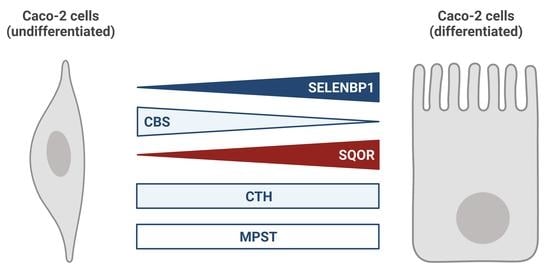
Altered Capacity for H2S Production during the Spontaneous Differentiation of Caco-2 Cells to Colonocytes Due to Reciprocal Regulation of CBS and SELENBP1
Abstract
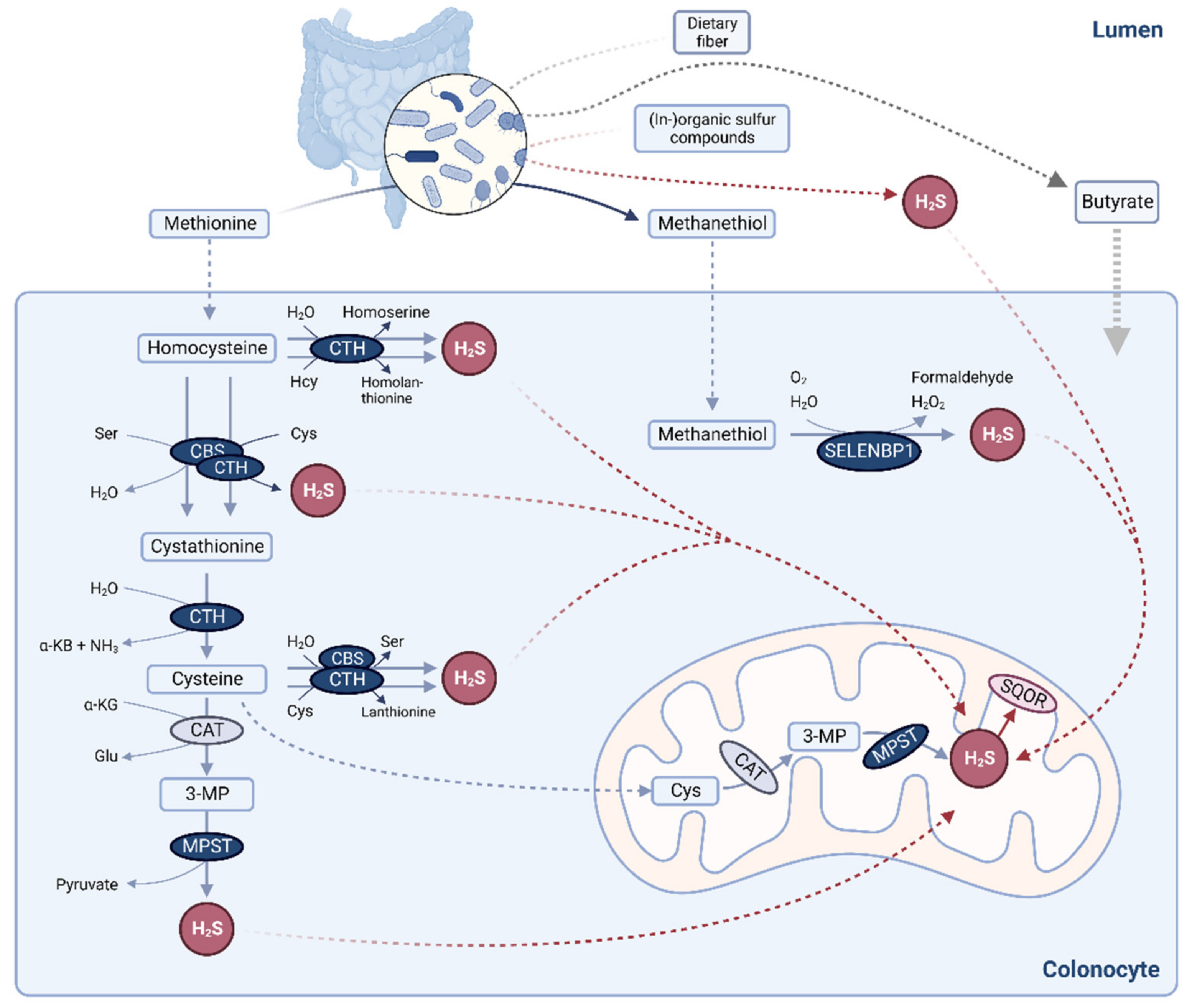
:1. Introduction
2. Materials and Methods
2.1. Culture and Differentiation of Caco-2 Cells
2.2. Assessment of SELENBP1 Promoter Activity
2.3. RNA Isolation and Real-Time RT-PCR (qRT-PCR) Analysis
2.4. SDS-PAGE and Immunoblot Analysis
2.5. Determination of H2S Production in Caco-2 Cell Lysates
2.6. Statistical Analysis
3. Results and Discussion
3.1. Alterations in mRNA Levels of H2S-Modulating Enzymes during Spontaneous Differentiation of Caco-2 Cells
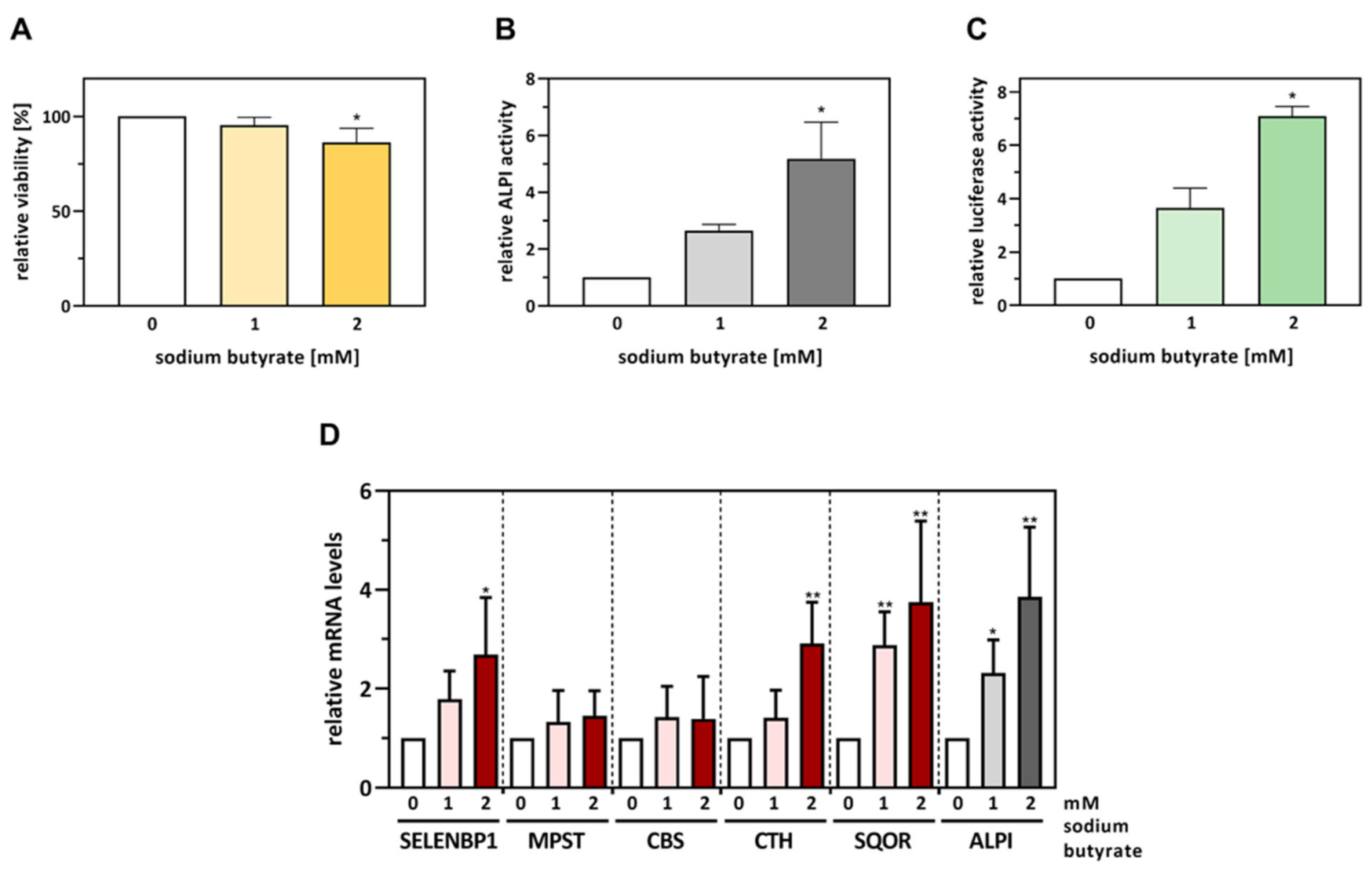
3.2. SELENBP1 Promoter Activity Is Stimulated in Caco-2 Cells Undergoing Butyrate-Induced Differentiation
3.3. The Effects of Spontaneous Differentiation on mRNA and Protein Levels of H2S-Modulating Enzymes Are Only Partially Mimicked by Butyrate Exposure
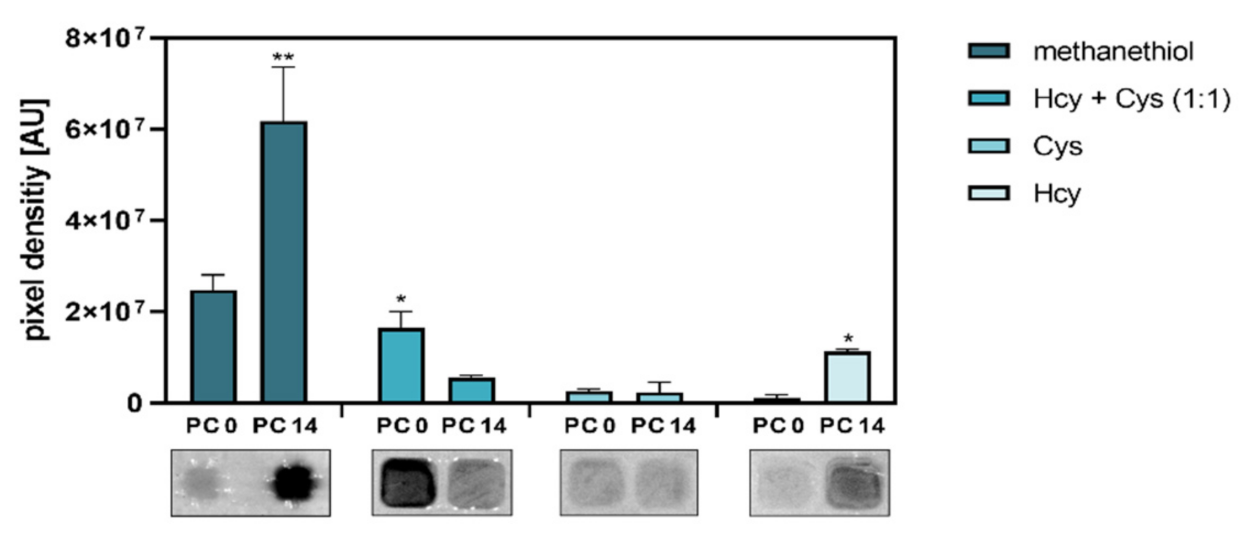
3.4. Differentiation of Caco-2 Cells Alters Their Capacity for Substrate-Dependent H2S Generation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical Biology of H2S Signaling through Persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef] [PubMed]
- Medani, M.; Collins, D.; Docherty, N.; Baird, A.; O’connell, P.R.; Winter, D.C. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm. Bowel Dis. 2011, 17, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, S.E.; Borland, G.; Carter, R.N.; Morton, N.M.; Selman, C. Hydrogen sulfide in ageing, longevity and disease. Biochem. J. 2021, 478, 3485–3504. [Google Scholar] [CrossRef] [PubMed]
- Barton, L.L.; Ritz, N.L.; Fauque, G.D.; Lin, H.C. Sulfur Cycling and the Intestinal Microbiome. Am. J. Dig. Dis. 2017, 62, 2241–2257. [Google Scholar] [CrossRef] [PubMed]
- Kabil, O.; Banerjee, R. Enzymology of H2S Biogenesis, Decay and Signaling. Antioxidants Redox Signal. 2014, 20, 770–782. [Google Scholar] [CrossRef]
