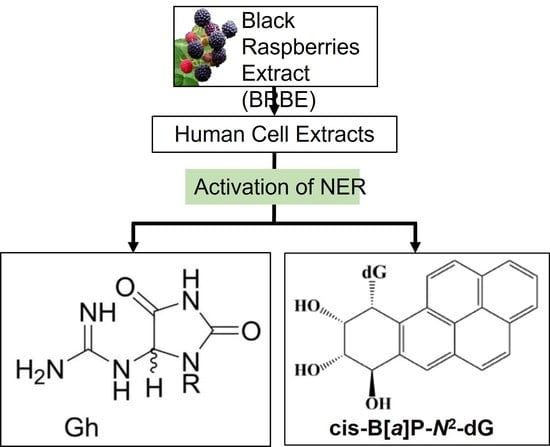
Treatment of Human HeLa Cells with Black Raspberry Extracts Enhances the Removal of DNA Lesions by the Nucleotide Excision Repair Mechanism
Abstract
1. Introduction
1.1. BRBE Preparation and Dosage
1.2. DNA Substrates
1.3. Cell Culture and BRBE Treatment
1.4. Mini-Whole Cell Extract Preparation and NER Excision Product Assays
1.5. Western Blots
2. Results
2.1. Effects of BRBE Pretreatment of HeLa Cells on the Repair of Oxidatively Generated Gh DNA Lesions
2.2. Effects of BRBE Pretreatment of Human Cells on the NER of Bulky BP-dG Adducts
2.3. Quantitative Comparisons of DNA Repair Yield Averages
2.4. Impact of BRBE Treatment on the Expression of the NER Factors XPA, XPB and XPD
3. Discussion
3.1. BRBE and Cancer Prevention
3.2. The BP-dG and Gh Lesions as Model Systems
3.3. Impact of BRBE on the Expression of NER Proteins
4. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- da Silva, S.D.; Hier, M.; Mlynarek, A.; Kowalski, L.P.; Alaoui-Jamali, M.A. Recurrent oral cancer: Current and emerging therapeutic approaches. Front. Pharmacol. 2012, 3, 149. [Google Scholar] [CrossRef] [PubMed]
- El-Bayoumy, K.; Chen, K.M.; Zhang, S.M.; Sun, Y.W.; Amin, S.; Stoner, G.; Guttenplan, J.B. Carcinogenesis of the Oral Cavity: Environmental Causes and Potential Prevention by Black Raspberry. Chem. Res. Toxicol. 2017, 30, 126–144. [Google Scholar] [CrossRef] [PubMed]
- Conney, A.H. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982, 42, 4875–4917. [Google Scholar]
- Straif, K.; Baan, R.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Cogliano, V. Carcinogenicity of polycyclic aromatic hydrocarbons. Lancet Oncol. 2005, 6, 931–932. [Google Scholar] [CrossRef]
- Jeffrey, A.M.; Jennette, K.W.; Blobstein, S.H.; Weinstein, I.B.; Beland, F.A.; Harvey, R.G.; Kasal, H.; Miura, I.; Nakanishi, K. Letter: Benzo[a]pyrene-nucleic acid derivative found in vivo: Structure of a benzo[a]pyrenetetrahydrodiol epoxide-guanosine adduct. J. Am. Chem. Soc. 1976, 98, 5714–5715. [Google Scholar] [CrossRef]
- Oghumu, S.; Casto, B.C.; Ahn-Jarvis, J.; Weghorst, L.C.; Maloney, J.; Geuy, P.; Horvath, K.Z.; Bollinger, C.E.; Warner, B.M.; Summersgill, K.F.; et al. Inhibition of Pro-inflammatory and Anti-apoptotic Biomarkers during Experimental Oral Cancer Chemoprevention by Dietary Black Raspberries. Front. Immunol. 2017, 8, 1325. [Google Scholar] [CrossRef]
- Mallery, S.R.; Tong, M.; Shumway, B.S.; Curran, A.E.; Larsen, P.E.; Ness, G.M.; Kennedy, K.S.; Blakey, G.H.; Kushner, G.M.; Vickers, A.M.; et al. Topical application of a mucoadhesive freeze-dried black raspberry gel induces clinical and histologic regression and reduces loss of heterozygosity events in premalignant oral intraepithelial lesions: Results from a multicentered, placebo-controlled clinical trial. Clin. Cancer Res. 2014, 20, 1910–1924. [Google Scholar] [CrossRef]
- Kresty, L.A.; Mallery, S.R.; Stoner, G.D. Black raspberries in cancer clinical trials: Past, present and future. J. Berry Res. 2016, 6, 251–261. [Google Scholar] [CrossRef]
- Casto, B.C.; Kresty, L.A.; Kraly, C.L.; Pearl, D.K.; Knobloch, T.J.; Schut, H.A.; Stoner, G.D.; Mallery, S.R.; Weghorst, C.M. Chemoprevention of oral cancer by black raspberries. Anticancer Res. 2002, 22, 4005–4015. [Google Scholar]
- Geacintov, N.E.; Broyde, S. Repair-Resistant DNA Lesions. Chem. Res. Toxicol. 2017, 30, 1517–1548. [Google Scholar] [CrossRef]
