Abstract
Bacterial genomes contain numerous insertion sequences (ISs) as transposable elements involved in actions such as the sequestration, transmission, mutation and activation of genes that can influence the responsive capacity of the organism to environmental challenges. To date, at least 30 IS families have been identified. In this review, we describe how certain ISs are transposed to carotenoid biosynthesis genes, such as phytoene synthase and phytoene desaturase, when radiation-resistant Deinococcus geothermalis with a redox imbalance and a targeted gene disruption mutation is exposed to oxidative stressors, such as gamma-irradiation, dielectric bilayer discharge plasma and hydrogen peroxide. We also explain the genetic features of IS elements, spontaneous mutation and various stress responses, including nutrient limitation, and physicochemical and oxidative stress, associated with the active transposition of bacterial ISs. Based on the current knowledge, we posit that the redox signalling mechanism inducing IS transposition involves redox sensing and redox switching for the activation of transposase expression and its activity.
1. Introduction
Since the discovery of insertion sequences (ISs; also called IS elements) in the late 1960s, the number and diversity of prokaryotic ISs have increased enormously. At present, more than 4600 different ISs in 29 families have been identified and deposited in the ISfinder platform (www-is.biotoul.fr accessed on 15 October 2021) database [1,2]. ISs are integrated as single or multiple copies throughout most bacterial chromosomes, including plasmids, as revealed in genome announcements and recent mobilome studies [3,4,5]. Intragenic transposition, and similarly, intergenic transposition of IS elements can cause gene inactivation. Furthermore, intergenic transposition of IS elements can also affect the expression of neighbouring genes. This is typically mediated by de-repression or by the introduction of a partial or complete promoter located within the IS.
Deinococcus geothermalis DSM 11300T is a radiation-resistant bacterium. Its genome contains 73 full-length IS elements from the main chromosome and two mega plasmids. Sixty-eight ISs were extracted to implicate earlier genome states as follows: Forty-two of the IS are integrated into intergenic regions, two in promoter regions and nineteen in open-reading frames (ORFs; e.g., of genes encoding enzymes and ABC transporters), and five in IS elements. Five ISs (e.g., IS200/IS605) are not extracted in silico because of the single-strand DNA (ssDNA) intermediate transposition without direct repeat sequences (DR) [6]. Disrupted genes could be reassembled, and the order of IS integration across “hot spots” could be determined through in silico correction of mutations caused by the IS element [6]. Subsequent syntenic analysis between Deinococcus sp. strain S9 from the microbial mat deposits of hot springs in the Himalaya ranges and D. geothermalis from the hot spring at Agnano in Italy revealed over 95% identical amino acid sequences of most genes in the gene arrangement of the longest contig 15 of strain S9. However, different IS elements and additional genes were integrated into the particular region of genomes in both species [6]. Thus, transpositions of IS elements are a major source of genomic plasticity and play a key role in shaping bacterial genomes.
Nowadays, ISs are easily detected and confirmed by comparative analysis of genome sequences using IS determination platforms, such as ISMapper [7], ISQuest [8], OASIS [9], and especially the pioneering database ISfinder [1]. These computational IS-finding platforms provide an IS framework using basic DNA sequence matching in the border regions of IS elements, such as the terminal inverted repeats (TIRs) and the functionally important transposase (Tpase or Tnp) gene, based on amino acid sequence similarity and conserved motifs, like the DDE-motif [2]. As reports accumulated, extracting the information became extremely difficult, so new IS nomenclature and classification had to be established. The ISfinder platform was designed and implemented to maintain a coherent annotation and is now functioning as a reference centre. It assigned nomenclature resembling that used for restriction enzymes: The first letter of the genus is followed by the first two letters of the species and a number (e.g., ISBce1 for Bacillus cereus; ISDge1 for D. geothermalis) [2]. According to ISfinder, the D. geothermalis genome contains 19 IS types belonging to 9 IS families.