- Pol, A.; Renkema, G.H.; Tangerman, A.; Winkel, E.G.; Engelke, U.F.; De Brouwer, A.P.M.; Lloyd, K.C.; Araiza, R.S.; Heuvel, L.V.D.; Omran, H.; et al. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018, 50, 120–129. [Google Scholar] [CrossRef]
- Philipp, T.M.; Will, A.; Richter, H.; Winterhalter, P.R.; Pohnert, G.; Steinbrenner, H.; Klotz, L.-O. A coupled enzyme assay for detection of selenium-binding protein 1 (SELENBP1) methanethiol oxidase (MTO) activity in mature enterocytes. Redox Biol. 2021, 43, 101972. [Google Scholar] [CrossRef]
- Li, T.; Yang, W.; Li, M.; Byun, D.-S.; Tong, C.; Nasser, S.; Zhuang, M.; Arango, D.; Mariadason, J.M.; Augenlicht, L.H. Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Mol. Nutr. Food Res. 2008, 52, 1289–1299. [Google Scholar] [CrossRef]
- Portune, K.J.; Beaumont, M.; Davila, A.-M.; Tomé, D.; Blachier, F.; Sanz, Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends Food Sci. Technol. 2016, 57, 213–232. [Google Scholar] [CrossRef]
- Levitt, M.D.; Furne, J.; Springfield, J.; Suarez, F.; DeMaster, E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J. Clin. Investig. 1999, 104, 1107–1114. [Google Scholar] [CrossRef] [Green Version]
- Suarez, F.L.; Springfield, J.; Levitt, M.D. Identification of gases responsible for the odour of human flatus and evaluation of a device purported to reduce this odour. Gut 1998, 43, 100–104. [Google Scholar] [CrossRef]
- Goubern, M.; Andriamihaja, M.; Nübel, T.; Blachier, F.; Bouillaud, F. Sulfide, the first inorganic substrate for human cells. FASEB J. 2007, 21, 1699–1706. [Google Scholar] [CrossRef]
- Libiad, M.; Vitvitsky, V.; Bostelaar, T.; Bak, D.W.; Lee, H.-J.; Sakamoto, N.; Fearon, E.; Lyssiotis, C.A.; Weerapana, E.; Banerjee, R. Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 2019, 294, 12077–12090. [Google Scholar] [CrossRef]
- Landry, A.P.; Ballou, D.P.; Banerjee, R. Hydrogen Sulfide Oxidation by Sulfide Quinone Oxidoreductase. ChemBioChem 2021, 22, 949–960. [Google Scholar] [CrossRef]
- Engle, M.J.; Goetz, G.S.; Alpers, D.H. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J. Cell. Physiol. 1998, 174, 362–369. [Google Scholar] [CrossRef]
