The Influence of Coronary Artery Disease in the Development of Aortic Stenosis and the Importance of the Albumin Redox State
Abstract
1. Introduction
2. Materials and Methods
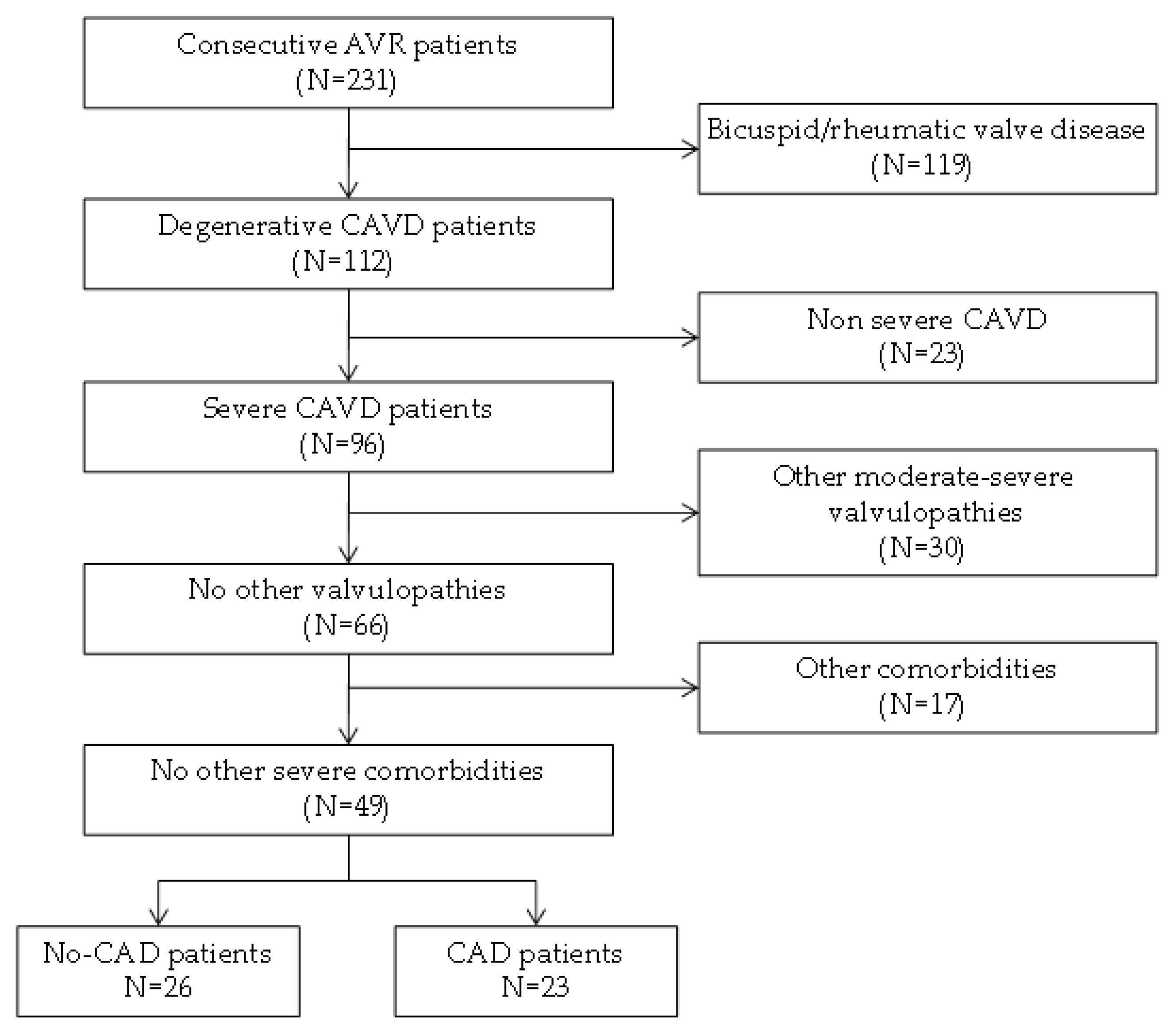
2.1. Patient Selection
2.2. Proteomics Overview
2.2.1. Tissue Sample Preparation and Isobaric Labeling
2.2.2. Protein Identification and Quantification
2.2.3. Verification: Targeted Proteomics
2.2.4. Verification: Western Blotting
2.3. Functional Annotation Clustering
2.4. Statistical Analysis
3. Results
3.1. Protein Functional Annotation
3.2. Verification Phase
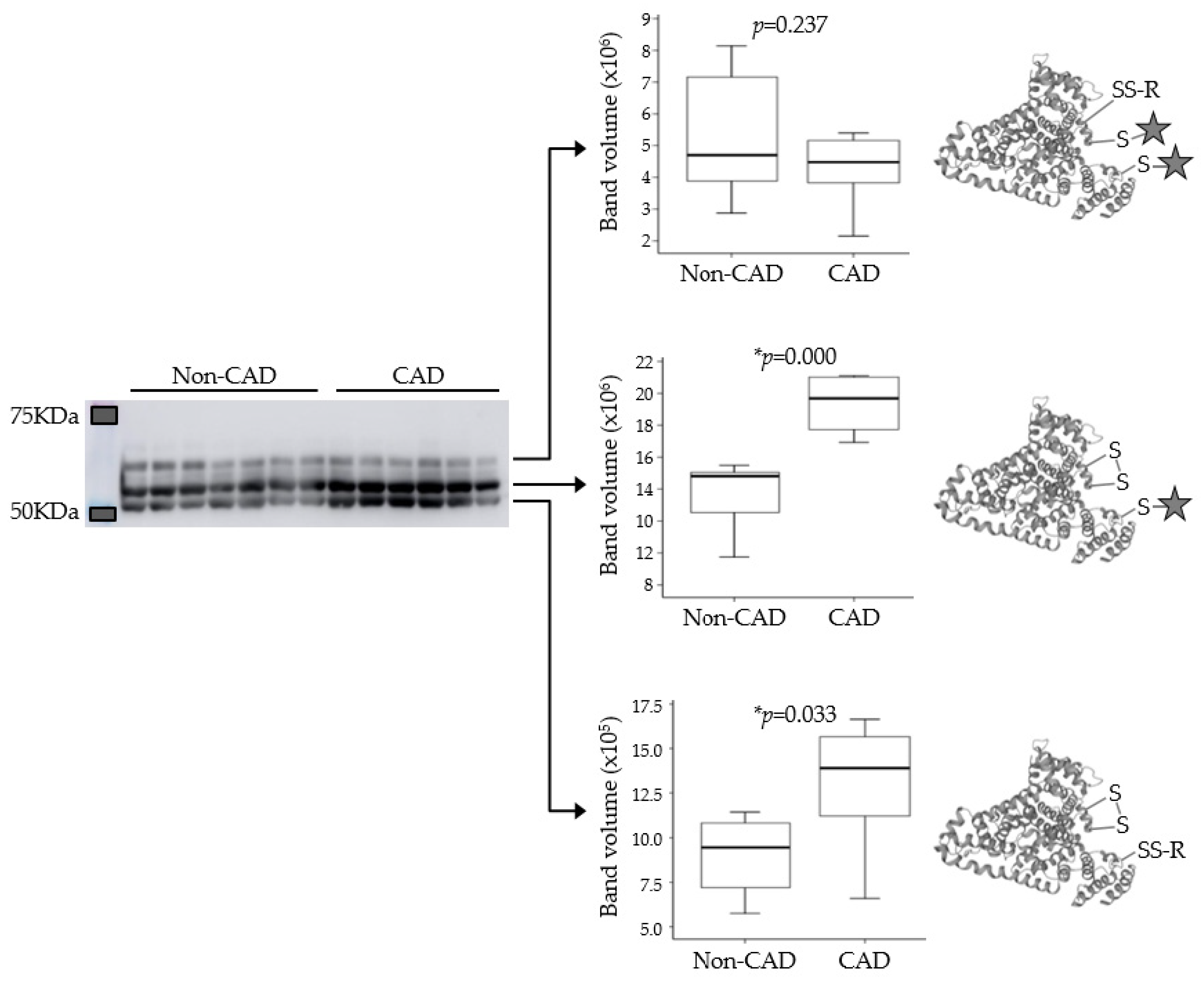
Targeted Proteomics (PRM) and Immunodetection (Western Blot)
3.3. Analysis of Cysteine Oxidation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pawade, T.; Sheth, T.; Guzzetti, E.; Dweck, M.R.; Clavel, M.A. Why and how to measure aortic valve calcification in patients with aortic stenosis. JACC Cardiovasc. Imaging 2019, 12, 1835–1848. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, H.; Falk, V.; Bax, J.J.; De Bonis, M.; Hamm, C.; Holm, P.J.; Iung, B.; Lancellotti, P.; Lansac, E.; Muñoz, D.R.; et al. 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur. Heart J. 2017, 38, 2739–2791. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.A.; Otto, C.M.; Bonow, R.O.; Carabello, B.A.; Erwin, J.P.; Guyton, R.A.; O’Gara, P.T.; Ruiz, C.E.; Skubas, N.J.; Sorajja, P.; et al. 2014 AHA/ACC Guideline for the management of patients with valvular heart disease: A report of the American College of Cardiology/american heart association task force on practice guidelines. Circulation 2014, 129, e521–e643. [Google Scholar] [CrossRef]
- Katz, R.; Wong, N.D.; Kronmal, R.; Takasu, J.; Shavelle, D.M.; Probstfield, J.L.; Bertoni, A.G.; Budoff, M.J.; O’Brien, K.D. Features of the metabolic syndrome and diabetes mellitus as predictors of aortic valve calcification in the Multi-Ethnic Study of Atherosclerosis. Circulation 2006, 113, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Meijboom, W.B.; Mollet, N.R.; Van Mieghem, C.A.; Kluin, J.; Weustink, A.C.; Pugliese, F.; Vourvouri, E.; Cademartiri, F.; Bogers, A.J.; Krestin, G.P.; et al. Pre-operative computed tomography coronary angiography to detect significant coronary artery disease in patients referred for cardiac valve surgery. J. Am. Coll. Cardiol. 2006, 48, 1658–1665. [Google Scholar] [CrossRef]
