Comparison of Growth Performance, Immunity, Antioxidant Capacity, and Liver Transcriptome of Calves between Whole Milk and Plant Protein-Based Milk Replacer under the Same Energy and Protein Levels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Animal, Study Design, and Diets
2.3. Determination of Growth Performance and Serum Indicators
2.4. Liver Biopsy
2.5. Total RNA Extraction and Sequencing
2.6. Validation of RNA-Seq Data by Quantitative Real-Time PCR (qRT-PCR)
2.7. Statistical Analyses
3. Results
3.1. Intake and Growth Performance
3.2. Serum Indicators
3.3. Mapping Summary Statistics
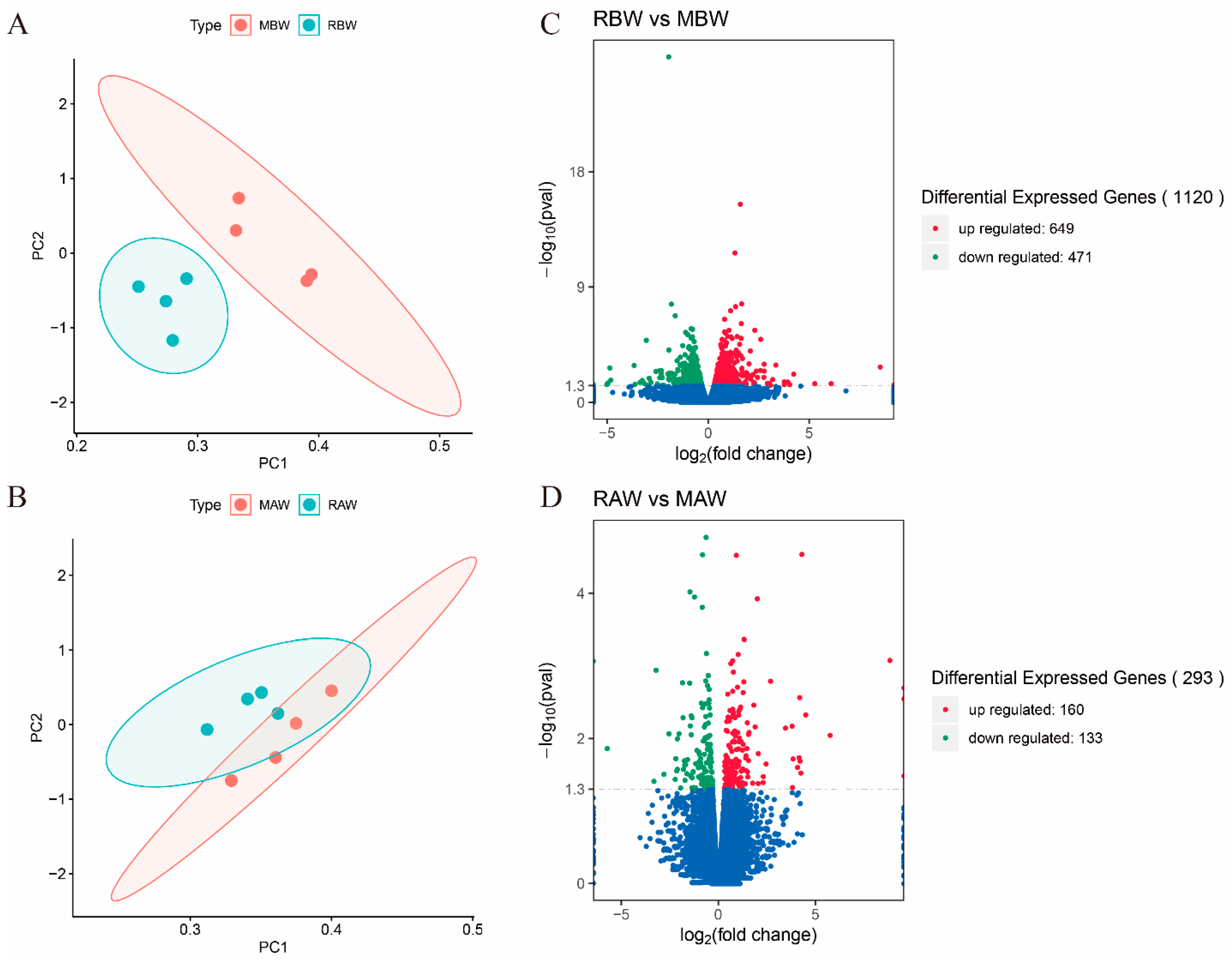
3.4. Differentially Expressed Genes (DEGs)
3.5. Enrichment Analysis of the DEGs
3.6. Protein-Protein Interaction Analysis of DEGs Using STRING
3.7. Correlation Analysis between Hub-Genes among DEGs and Apparent Indicators
3.8. Validation of RNA-Seq Data by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, M.A.; Bach, A.; Weary, D.M.; von Keyserlingk, M.A.G. Invited review: Transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 2016, 99, 885–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soberon, F.; Raffrenato, E.; Everett, R.W.; Van Amburgh, M.E. Preweaning milk replacer intake and effects on long-term productivity of dairy calves. J. Dairy Sci. 2012, 95, 783–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, Q.; Zhang, R.; Fu, T. Review of Strategies to Promote Rumen Development in Calves. Animals 2019, 9, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.J.; Hwang, G.H.; Saito, A.; Vermeire, D.A.; Drackley, J.K. Assessment of the effect of methionine supplementation and inclusion of hydrolyzed wheat protein in milk protein-based milk replacers on the performance of intensively fed Holstein calves. J. Dairy Sci. 2016, 99, 6324–6333. [Google Scholar] [CrossRef]
- Ghorbani, G.R.; Kowsar, R.; Alikhani, M.; Nikkhah, A. Soymilk as a novel milk replacer to stimulate early calf starter intake and reduce weaning age and costs. J. Dairy Sci. 2007, 90, 5692–5697. [Google Scholar] [CrossRef] [Green Version]
- Soberon, M.A.; Liu, R.H.; Cherney, D.J. Short communication: Antioxidant activity of calf milk replacers. J. Dairy Sci. 2012, 95, 2703–2706. [Google Scholar] [CrossRef]
- Petit, H.V.; Ivan, M.; Brisson, G.J. Digestibility measured by fecal and ileal collection in preruminant calves fed a clotting or a nonclotting milk replacer. J. Dairy Sci. 1989, 72, 123–128. [Google Scholar] [CrossRef]
- Ansia, I.; Drackley, J.K. Graduate Student Literature Review: The past and future of soy protein in calf nutrition. J. Dairy Sci. 2020, 103, 7625–7638. [Google Scholar] [CrossRef]
- Lagrange, V.; Whitsett, D.; Burris, C. Global market for dairy proteins. J. Food Sci. 2015, 80 (Suppl. 1), A16–A22. [Google Scholar] [CrossRef]
- Raeth, M.; Chester-Jones, H.; Ziegler, D.; Ziegler, B.; Schimek, D.; Cook, D.L.; Golombeski, G.; Grove, A.V. Pre- and postweaning performance and health of dairy calves fed milk replacers with differing protein sources. Prof. Anim. Sci. 2016, 32, 833–841. [Google Scholar] [CrossRef]