- Mariadason, J.M.; Arango, D.; Corner, G.A.; Arañes, M.J.; Hotchkiss, K.A.; Yang, W.; Augenlicht, L.H. A gene expression profile that defines colon cell maturation in vitro. Cancer Res. 2002, 62, 4791–4804. [Google Scholar]
- Sääf, A.M.; Halbleib, J.M.; Chen, X.; Yuen, S.T.; Leung, S.Y.; Nelson, W.J.; Brown, P.O. Parallels between Global Transcriptional Programs of Polarizing Caco-2 Intestinal Epithelial Cells In Vitro and Gene Expression Programs in Normal Colon and Colon Cancer. Mol. Biol. Cell 2007, 18, 4245–4260. [Google Scholar] [CrossRef]
- Klepinina, L.; Klepinin, A.; Truu, L.; Chekulayev, V.; Vija, H.; Kuus, K.; Teino, I.; Pook, M.; Maimets, T.; Kaambre, T. Colon cancer cell differentiation by sodium butyrate modulates metabolic plasticity of Caco-2 cells via alteration of phosphotransfer network. PLoS ONE 2021, 16, e0245348. [Google Scholar] [CrossRef]
- Mariadason, J.M.; Velcich, A.; Wilson, A.J.; Augenlicht, L.H.; Gibson, P.R. Resistance to butyrate-induced cell differentiation and apoptosis during spontaneous Caco-2 cell differentiation. Gastroenterology 2001, 120, 889–899. [Google Scholar] [CrossRef]
- Mariadason, J.M.; Rickard, K.L.; Barkla, D.H.; Augenlicht, L.H.; Gibson, P.R. Divergent phenotypic patterns and commitment to apoptosis of Caco-2 cells during spontaneous and butyrate-induced differentiation. J. Cell. Physiol. 2000, 183, 347–354. [Google Scholar] [CrossRef]
- Speckmann, B.; Bidmon, H.-J.; Pinto, A.; Anlauf, M.; Sies, H.; Steinbrenner, H. Induction of Glutathione Peroxidase 4 Expression during Enterocytic Cell Differentiation. J. Biol. Chem. 2011, 286, 10764–10772. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.W.; Tsui, S.K.; Liew, C.; Lee, C.; Waye, M.M.; Fung, K. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. J. Cell. Biochem. 1997, 64, 217–224. [Google Scholar] [CrossRef]
- Tsitsipatis, D.; Gopal, K.; Steinbrenner, H.; Klotz, L.-O. FOXO1 cysteine-612 mediates stimulatory effects of the coregulators CBP and PGC1α on FOXO1 basal transcriptional activity. Free Radic. Biol. Med. 2018, 118, 98–107. [Google Scholar] [CrossRef]