- Buterin, T.; Hess, M.T.; Luneva, N.; Geacintov, N.E.; Amin, S.; Kroth, H.; Seidel, A.; Naegeli, H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000, 60, 1849–1856. [Google Scholar] [PubMed]
- Hess, M.T.; Gunz, D.; Luneva, N.; Geacintov, N.E.; Naegeli, H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997, 17, 7069–7076. [Google Scholar] [CrossRef]
- Cosman, M.; Fiala, R.; Hingerty, B.E.; Amin, S.; Geacintov, N.E.; Broyde, S.; Patel, D.J. Solution conformation of the (+)-cis-anti-[BP] dG adduct opposite a deletion site in a DNA duplex: Intercalation of the covalently attached benzo [a] pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove. Biochemistry 1994, 33, 11518–11527. [Google Scholar] [CrossRef]
- Mocquet, V.; Kropachev, K.; Kolbanovskiy, M.; Kolbanovskiy, A.; Tapias, A.; Cai, Y.; Broyde, S.; Geacintov, N.E.; Egly, J.M. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo [a] pyrenyl-DNA lesions. EMBO J. 2007, 26, 2923–2932. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.; Di Mascio, P.; Wagner, J.R. Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 2017, 107, 13–34. [Google Scholar] [CrossRef]
- Wei, Y.; Han, I.K.; Hu, M.; Shao, M.; Zhang, J.J.; Tang, X. Personal exposure to particulate PAHs and anthraquinone and oxidative DNA damages in humans. Chemosphere 2010, 81, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Muller, J.G.; Dlouhy, A.C.; Burrows, C.J. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine to hydantoin products: Electrostatics, base stacking, and base pairing. J. Am. Chem. Soc. 2012, 134, 15091–15102. [Google Scholar] [CrossRef]
- Mangerich, A.; Knutson, C.G.; Parry, N.M.; Muthupalani, S.; Ye, W.; Prestwich, E.; Cui, L.; McFaline, J.L.; Mobley, M.; Ge, Z.; et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E1820–E1829. [Google Scholar] [CrossRef]
- Shafirovich, V.; Kropachev, K.; Anderson, T.; Liu, Z.; Kolbanovskiy, M.; Martin, B.D.; Sugden, K.; Shim, Y.; Chen, X.; Min, J.H.; et al. Base and nucleotide excision repair of oxidatively generated guanine lesions in DNA. J. Biol. Chem. 2016, 291, 5309–5319. [Google Scholar] [CrossRef]
- Peiffer, D.S.; Zimmerman, N.P.; Wang, L.S.; Ransom, B.W.; Carmella, S.G.; Kuo, C.T.; Siddiqui, J.; Chen, J.H.; Oshima, K.; Huang, Y.W.; et al. Chemoprevention of esophageal cancer with black raspberries, their component anthocyanins, and a major anthocyanin metabolite, protocatechuic acid. Cancer Prev. Res. 2014, 7, 574–584. [Google Scholar] [CrossRef]
- Guttenplan, J.B.; Chen, K.M.; Sun, Y.W.; Lajara, B.; Shalaby, N.A.E.; Kosinska, W.; Benitez, G.; Gowda, K.; Amin, S.; Stoner, G.; et al. Effects of Black Raspberry Extract and Berry Compounds on Repair of DNA Damage and Mutagenesis Induced by Chemical and Physical Agents in Human Oral Leukoplakia and Rat Oral Fibroblasts. Chem. Res. Toxicol. 2017, 30, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, J.B.; Chen, K.M.; Sun, Y.W.; Kosinska, W.; Zhou, Y.; Kim, S.A.; Sung, Y.; Gowda, K.; Amin, S.; Stoner, G.D.; et al. Effects of Black Raspberry Extract and Protocatechuic Acid on Carcinogen-DNA Adducts and Mutagenesis, and Oxidative Stress in Rat and Human Oral Cells. Cancer Prev. Res. 2016, 9, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Kolbanovskiy, A.; Zhuang, P.; Chen, J.; Krzeminski, J.; Amin, S.; Geacintov, N.E. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N6-adenine adducts: Unusual thermal stabilization of modified DNA duplexes. Chem. Res. Toxicol. 2002, 15, 249–261. [Google Scholar] [CrossRef]