- Wadhave, A.A.; Jadhav, A.I.; Arsul, V.A. Plant Proteins Applications: A Review. World J. Pharm. Pharm. Sci. 2014, 3, 702–712. [Google Scholar]
- Kertz, A.F.; Hill, T.M.; Quigley, J.D., 3rd; Heinrichs, A.J.; Linn, J.G.; Drackley, J.K. A 100-Year Review: Calf nutrition and management. J. Dairy Sci. 2017, 100, 10151–10172. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Khan, M.A.; Lee, W.S.; Yang, S.H.; Kim, S.B.; Ki, K.S.; Kim, H.S.; Ha, J.K.; Choi, Y.J. Influence of equalizing the gross composition of milk replacer to that of whole milk on the performance of Holstein calves. J. Anim. Sci. 2009, 87, 1129–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welboren, A.C.; Hatew, B.; López-Campos, O.; Cant, J.P.; Leal, L.N.; Martín-Tereso, J.; Steele, M.A. Effects of energy source in milk replacer on glucose metabolism of neonatal dairy calves. J. Dairy Sci. 2021, 104, 5009–5020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, W.B.; Bi, Y.L.; Tu, Y.; Beckers, Y.; Du, H.C.; Diao, Q.Y. Early Feeding Regime of Waste Milk, Milk, and Milk Replacer for Calves Has Different Effects on Rumen Fermentation and the Bacterial Community. Animals 2019, 9, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; Tu, Y.; Si, B.; Xu, G.; Guo, J.; Guo, F.; Yang, C.; Diao, Q. Effects of protein sources for milk replacers on growth performance and serum biochemical indexes of suckling calves. Anim. Nutr. 2015, 1, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Silano, M.; De Vincenzi, M. Bioactive antinutritional peptides derived from cereal prolamins: A review. Nahrung 1999, 43, 175–184. [Google Scholar] [CrossRef]
- Komatsu, S.; Kajiwara, H.; Hirano, H. A rice protein library: A data-file of rice proteins separated by two-dimensional electrophoresis. Appl. Genet. 1993, 86, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Chen, Y.Y.; Wu, C.T.; Yu, C.C.; Liao, H.F. Prolamin, a rice protein, augments anti-leukaemia immune response. J. Cereal Sci. 2010, 51, 189–197. [Google Scholar] [CrossRef]
- Kadarmideen, H.N. Genomics to systems biology in animal and veterinary sciences: Progress, lessons and opportunities. Livest. Sci. 2014, 166, 232–248. [Google Scholar] [CrossRef] [Green Version]
- Suravajhala, P.; Kogelman, L.J.; Kadarmideen, H.N. Multi-omic data integration and analysis using systems genomics approaches: Methods and applications in animal production, health and welfare. Genet. Sel. Evol. 2016, 48, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K.; Robinson, G.J.; Lundberg, A.E.; Bartlett, P.F.; Wray, N.R.; et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS ONE 2014, 9, e103207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Li, Y.; Diao, Q.; Bi, Y.; Tu, Y. The crucial role of lysine in the hepatic metabolism of growing Holstein dairy heifers as revealed by LC-MS-based untargeted metabolomics. Anim. Nutr. 2021, 7, 1152–1161. [Google Scholar] [CrossRef]