- Tauber, S.; Sieckmann, M.; Erler, K.; Stahl, W.; Klotz, L.-O.; Steinbrenner, H. Activation of Nrf2 by Electrophiles Is Largely Independent of the Selenium Status of HepG2 Cells. Antioxidants 2021, 10, 167. [Google Scholar] [CrossRef]
- Dydensborg, A.B.; Herring, E.; Auclair, J.; Tremblay, E.; Beaulieu, J.-F. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am. J. Physiol. Liver Physiol. 2006, 290, G1067–G1074. [Google Scholar] [CrossRef]
- Fosang, A.; Colbran, R. Transparency Is the Key to Quality. J. Biol. Chem. 2015, 290, 29692–29694. [Google Scholar] [CrossRef]
- Elhodaky, M.; Diamond, A.M. Selenium-Binding Protein 1 in Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3437. [Google Scholar] [CrossRef]
- Sen, N. Functional and Molecular Insights of Hydrogen Sulfide Signaling and Protein Sulfhydration. J. Mol. Biol. 2017, 429, 543–561. [Google Scholar] [CrossRef]
- Matsumoto, H.; Erickson, R.H.; Gum, J.R.; Yoshioka, M.; Gum, E.; Kim, Y.S. Biosynthesis of Alkaline Phosphatase During Differentiation of the Human Colon Cancer Cell Line Caco-2. Gastroenterology 1990, 98, 1199–1207. [Google Scholar] [CrossRef]
- Mimoun, S.; Andriamihaja, M.; Chaumontet, C.; Atanasiu, C.; Benamouzig, R.; Blouin, J.M.; Tomé, D.; Bouillaud, F.; Blachier, F. Detoxification of H2S by Differentiated Colonic Epithelial Cells: Implication of the Sulfide Oxidizing Unit and of the Cell Respiratory Capacity. Antioxidants Redox Signal. 2012, 17, 1–10. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, R.; Bei, L.; Hu, Z.; Yang, X.; Song, T.; Chen, L.; Meng, H.; Niu, G.; Ke, C. SELENBP1 inhibits progression of colorectal cancer by suppressing epithelial–mesenchymal transition. Open Med. 2022, 17, 1390–1404. [Google Scholar] [CrossRef] [PubMed]