- Di Minno, M.N.D.; Poggio, P.; Conte, E.; Myasoedova, V.; Songia, P.; Mushtaq, S.; Cavallotti, L.; Moschetta, D.; Di Minno, A.; Spadarella, G.; et al. Cardiovascular morbidity and mortality in patients with aortic valve calcification: A systematic review and meta-analysis. J. Cardiovasc. Comput. Tomogr. 2019, 13, 190–195. [Google Scholar] [CrossRef]
- Sankaramangalam, K.; Banerjee, K.; Kandregula, K.; Mohananey, D.; Parashar, A.; Jones, B.M.; Jobanputra, Y.; Mick, S.; Krishnaswamy, A.; Svensson, L.G.; et al. Impact of coronary artery disease on 30-day and 1-year mortality in patients undergoing transcatheter aortic valve replacement: A meta-analysis. J. Am. Heart Assoc. 2017, 6, e006092. [Google Scholar] [CrossRef]
- Ross, R. Atherosclerosis—An inflammatory disease. N. Engl. J. Med. 1999, 340, 115–126. [Google Scholar] [CrossRef]
- Otto, C.M.; Kuusisto, J.; Reichenbach, D.D.; Gown, A.M.; O’Brien, K.D. Characterization of the early lesion of ‘degenerative’ valvular aortic stenosis. Histological and immunohistochemical studies. Circulation 1994, 90, 844–853. [Google Scholar] [CrossRef]
- O’Brien, K.D.; Reichenbach, D.D.; Marcovina, S.M.; Kuusisto, J.; Alpers, C.E.; Otto, C.M. Apolipoproteins B, (a), and E accumulate in the morphologically early lesion of ‘degenerative’ valvular aortic stenosis. Arter. Thromb. Vasc. Biol. 1996, 16, 523–532. [Google Scholar] [CrossRef]
- Novaro, G.; Griffin, B. Calcific aortic stenosis: Another face of atherosclerosis? Cleve. Clin. J. Med. 2003, 70, 1–7. [Google Scholar] [CrossRef][Green Version]
- van der Wal, A.C.; Becker, A.E.; van der Loos, C.M.; Das, P.K. Site of intimal rupture or erosion of thrombosed coronary atherosclerotic plaques is characterized by an inflammatory process irrespective of the dominant plaque morphology. Circulation 1994, 89, 36–44. [Google Scholar] [CrossRef]
- Naghavi, M.; Libby, P.; Falk, E.; Casscells, S.W.; Litovsky, S.; Rumberger, J.; Badimon, J.J.; Stefanadis, C.; Moreno, P.; Pasterkamp, G.; et al. From vulnerable plaque to vulnerable patient: A call for new definitions and risk assessment strategies: Part I. Circulation 2003, 108, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Vichova, T.; Motovska, Z. Oxidative stress: Predictive marker for coronary artery disease. Exp. Clin. Cardiol. 2013, 18, e88–e91. [Google Scholar]