Here, we describe how certain ISs are transposed to carotenoid biosynthesis genes, such as phytoene synthase and phytoene desaturase, when radiation-resistant D. geothermalis with redox-imbalance and targeted gene disruption mutations is exposed to oxidative stressors, such as gamma-irradiation, dielectric bilayer discharge plasma and hydrogen peroxide. We also briefly summarise the current knowledge of IS transposition including the genetic features of IS elements, spontaneous mutation and various stress responses, including nutrient limitation, and physicochemical and oxidative stress, associated with the active transposition of bacterial ISs.
2. Structural Properties of IS Elements and Functional Aspects
Classical IS elements are between 0.7 and 3.0 kb in length, with one or two ORFs for Tpase and its related protein. Many ISs also contain auxiliary genes that control their transposition. The entire length of the IS is terminated by flanking (im)perfect TIR sequences. Tpase catalyses the DNA cleavages and strand transfers leading to IS transposition [2,10,11]. For example, the IS1 family classically has two ORFs, InsA and InsB’. InsA negatively controls the expression of the InsAB’-fused ORF for an actual Tpase [12,13]. ISs also often generate short flanking DR sequences at the integration site using the cohesive end of the DNA. Strictly speaking, these DR sequences are not IS components but are key factors for classifying IS elements because some IS elements generate completely conserved DR sequences, some produce variable sequences, and some do not produce DR sequences [2,6]. Although IS classification is based on a variety of characteristics, the amino acid sequence similarity of their Tpases is the principal parameter used for classification [1,14,15,16].
Tpase can be separated into DDE (Asp-Asp-Glu), DEDD (Asp-Glu-Asp-Asp), HUH (His-U-His, U represents a hydrophobic residue), tyrosine (Y), and serine (S) types following the conserved amino acids in the catalytic sites, which affects the chemistry used in breaking and rejoining DNA during transposition. ISs carrying the DDE-type Tpase represent the majority of presently reported IS elements [2]. In D. geothermalis, most IS elements are DDE-type, except for ISDge10, ISDge18 and ISDge19 of the IS200/IS605 family, which use HUH-type Tpase (ISfinder). DDE-type Tpases coordinate essential metal ions and use hydroxyl ions, OH− (e.g., H2O), as a nucleophile in a transesterification reaction [2,10]. They do not form covalent Tpase-DNA intermediates during the transposition process. HUH-type Tpases use tyrosine as a nucleophile and generate a transitory covalent 5′ tyrosine-DNA transposition intermediate. HUH-type Tpases are widespread single-strand nucleases [17,18,19] that also include Rep (replication) proteins involved in bacteriophage and plasmid rolling circle replication, as well as relaxases or Mob (mobilisation) proteins involved in conjugative plasmid transfer [20]. Serine Tpase and tyrosine Tpase are relatively minor types.
At present, IS elements in ISfinder are grouped into approximately 30 families, many of which are further divided into subgroups based on shared characteristics. According to Siguier et al., although there are some very well-defined homogeneous families (such as IS3, IS30 and IS256; IS6 and IS26) and others that have been redefined over time as ever more IS are identified (e.g., IS4 and IS5), there is no “quantitative” measure of the weight of each of the criteria used to define a family [2,21].