- Kargar, S.; Kanani, M. Reconstituted versus dry alfalfa hay in starter feed diets of Holstein dairy calves: Effects on growth performance, nutrient digestibility, and metabolic indications of rumen development. J. Dairy Sci. 2019, 102, 4051–4060. [Google Scholar] [CrossRef]
- Kong, F.; Bi, Y.; Wang, B.; Cui, K.; Li, Y.; Fu, T.; Diao, Q.; Tu, Y. Integrating RNA-sequencing and untargeted LC-MS metabolomics to evaluate the effect of lysine deficiency on hepatic functions in Holstein calves. Amino Acids 2020, 52, 781–792. [Google Scholar] [CrossRef]
- Flaga, J.; Korytkowski, Ł.; Górka, P.; Kowalski, Z.M. Age-related changes in mRNA expression of selected surface receptors in lymphocytes of dairy calves. Pol. J. Vet. Sci. 2018, 21, 213–216. [Google Scholar] [CrossRef]
- Wang, S.; Diao, Q.Y.; Hu, F.M.; Bi, Y.L.; Piao, M.Y.; Jiang, L.S.; Sun, F.; Li, H.; Tu, Y. Development of ruminating behavior in Holstein calves between birth and 30 days of age. J. Dairy Sci. 2022, 105, 572–584. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Brohée, S.; van Helden, J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinform. 2006, 7, 488. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasper, J.; Weary, D.M. Effects of ad libitum milk intake on dairy calves. J. Dairy Sci. 2002, 85, 3054–3058. [Google Scholar] [CrossRef]
- Blum, J.W.; Baumrucker, C.R. Colostral and milk insulin-like growth factors and related substances: Mammary gland and neonatal (intestinal and systemic) targets. Domest. Anim. Endocrinol. 2002, 23, 101–110. [Google Scholar] [CrossRef]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019, 20, 172. [Google Scholar] [CrossRef] [Green Version]
- Gerber, H.P.; Ferrara, N. Angiogenesis and Bone Growth. Trends Cardiovasc. Med. 2000, 10, 223–228. [Google Scholar] [CrossRef]
- Seegraber, F.J.; Morrill, J.L. Effect of protein source in calf milk replacers on morphology and absorptive ability of small intestine. J. Dairy Sci. 1986, 69, 460–469. [Google Scholar] [CrossRef]
- Freedman, S.F.; Anderson, P.J.; Epstein, D.L. Superoxide dismutase and catalase of calf trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 1985, 26, 1330–1335. [Google Scholar]
- Gaetani, G.D.; Parker, J.C.; Kirkman, H.N. Intracellular restraint: A new basis for the limitation in response to oxidative stress in human erythrocytes containing low-activity variants of glucose-6-phosphate dehydrogenase. Proc. Natl. Acad. Sci. USA 1974, 71, 3584–3587. [Google Scholar] [CrossRef] [Green Version]
- Schulz, J.B.; Lindenau, J.; Seyfried, J.; Dichgans, J. Glutathione, oxidative stress and neurodegeneration. Eur. J. Biochem. 2000, 267, 4904–4911. [Google Scholar] [CrossRef]