- Heistad, D.D.; Wakisaka, Y.; Miller, J.; Chu, Y.; Pena-Silva, R. Novel aspects of oxidative stress in cardiovascular diseases. Circ. J. 2009, 73, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Weiss, R.M.; Heistad, D.D. Calcific aortic valve stenosis: Methods, models, and mechanisms. Circ. Res. 2011, 108, 1392–1412. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K. Studies on free radicals, antioxidants, and co-factors. Clin. Interv. Aging 2007, 2, 219–236. [Google Scholar]
- Miller, J.D.; Chu, Y.; Brooks, R.M.; Richenbacher, W.E.; Peña-Silva, R.; Heistad, D.D. Dysregulation of antioxidant mechanisms contributes to increased oxidative stress in calcific aortic valvular stenosis in humans. J. Am. Coll. Cardiol. 2008, 52, 843–850. [Google Scholar] [CrossRef]
- Lee, S.E.; Sung, J.M.; Andreini, D.; Al-Mallah, M.H.; Budoff, M.J.; Cademartiri, F.; Chinnaiyan, K.; Choi, J.H.; Chun, E.J.; Conte, E.; et al. Association between aortic valve calcification progression and coronary atherosclerotic plaque volume progression in the PARADIGM registry. Radiology 2021, 300, 79–86. [Google Scholar] [CrossRef]
- Massera, D.; Buzkova, P.; Bortnick, A.E.; Owens, D.S.; Mao, S.; Li, D.; De Boer, I.H.; Kestenbaum, B.R.; Budoff, M.J.; Kizer, J.R. Bone mineral density and long-term progression of aortic valve and mitral annular calcification: The Multi-Ethnic Study of Atherosclerosis. Atherosclerosis 2021, 335, 126–134. [Google Scholar] [CrossRef]
- Razavi, A.C.; Cardoso, R.; Dzaye, O.; Budoff, M.; Thanassoulis, G.; Post, W.S.; Shah, S.; Berman, D.S.; Nasir, K.; Blaha, M.J.; et al. Risk markers for limited coronary artery calcium in persons with significant aortic valve calcium (from the Multi-ethnic Study of Atherosclerosis). Am. J. Cardiol. 2021, 156, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Bonzon-Kulichenko, E.; Camafeita, E.; López, J.A.; Gómez-Serrano, M.; Jorge, I.; Calvo, E.; Núñez, E.; Trevisan-Herraz, M.; Bagwan, N.; Bárcena, J.A.; et al. Improved integrative analysis of the thiol redox proteome using filter-aided sample preparation. J. Proteom. 2020, 214, 103624. [Google Scholar] [CrossRef] [PubMed]