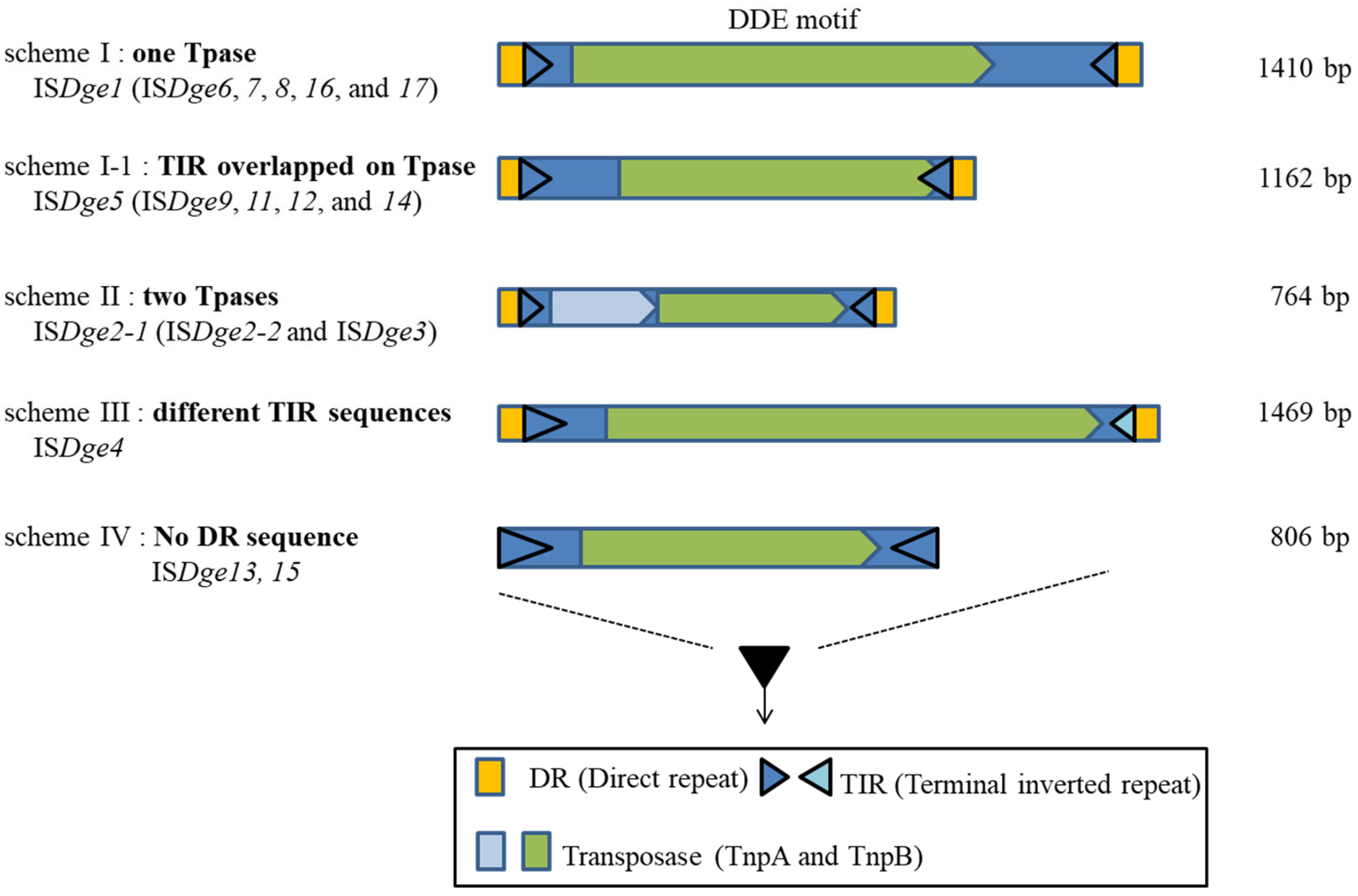
The IS200/IS605 family in the HUH superfamily is classified into five structural schemes based on the number of Tpase, Tpase composition and transcriptional direction of Tpase [19]. Thus, we proposed an upper level of IS family clustering and classify four structural schemes of majority DDE motif IS elements following the ORF (mainly Tpase) number, the identity of TIR sequence and DR sequence presence or absence in the case of D. geothermalis (Figure 1) [6].
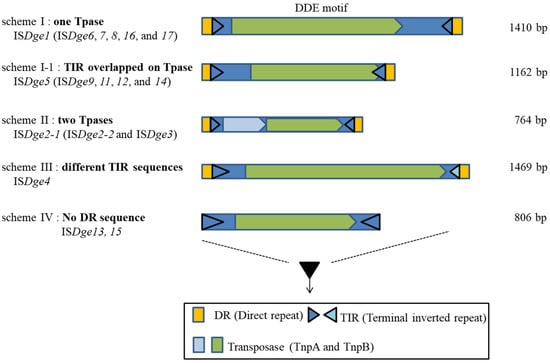
Figure 1.
Illustration of four schemes of DDE-type IS elements in Deinococcus geothermalis.