- Montagne, L.; Toullec, R.; Savidge, T.; Lallès, J.P. Morphology and enzyme activities of the small intestine are modulated by dietary protein source in the preruminant calf. Reprod. Nutr. Dev. 1999, 39, 455–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, I.; Cerbón, M.A.; Pasapera, A.M.; Gutiérrez-Sagal, R.; Garcia, G.A.; Orozco, C.; Camacho-Arroyo, I.; Anzaldua, R.; Pérez-Palacios, G. Molecular mechanisms of the antihormonal and antiimplantation effects of norethisterone and its A-ring reduced metabolites. Mol. Reprod. Dev. 1995, 40, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.J.; Tuffs, S.W.; Wee, B.A.; Seo, K.S.; Park, N.; Connelley, T.; Guinane, C.M.; Morrison, W.I.; Fitzgerald, J.R. Bovine Staphylococcus aureus Superantigens Stimulate the Entire T Cell Repertoire of Cattle. Infect. Immun. 2018, 86, e00505-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grasfeder, L.L.; Gaillard, S.; Hammes, S.R.; Ilkayeva, O.; Newgard, C.B.; Hochberg, R.B.; Dwyer, M.A.; Chang, C.Y.; McDonnell, D.P. Fasting-induced hepatic production of DHEA is regulated by PGC-1alpha, ERRalpha, and HNF4alpha. Mol. Endocrinol. 2009, 23, 1171–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salleh, M.S.; Mazzoni, G.; Höglund, J.K.; Olijhoek, D.W.; Lund, P.; Løvendahl, P.; Kadarmideen, H.N. RNA-Seq transcriptomics and pathway analyses reveal potential regulatory genes and molecular mechanisms in high- and low-residual feed intake in Nordic dairy cattle. BMC Genom. 2017, 18, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tizioto, P.C.; Coutinho, L.L.; Decker, J.E.; Schnabel, R.D.; Rosa, K.O.; Oliveira, P.S.; Souza, M.M.; Mourão, G.B.; Tullio, R.R.; Chaves, A.S.; et al. Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes. BMC Genom. 2015, 16, 242. [Google Scholar] [CrossRef] [Green Version]
- Santana, M.H.d.A.; Junior, G.A.; Cesar, A.S.; Freua, M.C.; Gomes, R.d.C.; da Luz, E.S.S.; Leme, P.R.; Fukumasu, H.; Carvalho, M.E.; Ventura, R.V.; et al. Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle. J. Appl. Genet. 2016, 57, 495–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Gui, L.S.; Xu, H.C.; Wu, S.; Zan, L.S. Identification of single nucleotide polymorphisms of the signal transducer and activator of transcription 3 gene (STAT3) associated with body measurement and carcass quality traits in beef cattle. Genet. Mol. Res. 2015, 14, 11242–11249. [Google Scholar] [CrossRef] [PubMed]
- da Costa, A.S.; Pires, V.M.; Fontes, C.M.; Prates, J.A.M. Expression of genes controlling fat deposition in two genetically diverse beef cattle breeds fed high or low silage diets. BMC Vet. Res. 2013, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Sumner-Thomson, J.M.; Vierck, J.L.; McNamara, J.P. Differential expression of genes in adipose tissue of first-lactation dairy cattle. J. Dairy Sci. 2011, 94, 361–369. [Google Scholar] [CrossRef]