- Corbacho-Alonso, N.; Baldán-Martín, M.; López, J.A.; Rodríguez-Sánchez, E.; Martínez, P.J.; Mourino-Alvarez, L.; Sastre-Oliva, T.; Cabrera, M.; Calvo, E.; Padial, L.R.; et al. Cardiovascular risk stratification based on oxidative stress for early detection of pathology. Antioxid. Redox Signal 2021, 35, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Bartolome, S.; Navarro, P.; Martin-Maroto, F.; Lopez-Ferrer, D.; Ramos-Fernandez, A.; Villar, M.; Garcia-Ruiz, J.P.; Vazquez, J. Properties of average score distributions of SEQUEST. Mol. Cell. Proteom. 2008, 7, 1135–1145. [Google Scholar] [CrossRef]
- Navarro, P.; Vazquez, J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 2009, 8, 1792–1796. [Google Scholar] [CrossRef]
- Bonzon-Kulichenko, E.; Garcia-Marques, F.; Trevisan-Herraz, M.; Vazquez, J. Revisiting peptide identification by high-accuracy mass spectrometry: Problems associated with the use of narrow mass precursor Windows. J. Proteome Res. 2015, 14, 700–710. [Google Scholar] [CrossRef]
- Trevisan-Herraz, M.; Bagwan, N.; García-Marqués, F.; Rodriguez, J.M.; Jorge, I.; Ezkurdia, I.; Bonzon-Kulichenko, E.; Vázquez, J. SanXoT: A modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics 2019, 35, 1594–1596. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. Activities at the Universal Protein Resource (UniProt). Nucleic. Acids. Res. 2014, 42, D191–D198. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic. Acids. Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Nuñez, E.; Martinez-Acedo, P.; Perez-Hernandez, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef]
- Bouchareb, R.; Boulanger, M.C.; Tastet, L.; Mkannez, G.; Nsaibia, M.J.; Hadji, F.; Dahou, A.; Messadeq, Y.; Arsenault, B.J.; Pibarot, P.; et al. Activated platelets promote an osteogenic programme and the progression of calcific aortic valve stenosis. Eur. Heart J. 2019, 40, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Kanda, H.; Yamakuchi, M.; Matsumoto, K.; Mukaihara, K.; Shigehisa, Y.; Tachioka, S.; Okawa, M.; Takenouchi, K.; Oyama, Y.; Hashiguchi, T.; et al. Dynamic changes in platelets caused by shear stress in aortic valve stenosis. Clin. Hemorheol. Microcirc. 2021, 77, 71–81. [Google Scholar] [CrossRef] [PubMed]
- von Hundelshausen, P.; Schmitt, M.M. Platelets and their chemokines in atherosclerosis-clinical applications. Front. Physiol. 2014, 5, 294. [Google Scholar] [CrossRef] [PubMed]