There are two typical IS structures distinguished by the number of ORFs (scheme I–II) and two distinct structures distinguished by the TIR and DR sequence patterns (scheme III–IV). Scheme I ISs have a single Tpase gene and constant DR sequences. Scheme I-1 has a single Tpase gene and an overlapping right TIR sequence in the ORF region of the Tpase. Scheme II ISs have two Tpases-related genes. Scheme III has a typical structural composition, but, interestingly, the TIR sequence is not identical. Scheme IV ISs do not have the two DR sequences in the IS border region.
When this scheme was adopted to DDE-motif IS family classification in D. geothermalis, IS4, IS5, IS630, IS701 and IS982 belonged to scheme I, the IS1 family belonged to scheme II, IS66 belonged to scheme III and the IS6 family belonged to scheme IV. The IS6/IS26 family has a rule in their TIR sequence, such as “GG-4N-G”, which means that the first two nucleotides are “GG” and the seventh nucleotide is also “G”. The IS6/IS26 family included ISDge13 and ISDge15 of D. geothermalis [22]. It was proposed that all the collected IS6 and IS26 families of bacterial and archaeal genomes encompassed a single family. However, ISDge13 has a duplicated “GG-4N-G” conserved sequence in the TIR sequence because of target site duplication: Either the DR sequence is not detected, or the DR sequence is eight nucleotides in length (ISfinder). Interestingly, there were different DR sequence absence ratios of IS6 family members in 88% of Gram-negative bacteria, 60.6% of Gram-positive bacteria and 35.1% of archaea, according to Harmer and Hall’s data [22]. Recently, Varani et al. also reviewed the IS6 family, specifically, the Tpase size compositions and the structures of 160 IS6 family members important in generating multiple antibiotic-resistance genes in bacteria and archaea [23]. Additional transposition experiments are needed to detect the transposition events in IS6/IS26 family members for further classification of these IS elements within prokaryotic genomes. The next section looks briefly at the transposition mechanisms of IS elements.
3. IS Transpositional Procedures and Trigger Factors
In general, IS transposition processing is dependent on the type of Tpase in the IS family. Most DDE-motif Tpases have a “copy-and-paste” mechanism [2,10,11]. In the case of D. geothermalis, the active transpositions of several IS elements of the IS1, IS5, IS6 and IS701 families perform this “copy-and-paste” or a replicative mode of transposition [24,25,26]. Briefly, the expressed Tpase bound to itself on the end of the IS element and cleavage covalent bond between adjacent nucleotides and the template DNA synthesised nascent strand in both strands. The excised double-stranded DNA-Tpase complexes bind to the target site and integrate in a manner similar to the sticky ends produced by restriction enzymes [2]. The sticky ends for novel integration became DR sequences in the border region of IS elements. There are several other modes of active transposition besides the “copy-and-paste”. For example, the IS4 family of IS10/IS50 shows the “cut-and-paste” mode, and IS91/Tn916 shows no DR sequence production and performed a rolling-circle mechanism [2].
For IS200/IS605 family members, the transposition mechanism is well understood due to a combination of genetic, biochemical and structural studies [27]. Briefly, it can be described as a single-strand “peel-and-paste” mechanism in which the IS is excised as a single strand (the “top” strand) from the lagging strand template of the donor molecule to form a single-strand transposon circle and then is inserted into a single-strand target at the replication fork [19]. The IS200/IS605 family members include subterminal secondary structures recognised by TnpA. The cleavage sites occur a short distance of 5′ to the left and 3′ to the right of the structure. These are not directly recognised by TnpA but form a complex set of interactions with the internal sequence that permits their cleavage. Transposition occurs by insertion of the left end 3′ to a specific tetra- or pentanucleotide essential for excision and further transposition. Insertion does not generate a DR and occurs preferentially in the lagging strand template of the replication fork. For example, IS608 from Helicobacter pylori always inserts 3′ into a TTAC tetranucleotide, and ISDra2 specifically integrates 3′ into a TTGAT pentanucleotide. This results in a clear orientation bias at the genome level, reflecting the direction of replication of the target replicon. The success of these IS elements is related to the interplay between Y1 HUH Tpase (an ssDNA endonuclease that contains only one tyrosine active site) and replication fork-associated factors and can be detected in numerous bacterial genomes. [18,19,20,27,28]. Moreover, ISDra2 transposition is intimately associated with the gamma-irradiation-induced genome fragmentation and reassembly of D. radiodurans. In this section, we provide a summary of IS transposition studies in various species of prokaryotes and some interesting perspectives.