- Lee, K.; Kerner, J.; Hoppel, C.L. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J. Biol. Chem. 2011, 286, 25655–25662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenzie, M.; Lazarou, M.; Thorburn, D.R.; Ryan, M.T. Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal. Biochem. 2007, 364, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Menezes, M.J.; Riley, L.G.; Christodoulou, J. Mitochondrial respiratory chain disorders in childhood: Insights into diagnosis and management in the new era of genomic medicine. Biochim. Biophys. Acta 2014, 1840, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Steffen, W.; Gemperli, A.C.; Cvetesic, N.; Steuber, J. Organelle-specific expression of subunit ND5 of human complex I (NADH dehydrogenase) alters cation homeostasis in Saccharomyces cerevisiae. FEMS Yeast Res. 2010, 10, 648–659. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Zhang, J.; Tsai, C.W.; Orlando, B.J.; Rodriguez, M.; Xu, Y.; Liao, M.; Tsai, M.F.; Feng, L. Structure and mechanism of the mitochondrial Ca(2+) uniporter holocomplex. Nature 2020, 582, 129–133. [Google Scholar] [CrossRef]
- McCabe, M.; Waters, S.; Morris, D.; Kenny, D.; Lynn, D.; Creevey, C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genom. 2012, 13, 193. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.; Li, F.; Feng, X.; Yang, H.; Han, L.; Fan, Y.; Nie, H.; Wang, Z.; Wang, F.; Zhang, Y. Influences of different dietary energy level on sheep testicular development associated with AMPK/ULK1/autophagy pathway. Theriogenology 2018, 108, 362–370. [Google Scholar] [CrossRef]
- Bartlett, K.S.; McKeith, F.K.; VandeHaar, M.J.; Dahl, G.E.; Drackley, J.K. Growth and body composition of dairy calves fed milk replacers containing different amounts of protein at two feeding rates. J. Anim. Sci. 2006, 84, 1454–1467. [Google Scholar] [CrossRef]
- Tikofsky, J.N.; Van Amburgh, M.E.; Ross, D.A. Effect of varying carbohydrate and fat content of milk replacer on body composition of Holstein bull calves. J. Anim. Sci. 2001, 79, 2260–2267. [Google Scholar] [CrossRef]
- Hu, F.M.; Dong, L.F.; Bi, Y.L.; Ma, J.N.; Wang, B.; Diao, Q.Y.; Tu, Y. Effects of Different Fatty Acid Sources in Milk Replacer on Growth Performance, Digestion and Metabolism of Sucking Calves. Chin. J. Anim. Nutr. 2018, 30, 1736–1747. [Google Scholar] [CrossRef]
- Coutinho, S.R.; With, E.; Rehfeld, J.F.; Kulseng, B.; Truby, H.; Martins, C. The impact of rate of weight loss on body composition and compensatory mechanisms during weight reduction: A randomized control trial. Clin. Nutr. 2018, 37, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Wang, B.; Ma, T.; Si, B.W.; Zhang, N.F.; Tu, Y.; Diao, Q.Y. Effects of dietary protein restriction followed by realimentation on growth performance and liver transcriptome alterations of lamb. Sci. Rep. 2018, 8, 15185. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.; Therkildsen, M.; Byrne, D.V. Effects of a compensatory growth strategy on sensory and physical properties of meat from young bulls. Meat Sci. 2006, 74, 628–643. [Google Scholar] [CrossRef] [PubMed]