- Nording, H.M.; Seizer, P.; Langer, H.F. Platelets in inflammation and atherogenesis. Front. Immunol. 2015, 6, 98. [Google Scholar] [CrossRef]
- Del Conde, I.; Crúz, M.A.; Zhang, H.; López, J.A.; Afshar-Kharghan, V. Platelet activation leads to activation and propagation of the complement system. J. Exp. Med. 2005, 201, 871–879. [Google Scholar] [CrossRef]
- Eriksson, O.; Mohlin, C.; Nilsson, B.; Ekdahl, K.N. The human platelet as an innate immune cell: Interactions between activated platelets and the complement system. Front. Immunol. 2019, 10, 1590. [Google Scholar] [CrossRef]
- Ter Weeme, M.; Vonk, A.B.; Kupreishvili, K.; van Ham, M.; Zeerleder, S.; Wouters, D.; Stooker, W.; Eijsman, L.; Van Hinsbergh, V.W.; Krijnen, P.A.; et al. Activated complement is more extensively present in diseased aortic valves than naturally occurring complement inhibitors: A sign of ongoing inflammation. Eur. J. Clin. Invest. 2010, 40, 4–10. [Google Scholar] [CrossRef]
- Ge, X.; Xu, C.; Liu, Y.; Zhu, K.; Zeng, H.; Su, J.; Huang, J.; Ji, Y.; Tan, Y.; Hou, Y. Complement activation in the arteries of patients with severe atherosclerosis. Int. J. Clin. Exp. Pathol. 2018, 11, 1–9. [Google Scholar] [PubMed]
- Patzelt, J.; Verschoor, A.; Langer, H.F. Platelets and the complement cascade in atherosclerosis. Front. Physiol. 2015, 6, 49. [Google Scholar] [CrossRef]
- Xu, H.; Shang, Q.; Chen, H.; Du, J.; Wen, J.; Li, G.; Shi, D.; Chen, K. ITIH4: A new potential biomarker of “Toxin Syndrome” in coronary heart disease patient identified with proteomic method. Evid.-Based Complementary Altern. Med. Ecam 2013, 2013, 360149. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rojas, T.; Mourino-Alvarez, L.; Gil-Dones, F.; de la Cuesta, F.; Rosello-Lleti, E.; Laborde, C.M.; Rivera, M.; Lopez-Almodovar, L.F.; Lopez, J.A.; Akerstrom, F.; et al. A clinical perspective on the utility of alpha 1 antichymotrypsin for the early diagnosis of calcific aortic stenosis. Clin. Proteom. 2017, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Hribal, M.; Procopio, T.; Petta, S.; Sciacqua, A.; Grimaudo, S.; Pipitone, R.; Perticone, F.; Sesti, G. Insulin-like growth factor-I, inflammatory proteins, and fibrosis in subjects with nonalcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2013, 98, E304–E308. [Google Scholar] [CrossRef] [PubMed]