3.1. IS Transposition in Deinococcus-Thermus
Although the genome of the hyperthermophilic bacterium Thermus scotoductus contains the highest IS copies in terms of the genome size, the genomes of 3 of the 28 Thermus spp. (Thermus thermophilus HB8, HB27 and NAR1) contain a broad spectrum of eight IS family members: IS3, IS4, IS5, IS1634, IS256, IS630, IS701 and IS110 [3]. Interestingly, ISTth3 of the IS1634 family, ISTth4 of the IS256 family, ISTth7 of the IS5 family and IS1000A of the IS110 family transpose at a relatively high frequency in Thermus spp. The functional roles of ISTth7, which is actively transposed by streptomycin, were identified by Gregory and Dahlberg [29]. ISTth7 inserted in the rsmG gene encoding a methyltransferase that produces m7G527 in the 16S rRNA, resulting in the streptomycin resistance phenotype. ISTth7 is widely distributed in the genomes of T. thermophilus strains in the form of a potentially activated full-length IS element [3].
When D. radiodurans was exposed to gamma-irradiation, ISDra2 of the IS200/IS605 family was inserted into the thyA gene encoding a thymidylate synthase, which was selected under appropriate conditions on a trimethoprim (100 µg/mL) plate [17,18,19,27,30].
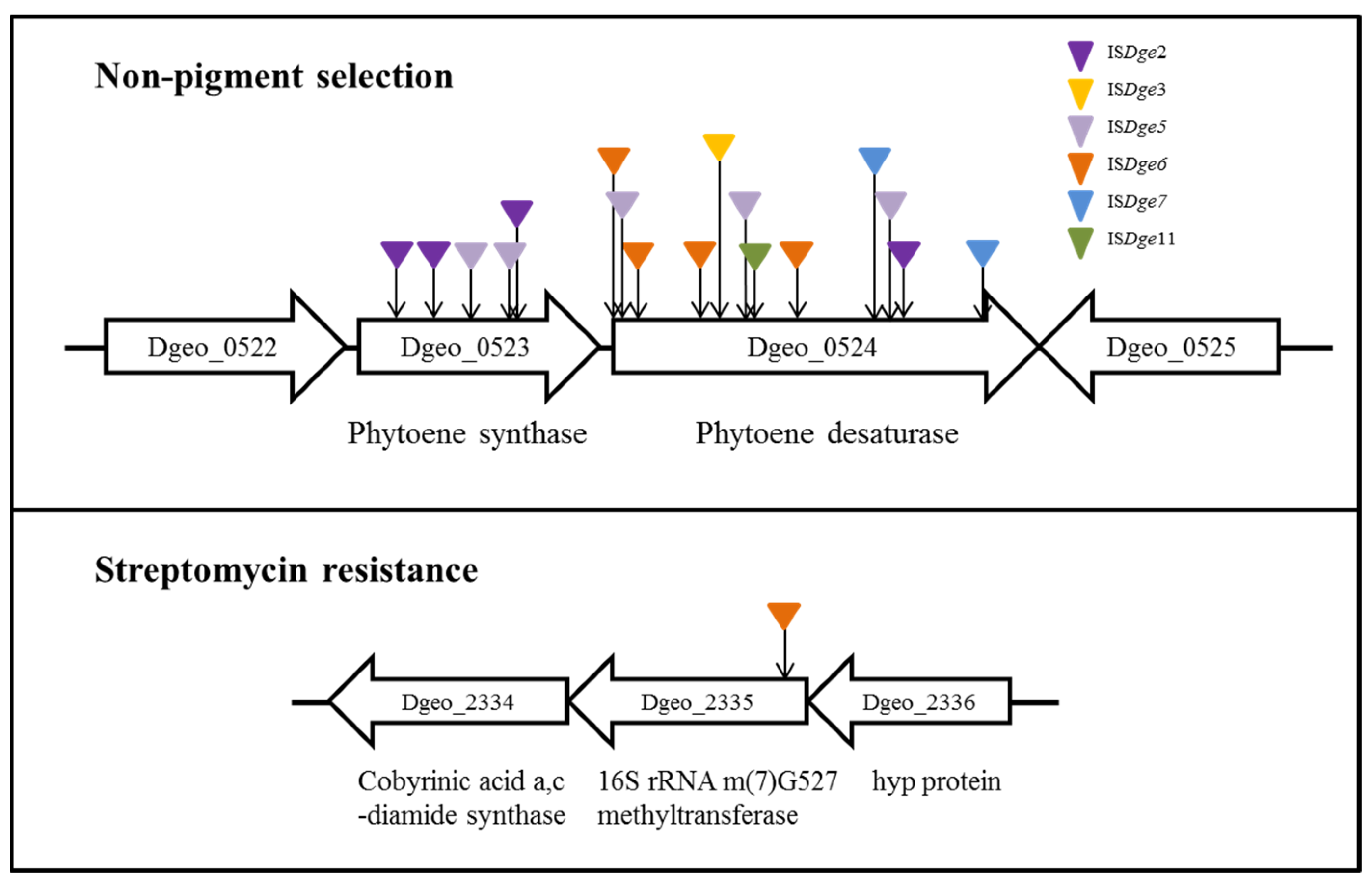