- Kolbanovskiy, M.; Shim, Y.; Min, J.H.; Geacintov, N.E.; Shafirovich, V. Inhibition of Excision of Oxidatively Generated Hydantoin DNA Lesions by NEIL1 by the Competitive Binding of the Nucleotide Excision Repair Factor XPC-RAD23B. Biochemistry 2020, 59, 1728–1736. [Google Scholar] [CrossRef] [PubMed]
- Kolbanovskiy, M.; Aharonoff, A.; Sales, A.H.; Geacintov, N.E.; Shafirovich, V. Remarkable Enhancement of Nucleotide Excision Repair of a Bulky Guanine Lesion in a Covalently Closed Circular DNA Plasmid Relative to the Same Linearized Plasmid. Biochemistry 2020, 59, 2842–2848. [Google Scholar] [CrossRef]
- Smeaton, M.B.; Miller, P.S.; Ketner, G.; Hanakahi, L.A. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007, 35, e152. [Google Scholar] [CrossRef][Green Version]
- Reardon, J.T.; Sancar, A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006, 408, 189–213. [Google Scholar] [CrossRef]
- Kolbanovskiy, M.; Aharonoff, A.; Sales, A.H.; Geacintov, N.E.; Shafirovich, V. Base and Nucleotide Excision Repair Pathways in DNA Plasmids Harboring Oxidatively Generated Guanine Lesions. Chem. Res. Toxicol. 2021, 34, 154–160. [Google Scholar] [CrossRef]
- Shafirovich, V.; Kropachev, K.; Kolbanovskiy, M.; Geacintov, N.E. Excision of Oxidatively Generated Guanine Lesions by Competing Base and Nucleotide Excision Repair Mechanisms in Human Cells. Chem. Res. Toxicol. 2019, 32, 753–761. [Google Scholar] [CrossRef]
- Scharer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef]
- Kropachev, K.; Kolbanovskii, M.; Cai, Y.; Rodriguez, F.; Kolbanovskii, A.; Liu, Y.; Zhang, L.; Amin, S.; Patel, D.; Broyde, S.; et al. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: Insights into the structural factors that favor dual incisions. J. Mol. Biol. 2009, 386, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Kropachev, K.; Kolbanovskiy, M.; Liu, Z.; Cai, Y.; Zhang, L.; Schwaid, A.G.; Kolbanovskiy, A.; Ding, S.; Amin, S.; Broyde, S.; et al. Adenine-DNA adducts derived from the highly tumorigenic Dibenzo[a,l]pyrene are resistant to nucleotide excision repair while guanine adducts are not. Chem. Res. Toxicol. 2013, 26, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Tsutakawa, S.E.; Tsai, C.L.; Yan, C.; Bralic, A.; Chazin, W.J.; Hamdan, S.M.; Scharer, O.D.; Ivanov, I.; Tainer, J.A. Envisioning how the prototypic molecular machine TFIIH functions in transcription initiation and DNA repair. DNA Repair 2020, 96, 102972. [Google Scholar] [CrossRef] [PubMed]
- Feher, K.M.; Kolbanovskiy, A.; Durandin, A.; Shim, Y.; Min, J.H.; Lee, Y.C.; Shafirovich, V.; Mu, H.; Broyde, S.; Geacintov, N.E. The DNA damage-sensing NER repair factor XPC-RAD23B does not recognize bulky DNA lesions with a missing nucleotide opposite the lesion. DNA Repair 2020, 96, 102985. [Google Scholar] [CrossRef] [PubMed]
- Cheon, N.Y.; Kim, H.S.; Yeo, J.E.; Scharer, O.D.; Lee, J.Y. Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 2019, 47, 8337–8347. [Google Scholar] [CrossRef]
- Pahlke, G.; Ahlberg, K.; Oertel, A.; Janson-Schaffer, T.; Grabher, S.; Mock, H.P.; Matros, A.; Marko, D. Antioxidant Effects of Elderberry Anthocyanins in Human Colon Carcinoma Cells: A Study on Structure-Activity Relationships. Mol. Nutr. Food Res. 2021, 65, e2100229. [Google Scholar] [CrossRef]