- Keady, S.M.; Waters, S.M.; Hamill, R.M.; Dunne, P.G.; Keane, M.G.; Richardson, R.I.; Kenny, D.A.; Moloney, A.P. Compensatory growth in crossbred Aberdeen Angus and Belgian Blue steers: Effects on the colour, shear force and sensory characteristics of longissimus muscle. Meat Sci. 2017, 125, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, J.G.; Forsén, T.; Tuomilehto, J.; Winter, P.D.; Osmond, C.; Barker, D.J. Catch-up growth in childhood and death from coronary heart disease: Longitudinal study. BMJ 1999, 318, 427–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibáñez, L.; Lopez-Bermejo, A.; Diaz, M.; de Zegher, F. Catch-up growth in girls born small for gestational age precedes childhood progression to high adiposity. Fertil. Steril. 2011, 96, 220–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silveira, P.P.; Pokhvisneva, I.; Gaudreau, H.; Rifkin-Graboi, A.; Broekman, B.F.P.; Steiner, M.; Levitan, R.; Parent, C.; Diorio, J.; Meaney, M.J. Birth weight and catch up growth are associated with childhood impulsivity in two independent cohorts. Sci. Rep. 2018, 8, 13705. [Google Scholar] [CrossRef]


| Treatment | ||
|---|---|---|
| Items | Milk Powder 1 | Milk Replacer 2 |
| Chemical composition (% of DM except for Dry matter; mean ± SD) | ||
| Dry matter | 96.76 ± 0.61 | 96.32 |
| Gross energy MJ/kg | 24.81 ± 0.41 | 24.62 |
| Crude Protein | 27.01 ± 0.36 | 26.93 |
| Ether extract | 30.67 ± 0.53 | 16.28 |
| Ash | 5.58 ± 0.05 | 5.53 |
| Calcium | 1.06 ± 0.03 | 1.02 |
| Phosphorus | 0.75 ± 0.03 | 0.77 |
| Treatment (T) 1 | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|
| Items 4 | MR | M | SEM | T | Days (D) 2 | T × D | P 3 | T × P |
| Pre-weaning (0–70 d) | ||||||||
| DMI of starter feed, kg/d | 0.23 | 0.16 | 0.02 | 0.009 | <0.001 | 0.601 | ||
| DMI of liquid feed, kg/d | 1.00 | 1.05 | 0.01 | 0.043 | <0.001 | 0.622 | ||
| Total DMI 4, kg/d | 1.23 | 1.22 | 0.02 | 0.513 | <0.001 | 0.742 | ||
| ADG, kg/d | 0.83 | 0.92 | 0.02 | 0.007 | <0.001 | 0.461 | ||
| Feed efficiency 5 | 0.67 | 0.76 | 0.03 | <0.001 | <0.001 | 0.731 | ||
| Post-weaning (70–98 d) | ||||||||
| DMI of starter feed, kg/d | 2.79 | 2.28 | 0.10 | <0.001 | <0.001 | 0.231 | ||
| ADG, kg/d | 1.09 | 0.97 | 0.03 | 0.024 | <0.001 | 0.814 | ||
| Feed efficiency | 0.39 | 0.43 | 0.01 | 0.006 | <0.001 | 0.731 | ||
| Overall (0–98 d) | ||||||||
| Total DMI, kg/d | 1.68 | 1.51 | 0.03 | 0.002 | <0.001 | 0.004 | ||
| ADG, kg/d | 0.91 | 0.94 | 0.02 | 0.334 | <0.001 | 0.193 | ||
| Feed efficiency | 0.45 | 0.54 | 0.01 | <0.001 | <0.001 | 0.241 | ||
| Body weight, kg | ||||||||
| Initial (d 0) | 41.4 | 41.9 | 1.65 | 0.781 | ||||
| Weaning (d 70) | 98.2 | 107.4 | 2.83 | 0.004 | ||||
| Final (d 98) | 128.1 | 132.7 | 3.68 | 0.225 | ||||
| Skeletal growth | ||||||||
| Withers height, cm | ||||||||
| Initial (d 0) | 77.7 | 76.0 | 0.93 | 0.072 | ||||
| Weaning (d 70) | 94.6 | 97.2 | 1.13 | 0.026 | ||||
| Final (d 98) | 98.7 | 101.3 | 1.30 | 0.055 | ||||
| Heart girth, cm | ||||||||
| Initial (d 0) | 77.3 | 76.1 | 0.98 | 0.233 | ||||
| Weaning (d 70) | 107.1 | 109.9 | 1.10 | 0.018 | ||||
| Final (d 98) | 116.9 | 118.2 | 1.35 | 0.341 | ||||
| Body length, cm | ||||||||
| Initial (d 0) | 69.8 | 69.4 | 1.06 | 0.702 | ||||
| Weaning (d 70) | 97.3 | 99.4 | 1.04 | 0.131 | ||||