- Desai, D.; Faubion, W.A.; Sandborn, W.J. Review article: Biological activity markers in inflammatory bowel disease. Aliment. Pharm. 2007, 25, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Jadhav, A.; Hassan, A.; Meng, Q. Acute phase reactants as novel predictors of cardiovascular disease. ISRN Inflamm 2012, 6, 953461. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, A.; Epistolato, M.C.; Gianetti, J.; Castagnini, M.; Sassi, C.; Ceravolo, R.; Bevilacqua, S.; Glauber, M.; Biagini, A.; Tanganelli, P. Biological features (inflammation and neoangiogenesis) and atherosclerotic risk factors in carotid plaques and calcified aortic valve stenosis: Two different sites of the same disease? Am. J. Clin. Pathol. 2006, 126, 494–502. [Google Scholar] [CrossRef]
- Eaton, P. Protein thiol oxidation in health and disease: Techniques for measuring disulfides and related modifications in complex protein mixtures. Free. Radic. Biol. Med. 2006, 40, 1889–1899. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Zinellu, A.; Nieddu, G.; De Muro, P.; Carru, C.; Spirito, R.; Guarino, A.; Piredda, F.; Formato, M. Human serum albumin Cys34 oxidative modifications following infiltration in the carotid atherosclerotic plaque. Oxidative Med. Cell. Longev. 2014, 2014, 690953. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Formato, M. Oxidative modifications in advanced atherosclerotic plaques: A focus on in situ protein sulfhydryl group oxidation. Oxidative Med. Cell. Longev. 2020, 2020, 6169825. [Google Scholar] [CrossRef]
- Nakashima, F.; Shibata, T.; Kamiya, K.; Yoshitake, J.; Kikuchi, R.; Matsushita, T.; Ishii, I.; Giménez-Bastida, J.A.; Schneider, C.; Uchida, K. Structural and functional insights into S-thiolation of human serum albumins. Sci. Rep. 2018, 8, 932. [Google Scholar] [CrossRef]
- Al-Harthi, S.; Lachowicz, J.I.; Nowakowski, M.E.; Jaremko, M.; Jaremko, Ł. Towards the functional high-resolution coordination chemistry of blood plasma human serum albumin. J. Inorg. Biochem. 2019, 198, 110716. [Google Scholar] [CrossRef]
