioneins in biological samples. In Marine Pollution: New Research; Tobias, N.H., Ed.; Nova Science Publishers Inc.: New York, NY, USA, 2008; Chapter 3; pp. 101–123. [Google Scholar]
- Santovito, G.; Piccinni, E.; Boldrin, F.; Irato, P. Comparative study on metal homeostasis and detoxification in two Antarctic teleosts. Comp. Biochem. Physiol. C 2012, 155, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Santovito, G.; Boldrin, F.; Irato, P. Metal and metallothionein distribution in different tissues of the Mediterranean clam Venerupis philippinarum during copper treatment and detoxification. Comp. Biochem. Physiol. C 2015, 174–175, 46–53. [Google Scholar] [CrossRef]
- Ferro, K.; Ferro, D.; Corrà, F.; Bakiu, R.; Santovito, G.; Kurtz, J. Cu, Zn SOD genes in Tribolium castaneum: Evolution, molecular characterisation and gene expression during immune priming. Front. Immunol. 2017, 8, 1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, F.; Lauro, F.M.; Grzymski, J.J.; Read, R.; Bakiu, R.; Santovito, G.; Luporini, P.; Vallesi, A. The anti-oxidant defense system of the marine polar ciliate Euplotes nobilii: Characterization of the msrB gene family. Biology 2017, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Garofalo, F.; Santovito, G.; Amelio, D. Morpho-functional effects of heat stress on the gills of Antarctic, T. bernacchii and C. hamatus. Mar. Poll. Bull. 2019, 141, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Bukola, D.; Zaid, A. Consequences of anthropogenic activities on fish and the aquatic environment. Poult. Fish. Wildl. Sci. 2015, 3, 138. [Google Scholar] [CrossRef]
- Dey, S.; Sidor, A.; O’Rourke, B. compartment-specific control of reactive oxygen species scavenging by antioxidant pathway enzymes. J. Biol. Chem. 2016, 291, 11185–11197. [Google Scholar] [CrossRef] [Green Version]
- Abbas, M.N.; Kausar, S.; Cui, H. The biological role of peroxiredoxins in innate immune responses of aquatic invertebrates. Fish Shellfish Immun. 2019, 89, 91–97. [Google Scholar] [CrossRef]
- Detienne, G.; de Haes, W.; Mergan, L.; Edwards, S.L.; Temmerman, L.; van Bael, S. Beyond ROS clearance: Peroxiredoxins in stress signaling and aging. Ageing Res. Rev. 2018, 44, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Fujii, J.; Ikeda, Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002, 7, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Kusakisako, K.; Hernandez, E.P.; Talactac, M.R.; Yoshii, K.; Umemiya-Shirafuji, R.; Fujisaki, K.; Tanaka, T. Peroxiredoxins are important for the regulation of hydrogen peroxide concentrations in ticks and tick cell line. Ticks Tick-Borne Dis. 2018, 9, 872–881. [Google Scholar] [CrossRef]
- Perkins, A.; Nelson, K.J.; Parsonage, D.; Poole, L.B.; Karplus, P.A. Peroxiredoxins: Guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015, 40, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakiu, R.; Santovito, G. New insights into the molecular evolution of metazoan peroxiredoxins. Acta Zool. Bulg 2015, 67, 305–317. [Google Scholar]
- Tolomeo, A.M.; Carraro, A.; Bakiu, R.; Toppo, S.; Place, S.P.; Ferro, D.; Santovito, G. Peroxiredoxin 6 from the Antarctic emerald rockcod: Molecular characterization of its response to warming. J. Comp. Physiol. 2016, 186B, 59–71. [Google Scholar] [CrossRef]
- Al-Asadi, S.; Malik, A.; Bakiu, R.; Santovito, G.; Schuller, K. Characterization of the peroxiredoxin 1 subfamily from Tetrahymena thermophila. Cell. Mol. Life Sci. 2019, 76, 4745–4768. [Google Scholar] [CrossRef]