- Dan, K.; Takada, A.; Kanaho, Y.; Kusumi, Y.; Banno, H. Inhibitory effect of black raspberry extract on AGE accumulation and degradation, and ROS production in HUVEC cells. Funct. Foods Health Dis. 2020, 10, 242–253. [Google Scholar] [CrossRef]
- Surh, Y.J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. [Google Scholar] [CrossRef]
- Maurya, D.K.; Salvi, V.P.; Nair, C.K. Radiation protection of DNA by ferulic acid under in vitro and in vivo conditions. Mol. Cell. Biochem. 2005, 280, 209–217. [Google Scholar] [CrossRef]
- Niture, S.K.; Velu, C.S.; Smith, Q.R.; Bhat, G.J.; Srivenugopal, K.S. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines. Carcinogenesis 2007, 28, 378–389. [Google Scholar] [CrossRef]
- Bjornson, K.P.; Amaratunga, M.; Moore, K.J.; Lohman, T.M. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry 1994, 33, 14306–14316. [Google Scholar] [CrossRef] [PubMed]
- Riedl, T.; Hanaoka, F.; Egly, J.M. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003, 22, 5293–5303. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Walle, T. Preferential induction of CYP1B1 by benzo [a] pyrene in human oral epithelial cells: Impact on DNA adduct formation and prevention by polyphenols. Carcinogenesis 2005, 26, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.; Walle, U.K.; Sedmera, D.; Klausner, M. Benzo [A] pyrene-induced oral carcinogenesis and chemoprevention: Studies in bioengineered human tissue. Drug Metab. Dispos. 2006, 34, 346–350. [Google Scholar] [CrossRef]
- Pegg, A.E.; Kanugula, S.; Loktionova, N. O6-Alkylguanine-DNA Alkyltransferase. In Chemical Carcinogenesis; Penning, T.M., Ed.; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Tyson, J.; Mathers, J.C. Dietary and genetic modulation of DNA repair in healthy human adults. Proc. Nutr. Soc. 2007, 66, 42–51. [Google Scholar] [CrossRef]
- Wang, H.; Cao, G.; Prior, R.L. Oxygen Radical Absorbing Capacity of Anthocyanins. J. Agric. Food Chem. 1997, 45, 304–309. [Google Scholar] [CrossRef]
- Johnson, D.E.; Bauman, J.E. When the Damage Is Done: Selecting Patients for Head and Neck Cancer Chemoprevention Trials. Cancer Prev. Res. 2017, 10, 489–490. [Google Scholar] [CrossRef][Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sales, A.H.; Kolbanovskiy, M.; Geacintov, N.E.; Chen, K.-M.; Sun, Y.-W.; El-Bayoumy, K. Treatment of Human HeLa Cells with Black Raspberry Extracts Enhances the Removal of DNA Lesions by the Nucleotide Excision Repair Mechanism. Antioxidants 2022, 11, 2110. https://doi.org/10.3390/antiox11112110
Sales AH, Kolbanovskiy M, Geacintov NE, Chen K-M, Sun Y-W, El-Bayoumy K. Treatment of Human HeLa Cells with Black Raspberry Extracts Enhances the Removal of DNA Lesions by the Nucleotide Excision Repair Mechanism. Antioxidants. 2022; 11(11):2110. https://doi.org/10.3390/antiox11112110
Chicago/Turabian StyleSales, Ana H., Marina Kolbanovskiy, Nicholas E. Geacintov, Kun-Ming Chen, Yuan-Wan Sun, and Karam El-Bayoumy. 2022. "Treatment of Human HeLa Cells with Black Raspberry Extracts Enhances the Removal of DNA Lesions by the Nucleotide Excision Repair Mechanism" Antioxidants 11, no. 11: 2110. https://doi.org/10.3390/antiox11112110
APA StyleSales, A. H., Kolbanovskiy, M., Geacintov, N. E., Chen, K.-M., Sun, Y.-W., & El-Bayoumy, K. (2022). Treatment of Human HeLa Cells with Black Raspberry Extracts Enhances the Removal of DNA Lesions by the Nucleotide Excision Repair Mechanism. Antioxidants, 11(11), 2110. https://doi.org/10.3390/antiox11112110