- Maciążek-Jurczyk, M.; Szkudlarek, A.; Chudzik, M.; Pożycka, J.; Sułkowska, A. Alteration of human serum albumin binding properties induced by modifications: A review. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2018, 188, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Cowell, S.J.; Newby, D.E.; Prescott, R.J.; Bloomfield, P.; Reid, J.; Northridge, D.B.; Boon, N.A. A randomized trial of intensive lipid-lowering therapy in calcific aortic stenosis. N. Engl. J. Med. 2005, 352, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Moura, L.M.; Ramos, S.F.; Zamorano, J.L.; Barros, I.M.; Azevedo, L.F.; Rocha-Gonçalves, F.; Rajamannan, N.M. Rosuvastatin affecting aortic valve endothelium to slow the progression of aortic stenosis. J. Am. Coll. Cardiol. 2007, 49, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Rossebø, A.B.; Pedersen, T.R.; Boman, K.; Brudi, P.; Chambers, J.B.; Egstrup, K.; Gerdts, E.; Gohlke-Bärwolf, C.; Holme, I.; Kesäniemi, Y.A.; et al. Intensive lipid lowering with simvastatin and ezetimibe in Aortic Stenosis. N. Engl. J. Med. 2008, 359, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.L.; Teo, K.; Dumesnil, J.G.; Ni, A.; Tam, J. Effect of Lipid lowering with rosuvastatin on progression of aortic stenosis: Results of the aortic stenosis progression observation: Measuring effects of rosuvastatin (ASTRONOMER) trial. Circulation 2010, 121, 306–314. [Google Scholar] [CrossRef]



| Clinical Characteristics | CAVD (n = 22) | CAVD + CAD (n = 22) | p |
|---|---|---|---|
| Age | 75.9 ± 7.6 | 77.6 ± 6.5 | 0.435 |
| Gender (%M/F) | 60/40 | 32/68 | 0.073 |
| %Obesity | 5 | 14 | 0.300 |
| %Hypertension | 77 | 82 | 0.712 |
| %Dyslipidemia | 68 | 73 | 0.744 |
| %Diabetes | 32 | 45 | 0.359 |
| %Smokers | 18 | 9 | 0.385 |
| Protein | Peptide Sequence | m/z | Charge (z) | Rt Start (min) | Rt Stop (min) |
|---|---|---|---|---|---|
| P01011 AACT | DYNLNDILLQLGIEEAFTSK | 1148.589 | 2 | 122.6 | 142.6 |
| DYNLNDILLQLGIEEAFTSK | 766.0618 | 3 | 122.6 | 142.6 | |
| EIGELYLPK | 531.2975 | 2 | 66.2 | 86.2 | |
| ITLLSALVETR | 608.369 | 2 | 92 | 112 | |
| NLAVSQVVHK | 547.8195 | 2 | 37 | 57 | |
| NLAVSQVVHK | 365.5487 | 3 | 36.8 | 56.8 | |
| P01024 CO3 | EYVLPSFEVIVEPTEK | 939.9904 | 2 | 96.2 | 116.2 |
| EYVLPSFEVIVEPTEK | 626.996 | 3 | 96.2 | 116.2 | |
| VEGTAFVIFGIQDGEQR | 933.4732 | 2 | 90 | 110 | |
| VEGTAFVIFGIQDGEQR | 622.6513 | 3 | 90 | 110 | |
| VHQYFNVELIQPGAVK | 921.4991 | 2 | 75.8 | 95.8 | |
| VHQYFNVELIQPGAVK | 614.6685 | 3 | 75.8 | 95.8 | |
| VPVAVQGEDTVQSLTQGDGVAK | 1099.569 | 2 | 67.8 | 87.8 | |
| VPVAVQGEDTVQSLTQGDGVAK | 733.3815 | 3 | 67.8 | 87.8 | |