- Tolomeo, A.M.; Carraro, A.; Bakiu, R.; Toppo, S.; Garofalo, F.; Pellegrino, D.; Gerdol, M.; Ferro, D.; Place, S.P.; Santovito, G. Molecular characterization of novel mitochondrial peroxiredoxins from the Antarctic emerald rockcod and their gene expression in response to environmental warming. Comp. Biochem. Physiol. 2019, C255, 108580. [Google Scholar] [CrossRef]
- Knoops, B.; Loumaye, E.; Van Der Eecken, V.; Knoops, B.; Loumaye, E.; Van Der Eecken, V. Evolution of the peroxiredoxins. In Peroxiredoxin Systems; Flohé, L., Harris, J.R., Eds.; Subcellular Biochemistry; Springer: Dordrecht, The Netherlands, 2007; Volume 44. [Google Scholar] [CrossRef]
- Henderson, D.; Huebner, C.; Markowitz, M.; Taube, N.; Harvanek, Z.M.; Jakob, U.; Knoefler, D. Do developmental temperatures affect redox level and lifespan in C. elegans through upregulation of peroxiredoxin? Redox. Biol. 2018, 14, 386–390. [Google Scholar] [CrossRef]
- Rhee, S.G.; Woo, H.A.; Kil, I.S.; Bae, S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 2012, 287, 4403–4410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poole, L.B.; Nelson, K.J. Distribution and features of the six classes of peroxiredoxins. Mol. Cells. 2016, 39, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Hoyle, N.P.; O’Neill, J.S. Oxidation–reduction cycles of peroxiredoxin proteins and non transcriptional aspects of timekeeping. Biochemistry 2015, 54, 184–193. [Google Scholar] [CrossRef]
- Wood, Z.A.; Schröder, E.; Robin Harris, J.; Poole, L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003, 28, 32–40. [Google Scholar] [CrossRef]
- Fisher, A.B. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase a 2 activities. Antioxid. Redox. Sign. 2011, 15, 838–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, A.; Karplus, P.A.; Poole, L.B. Typical 2-Cys Peroxiredoxins—Structures, mechanisms and functions. FEBS J. 2009, 276, 2469–2477. [Google Scholar] [CrossRef] [Green Version]
- Costa, P.M.; Chicano-Gálvez, E.; Caeiro, S.; Lobo, J.; Martins, M.; Ferreira, A.M.; Caetano, M.; Vale, C.; Alhama-Carmona, J.; Lopez-Barea, J. Hepatic proteome changes in Solea senegalensis exposed to contaminated estuarine sediments: A laboratory and in situ survey. Ecotoxicology 2012, 21, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yu, R.; Li, M.; Liu, L.; Zhang, K.; Wang, Y.; Li, B.; Zhang, L.; Song, G.; Zheng, X. Molecular cloning and characterization of two genes encoding peroxiredoxins from freshwater bivalve Anodonta woodiana: Antioxidative effect and immune defense. Fish. Shellfish. Immun. 2018, 82, 476–491. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.S.; Yu, X.M.; Abbas, M.N.; Li, C.S.; Chu, S.H.; Kausar, S.; Wang, T.T. Essential role of the peroxiredoxin 4 in Procambarus clarkii antioxidant defense and immune responses. Fish Shellfish Immun. 2018, 75, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef]
- Gallo, A.; Tosti, E. The ascidian Ciona intestinalis as model organism for ecotoxicological bioassays. J. Marine. Sci. Res. Dev. 2015, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Dardaillon, J.; Dauga, D.; Simion, P.; Faure, E.; Onuma, T.A.; DeBiasse, M.B.; Louis, A.; Nitta, K.R.; Naville, M.; Besnardeau, L. ANISEED 2019: 4D exploration of genetic data for an extended range of tunicates. Nucleic Acids Res. 2019, 48, D668–D675. [Google Scholar] [CrossRef] [PubMed]