| Final (d 98) | 105.5 | 106.5 | 1.73 | 0.584 | ||||
| Treatment (T) 1 | p-Value | |||||
|---|---|---|---|---|---|---|
| Items | MR | M | SEM | T | Days (D) 2 | T × D |
| ALT, U/L | ||||||
| pre-weaning (35–49 d) | 7.5 | 7.1 | 0.99 | 0.524 | 0.979 | 0.621 |
| post-weaning (84–98 d) | 19.9 | 15.2 | 1.05 | 0.035 | 0.837 | 0.179 |
| AST, U/L | ||||||
| pre-weaning (35–49 d) | 121.6 | 91.5 | 2.88 | 0.028 | 0.778 | 0.632 |
| post-weaning (84–98 d) | 183.2 | 148.7 | 4.23 | 0.036 | 0.081 | 0.712 |
| ALP, U/L | ||||||
| pre-weaning (35–49 d) | 159.6 | 149.4 | 7.93 | 0.205 | 0.730 | 0.453 |
| post-weaning (84–98 d) | 188.1 | 186.1 | 11.49 | 0.693 | 0.334 | 0.818 |
| IgG, g/L | ||||||
| pre-weaning (35–49 d) | 9.9 | 10.2 | 0.50 | 0.942 | 0.706 | 0.579 |
| post-weaning (84–98 d) | 12.7 | 12.3 | 0.44 | 0.994 | 0.206 | 0.765 |
| IgA, g/L | ||||||
| pre-weaning (35–49 d) | 1.3 | 1.6 | 0.04 | 0.571 | 0.002 | 0.442 |
| post-weaning (84–98 d) | 1.1 | 1.2 | 0.02 | 0.028 | 0.004 | 0.668 |
| IgM, g/L | ||||||
| pre-weaning (35–49 d) | 2.1 | 2.8 | 0.06 | 0.012 | 0.725 | 0.971 |
| post-weaning (84–98 d) | 2.1 | 2.7 | 0.06 | 0.001 | 0.819 | 0.715 |
| SOD, U/mL | ||||||
| pre-weaning (35–49 d) | 85.3 | 99.1 | 3.06 | 0.045 | 0.679 | 0.391 |
| post-weaning (84–98 d) | 128.3 | 125.8 | 2.26 | 0.278 | 0.138 | 0.442 |
| MDA, nmol/mL | ||||||
| pre-weaning (35–49 d) | 7.2 | 5.9 | 0.23 | 0.035 | 0.673 | 0.229 |
| post-weaning (84–98 d) | 4.7 | 4.2 | 0.11 | 0.015 | 0.714 | 0.814 |
| GSH-PX, U/mL | ||||||
| pre-weaning (35–49 d) | 824 | 837 | 26.48 | 0.595 | 0.115 | 0.415 |
| post-weaning (84–98 d) | 1173 | 1202 | 30.46 | 0.698 | <0.001 | 0.458 |
| T-AOC, U/mL | ||||||
| pre-weaning (35–49 d) | 6.2 | 7.1 | 0.16 | 0.005 | 0.541 | 0.818 |
| post-weaning (84–98 d) | 9.8 | 10.3 | 0.42 | 0.103 | 0.328 | 0.643 |
| CAT, U/mL | ||||||
| pre-weaning (35–49 d) | 8.6 | 8.5 | 0.33 | 0.596 | 0.333 | 0.992 |
| post-weaning (84–98 d) | 11.4 | 12.1 | 0.48 | 0.286 | 0.215 | 0.710 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Hu, F.; Diao, Q.; Li, S.; Tu, Y.; Bi, Y. Comparison of Growth Performance, Immunity, Antioxidant Capacity, and Liver Transcriptome of Calves between Whole Milk and Plant Protein-Based Milk Replacer under the Same Energy and Protein Levels. Antioxidants 2022, 11, 270. https://doi.org/10.3390/antiox11020270
Wang S, Hu F, Diao Q, Li S, Tu Y, Bi Y. Comparison of Growth Performance, Immunity, Antioxidant Capacity, and Liver Transcriptome of Calves between Whole Milk and Plant Protein-Based Milk Replacer under the Same Energy and Protein Levels. Antioxidants. 2022; 11(2):270. https://doi.org/10.3390/antiox11020270
Chicago/Turabian StyleWang, Shuo, Fengming Hu, Qiyu Diao, Shuang Li, Yan Tu, and Yanliang Bi. 2022. "Comparison of Growth Performance, Immunity, Antioxidant Capacity, and Liver Transcriptome of Calves between Whole Milk and Plant Protein-Based Milk Replacer under the Same Energy and Protein Levels" Antioxidants 11, no. 2: 270. https://doi.org/10.3390/antiox11020270