- Szabo, C.; Coletta, C.; Chao, C.; Módis, K.; Szczesny, B.; Papapetropoulos, A.; Hellmich, M.R. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 12474–12479. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.M.; Zatarain, J.R.; Nicholls, M.E.; Porter, C.; Widen, S.G.; Thanki, K.; Johnson, P.; Jawad, M.U.; Moyer, M.P.; Randall, J.W.; et al. Upregulation of Cystathionine-β-Synthase in Colonic Epithelia Reprograms Metabolism and Promotes Carcinogenesis. Cancer Res. 2017, 77, 5741–5754. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, Y.; Yang, X.; Jiang, Y. Selenium-binding protein 1 is associated with the degree of colorectal cancer differentiation and is regulated by histone modification. Oncol. Rep. 2014, 31, 2506–2514. [Google Scholar] [CrossRef]
- Yamagishi, K.; Onuma, K.; Chiba, Y.; Yagi, S.; Aoki, S.; Sato, T.; Sugawara, Y.; Hosoya, N.; Saeki, Y.; Takahashi, M.; et al. Generation of gaseous sulfur-containing compounds in tumour tissue and suppression of gas diffusion as an antitumour treatment. Gut 2012, 61, 554–561. [Google Scholar] [CrossRef]
- Funato, K.; Abe, T.; Kurita, R.; Watanabe, Y.; Nakamura, Y.; Miyata, S.; Furukawa, Y.; Satake, M. Identification of characteristic proteins at late-stage erythroid differentiation in vitro. Hum. Cell 2021, 34, 745–749. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Micoogullari, M.; Hoang, N.A.; Bergheim, I.; Klotz, L.-O.; Sies, H. Selenium-binding protein 1 (SELENBP1) is a marker of mature adipocytes. Redox Biol. 2019, 20, 489–495. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Singh, S.; Padovani, D.; Leslie, R.A.; Chiku, T.; Banerjee, R. Relative Contributions of Cystathionine β-Synthase and γ-Cystathionase to H2S Biogenesis via Alternative Trans-sulfuration Reactions. J. Biol. Chem. 2009, 284, 22457–22466. [Google Scholar] [CrossRef]
- Chiku, T.; Padovani, D.; Zhu, W.; Singh, S.; Vitvitsky, V.; Banerjee, R. H2S Biogenesis by Human Cystathionine γ-Lyase Leads to the Novel Sulfur Metabolites Lanthionine and Homolanthionine and Is Responsive to the Grade of Hyperhomocysteinemia. J. Biol. Chem. 2009, 284, 11601–11612. [Google Scholar] [CrossRef]
- Kožich, V.; Krijt, J.; Sokolová, J.; Melenovská, P.; Ješina, P.; Vozdek, R.; Majtán, T.; Kraus, J.P. Thioethers as markers of hydrogen sulfide production in homocystinurias. Biochimie 2016, 126, 14–20. [Google Scholar] [CrossRef]