- Franchi, N.; Boldrin, F.; Ballarin, L.; Piccinni, E. CiMT-1, an unusual chordate metallothionein gene in Ciona intestinalis genome: Structure and expression studies. J. Exp. Zool. Part A 2011, 315A, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Franchi, N.; Ferro, D.; Ballarin, L.; Santovito, G. Transcription of genes involved in glutathione biosynthesis in the solitary tunicate Ciona intestinalis exposed to metals. Aquat. Toxicol. 2012, 114–115, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Franchi, N.; Piccinni, E.; Ferro, D.; Basso, G.; Spolaore, B.; Santovito, G.; Ballarin, L. Characterization and transcription studies of a phytochelatin synthase gene from the solitary tunicate Ciona intestinalis exposed to cadmium. Aquat. Toxicol. 2014, 152, 47–56. [Google Scholar] [CrossRef]
- Ferro, D.; Franchi, N.; Mangano, V.; Bakiu, R.; Cammarata, M.; Parrinello, N.; Santovito, G.; Ballarin, L. Characterization and metal-induced gene transcription of two new copper zinc superoxide dismutases in the solitary ascidian Ciona intestinalis. Aquat. Toxicol. 2013, 140–141, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Ferro, D.; Franchi, N.; Bakiu, R.; Ballarin, L.; Santovito, G. Molecular characterization and metal induced gene expression of the novel glutathione peroxidase 7 from the chordate invertebrate Ciona robusta. Comp. Biochem. Phys. C 2018, 205, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Peronato, A.; Franchi, N.; Ballarin, L.; Bakiu, R.; Santovito, G. Stress granules in Ciona robusta: First evidences of tia-1-related nucleolysin and tristetraprolin gene expression under metal exposure. Comp. Biochem. Phys. C 2021, 243, 108977. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Winston, G.W.; Livingstone, D.R.; Lips, F. Oxygen reduction metabolism by the digestive gland of the common marine mussel, Mytilus edulis. J. Exp. Zool 1990, 255, 296–308. [Google Scholar] [CrossRef]
- Bacanskas, L.R.; Whitaker, J.; Di Giulio, R.T. Oxidative stress in two populations of killifish (Fundulus heteroclitus) with differing contaminant exposure histories. Mar. Environ. Res. 2004, 58, 597–601. [Google Scholar] [CrossRef]
- Stach, T.; Braband, A.; Podsiadlowski, L. Erosion of phylogenetic signal in tunicate mitochondrial genomes on different levels of analysis. Mol. Phylogenet. Evol. 2010, 55, 860–870. [Google Scholar] [CrossRef]
- Turbeville, J.M.C.; Schulz, J.R.; Raff, R.A. Deuterostome phylogeny and the sister group of the chordates: Evidence from molecules and morphology. Mol. Biol. Evol. 1994, 11, 648–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatzidimitriou, E.; Bisaccia, P.; Corrà, F.; Bonato, M.; Irato, P.; Manuto, L.; Toppo, S.; Bakiu, R.; Santovito, G. Copper/zinc superoxide dismutase from the crocodile icefish Chionodraco hamatus: Antioxidant defense at constant sub-zero temperature. Antioxidants 2020, 9, 325. [Google Scholar] [CrossRef]
- Ferro, D.; Bakiu, R.; Pucciarelli, S.; Miceli, C.; Vallesi, A.; Irato, P.; Santovito, G. Molecular characterization, protein-protein interaction network, and evolution of four glutathione peroxidases from Tetrahymena thermophila. Antioxidants 2020, 9, 949. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Heller, M.; Gasnier, F.; Luche, S.; Rey, C.; Aebersold, R.; Benahmed, M.; Louisot, P.; Lunardi, J. Proteomics analysis of cellular response to oxidative stress: Evidence for in vivo over-oxidation of peroxiredoxins at their active site. J. Biol. Chem. 2002, 277, 19396–19401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valero, Y.; Martínez-Morcillo, F.J.; Esteban, M.À.; Chaves-Pozo, E.; Cuesta, A. Fish peroxiredoxins and their role in immunity. Biology 2015, 4, 860–880. [Google Scholar] [CrossRef] [Green Version]