Among different D. geothermalis lineages, the following specific types of IS elements actively transposed to carotenoid biosynthesis genes (phytoene cyclase and phytoene desaturase) and an rsmG gene (conferring streptomycin resistance) with a replicative mode due to hydrogen peroxide-induced oxidative stress conditions: ISDge6 of the IS5 family in a LysR family regulator-disrupted mutant, ISDge5 of the IS701 family in a dps gene (encoding a chromosome stabilizer protein, Dps: DNA-binding protein from starved cells)-disrupted mutant strain and a cystine importer-disrupted mutants and ISDge11 of the IS4 family in the wild-type strain (Figure 2) [24,25,26]. Nevertheless, the scheme III transposition events of IS elements with different TIR sequences and the scheme IV transposition events of IS elements without DR sequences have not yet been found under the oxidative stress condition in WT and target gene-disrupted mutants [31]. If the transposition of particular IS elements is under the influence of unique target genes, it will be possible to explain and understand the states of IS induction for sensing and signalling environmental stress and genome evolutionary processes in prokaryotic genomes.
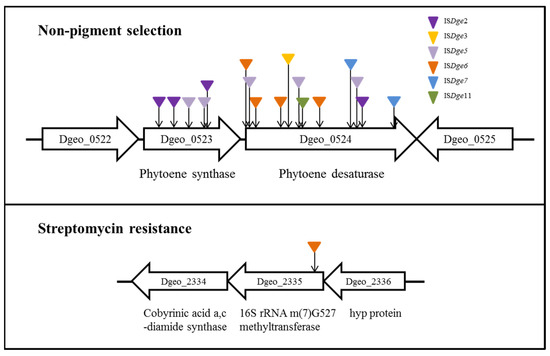
Figure 2.
Active transposition of IS elements in Deinococcus geothermalis under oxidative stress. The transposition events were isolated from two phenotypic selections.