| Gene | Gene-ID | Forward Primer | Reverse Primer |
|---|---|---|---|
| ALPI | NM_001631 | cctgccgttggaccttcac | acaaatctgcagtctctgggg |
| CAT | NM_002079 | gctgaccggattctgaccat | cgttgattcgaccacttggc |
| CBS | NM_000071 | gtttgagcggtgctgaactg | tatcctctggggaccccttc |
| CTH | NM_001902 | ccagcactcgggttttgaat | gtgctgccactgctttttca |
| MPST | NM_021126 | catggcggagccaggaag | gtagatcacgacgtgggtgg |
| RPLP0 | NM_001002 | acatgctcaacatctccccc | caggactcgtttgtacccgt |
| SELENBP1 | NM_003944 | cagcgcttctacaagaacga | tgatcaggcctggcattt |
| SQOR | NM_021199 | gaagggagcgaaggtttttgc | gtctctcactgggctcaaca |
| Antibody | Supplier | Catalog Number |
|---|---|---|
| rabbit anti-CBS | Cell Signaling Technology | 14,782 |
| rabbit anti-CTH | Sigma-Aldrich | HPA023300 |
| mouse anti-GAPDH | Sigma-Aldrich | G8795 |
| rabbit anti-MPST | Sigma-Aldrich | HPA001240 |
| mouse anti-SELENBP1 | Medical & Biological Laboratories (Nagoya, Japan) | M061-3 |
| rabbit anti-SQOR | Sigma-Aldrich | HPA017079 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scheller, A.S.; Philipp, T.M.; Klotz, L.-O.; Steinbrenner, H. Altered Capacity for H2S Production during the Spontaneous Differentiation of Caco-2 Cells to Colonocytes Due to Reciprocal Regulation of CBS and SELENBP1. Antioxidants 2022, 11, 1957. https://doi.org/10.3390/antiox11101957
Scheller AS, Philipp TM, Klotz L-O, Steinbrenner H. Altered Capacity for H2S Production during the Spontaneous Differentiation of Caco-2 Cells to Colonocytes Due to Reciprocal Regulation of CBS and SELENBP1. Antioxidants. 2022; 11(10):1957. https://doi.org/10.3390/antiox11101957
Chicago/Turabian StyleScheller, Anne Sophie, Thilo Magnus Philipp, Lars-Oliver Klotz, and Holger Steinbrenner. 2022. "Altered Capacity for H2S Production during the Spontaneous Differentiation of Caco-2 Cells to Colonocytes Due to Reciprocal Regulation of CBS and SELENBP1" Antioxidants 11, no. 10: 1957. https://doi.org/10.3390/antiox11101957