- Zito, E.; Melo, E.P.; Yang, Y.; Wahlander, A.; Neubert, T.A.; Ron, D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell 2010, 40, 787–797. [Google Scholar] [CrossRef] [Green Version]
- Zito, E. ERO1: A protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 2015, 83, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Tavender, T.J.; Sheppard, A.M.; Bulleid, N.J. Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 2008, 411, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Radyuk, S.N.; Klichko, V.I.; Michalak, K.; Orr, W.C. The effect of peroxiredoxin 4 on fly physiology is a complex interplay of antioxidant and signaling functions. FASEB J. 2013, 27, 1426–1438. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Shida, R.; Hasegawa, T.; Satoh, M.; Seko, Y.; Tohyama, C.; Kuroda, J.; Shibata, N.; Imura, N.; Himeno, S. Induction of hepatic metallothionein by trivalent cerium: Role of interleukin 6. Biol. Pharm. Bull. 2005, 28, 1859–1863. [Google Scholar] [CrossRef] [Green Version]
- Franchi, N.; Ballarin, L. Immunity in protochordates: The tunicate perspective. Front. Immunol. 2017, 8, 674. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free radicals in biology and medicine. Acta. Crystallogr. D 2017, 73, 384–385. [Google Scholar] [CrossRef]
- Sattin, G.; Bakiu, R.; Tolomeo, A.M.; Carraro, A.; Coppola, D.; Ferro, D.; Patarnello, T.; Santovito, G. Characterization and expression of a new cytoplasmic glutathione peroxidase 1 gene in the antarctic fish Trematomus bernacchii. Hydrobiologia 2015, 761, 363–372. [Google Scholar] [CrossRef]
- Waris, S.; Wilce, M.; Wilce, J. RNA recognition and stress granule formation by TIA proteins. Int. J. Mol. Sci. 2014, 15, 23377–23388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavut, A.; Raveh, D. Sequestration of highly expressed mrnas in cytoplasmic granules, p-bodies, and stress granules enhances cell viability. PLoS Genet. 2012, 8, e1002527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Treeck, B.; Parker, R. Principles of stress granules revealed by imaging approaches. CSH Perspect. Biol. 2019, 11, a033068. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drago, L.; Ferro, D.; Bakiu, R.; Ballarin, L.; Santovito, G. Typical 2-Cys Peroxiredoxins as a Defense Mechanism against Metal-Induced Oxidative Stress in the Solitary Ascidian Ciona robusta. Antioxidants 2022, 11, 93. https://doi.org/10.3390/antiox11010093
Drago L, Ferro D, Bakiu R, Ballarin L, Santovito G. Typical 2-Cys Peroxiredoxins as a Defense Mechanism against Metal-Induced Oxidative Stress in the Solitary Ascidian Ciona robusta. Antioxidants. 2022; 11(1):93. https://doi.org/10.3390/antiox11010093
Chicago/Turabian StyleDrago, Laura, Diana Ferro, Rigers Bakiu, Loriano Ballarin, and Gianfranco Santovito. 2022. "Typical 2-Cys Peroxiredoxins as a Defense Mechanism against Metal-Induced Oxidative Stress in the Solitary Ascidian Ciona robusta" Antioxidants 11, no. 1: 93. https://doi.org/10.3390/antiox11010093
APA StyleDrago, L., Ferro, D., Bakiu, R., Ballarin, L., & Santovito, G. (2022). Typical 2-Cys Peroxiredoxins as a Defense Mechanism against Metal-Induced Oxidative Stress in the Solitary Ascidian Ciona robusta. Antioxidants, 11(1), 93. https://doi.org/10.3390/antiox11010093