| P05155 IC1 | GVTSVSQIFHSPDLAIR | 913.9916 | 2 | 80.2 | 100.2 |
| GVTSVSQIFHSPDLAIR | 609.6635 | 3 | 80.2 | 100.2 | |
| LEDMEQALSPSVFK | 797.3951 | 2 | 78.9 | 98.9 | |
| LLDSLPSDTR | 558.7984 | 2 | 48.7 | 68.7 | |
| TNLESILSYPK | 632.8428 | 2 | 77.7 | 97.7 | |
| Q14624 ITIH4 | GPDVLTATVSGK | 572.8141 | 2 | 46.9 | 66.9 |
| LGVYELLLK | 524.3261 | 2 | 88.7 | 108.7 | |
| NGIDIYSLTVDSR | 726.8701 | 2 | 77 | 97 | |
| TGLLLLSDPDK | 586.3321 | 2 | 73.3 | 93.3 |
| Cluster | Function | Enrich. Score | N. of Terms | Proteins |
|---|---|---|---|---|
| 1 | Platelet degranulation and regulation of endopeptidase activity | 3.96 | 4 | IGHM, PON1, ITIH4, IC1, AACT, CO3 |
| 2 | Extracellular matrix; Golgi and lysosomal lumen | 2.90 | 4 | BGN, DCN, COFA1, LUM, FA9 |
| 3 | Extracellular matrix (structural) | 2.43 | 3 | BGN, DCN, COFA1, LUM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sastre-Oliva, T.; Corbacho-Alonso, N.; Albo-Escalona, D.; Lopez, J.A.; Lopez-Almodovar, L.F.; Vázquez, J.; Padial, L.R.; Mourino-Alvarez, L.; Barderas, M.G. The Influence of Coronary Artery Disease in the Development of Aortic Stenosis and the Importance of the Albumin Redox State. Antioxidants 2022, 11, 317. https://doi.org/10.3390/antiox11020317
Sastre-Oliva T, Corbacho-Alonso N, Albo-Escalona D, Lopez JA, Lopez-Almodovar LF, Vázquez J, Padial LR, Mourino-Alvarez L, Barderas MG. The Influence of Coronary Artery Disease in the Development of Aortic Stenosis and the Importance of the Albumin Redox State. Antioxidants. 2022; 11(2):317. https://doi.org/10.3390/antiox11020317
Chicago/Turabian StyleSastre-Oliva, Tamara, Nerea Corbacho-Alonso, Diego Albo-Escalona, Juan A. Lopez, Luis F. Lopez-Almodovar, Jesús Vázquez, Luis R. Padial, Laura Mourino-Alvarez, and Maria G. Barderas. 2022. "The Influence of Coronary Artery Disease in the Development of Aortic Stenosis and the Importance of the Albumin Redox State" Antioxidants 11, no. 2: 317. https://doi.org/10.3390/antiox11020317
APA StyleSastre-Oliva, T., Corbacho-Alonso, N., Albo-Escalona, D., Lopez, J. A., Lopez-Almodovar, L. F., Vázquez, J., Padial, L. R., Mourino-Alvarez, L., & Barderas, M. G. (2022). The Influence of Coronary Artery Disease in the Development of Aortic Stenosis and the Importance of the Albumin Redox State. Antioxidants, 11(2), 317. https://doi.org/10.3390/antiox11020317