3.2. IS Transposition in Gram-Negative Bacteria
Here, we focused on the well-defined transposition cycle of IS911. IS911 of the IS3 family was transposed to the phage cI repressor gene in Shigella dysenteriae phage λ lysogen by spontaneous insertion [32]. This IS911 element is found in the genome of various Gram-negative bacteria, including Escherichia coli K12. Scheme II IS911, containing DDE motif Tpase has an overlapping right TIR sequence in the Tpase ORF, may need to extend scheme II-1. Temperature-induced (42 °C) transposition of IS911 formed figure-eight molecules [33]. This transposition procedure is the “copy-out-paste-in” mechanism. Interestingly, IS911 expressed the fusion protein OrfAB by frameshifting through “slippery” lysine codons, and Tpase expression was controlled by the “junction” promoter that assembled in both TIRs following the circulation of IS911. The overall two-step IS911 transposition required the consecutive assembly of synaptic complex A (SCA) to start the process, leading to replication and circularisation of the transposing IS copy, and synaptic complex B (SCB) to ensure that the replicated copy was integrated into the target DNA. In the first step, OrfAB Tpase is bound to the TIRs, then assembled by end pairing, followed by one-end cleavage and a strand transfer to form figure-eight molecules for excision of the IS element. In the second step, the circular IS911 was generated by replication and target pairing with the involvement of the OrfAB complex through one-end cleavage and strand transfer, then inserted in the target loci. IS911 insertion is also possible at nontarget loci due to the collaboration between OrfAB and OrfA via the two-end cleavage and strand transfer [32,33].
Transposition procedures of ISs occurred in different ways dependent on the IS family in numerous Gram-negative bacteria, and the triggering factors of transposition are also varied. For example, Pseudomonas and Burkholderia respond to high temperatures, conjugation or oxidative stress resulting in active transposition of IS elements [34,35,36].
3.3. IS Transposition in Gram-Positive Bacteria
Transposition of various ISs, such as IS4, IS701, IS1634 and ISLre2, has been described in Geobacillus kaustophilus via sigX-dependent stress responses at elevated temperatures, under nutrient limitation for uracil prototrophs, C- and N-source starvation and antibiotics treatment [37]. A mobilome analysis of 102 genomes of Bacillus cereus sensu lato species revealed 16 IS families distributed in a species-dependent manner [4]. For example, IS982, IS630 and IS5 were uniquely located in Bacillus thuringiensis, whereas IS4 and IS3 were distributed among all analysed genomes. Moreover, Bacillus subtilis 168, which has no IS4Bsu1, exhibited IS4Bsu1 transpositions from B. subtilis (natto) when grown under a high temperature and competence-inducing conditions but not under an optimal temperature and nutrient-rich medium conditions [38].
3.4. IS Transposition in Archaea
Filée et al. reviewed archaeal IS diversity. Archaeal genomes contain numerous IS families, just like bacterial genomes [39]. Although genome sequence analysis has detected many IS elements, there are rare observations of IS transposition via an induction mechanism, except for spontaneous mutation [40]. Perhaps the archaeal active transposition of IS is a prominent case study in this evolutional research field [41].
3.5. Use of Transposon Mutagenesis
Typically, transposon (Tn) mutagenesis is a powerful tool for detecting the functional role of uncharacterised genes using random transposition selection by Tn. In Bifidobacterium longum, ISBlo11 of the IS3 family was detected by the sacB-based counterselection system and analysed for its activity in E. coli. The constructed E. coli−Bifidobacterium shuttle vector harbouring sacB was introduced into B. longum. ISBlo11 moved into sacB, and the sucrose-resistant phenotype was selected. IS-transposed clones in B. longum 105-A were selected by simple conjugation in E. coli and growth in 4% xylose and 1% glucose [42,43].
4. Inductive Signals of Active Transposition
In many IS elements, a single Tpase controls its own IS expression and transposition by its own promoter. However, in the case of IS elements consisting of two ORFs, for example, tnpA encodes a Tpase and tnpB encodes a repressor resulting in ISDra2, transposition is regulated by inhibiting excision and insertion of whole IS elements [18,19,27]. The expression level, protein stability and activity of a particular Tpase are important factors for the successful transposition of a particular IS. Together with different host factors, these Tpase-associated factors are controlled by transcriptional and translational regulation [44].
However, the question remains: What are the Tpase induction signalling pathways that sense environmental stressors? Some of the many possibilities are host factors, such as directly bound chromosomal DNA stabilisers; redox-sensing regulators, including reactive oxygen species (ROS)-sensing; particular signal transduction pathways; intracellular low-molecular-weight (LMW) compounds, such as thiols and redoxins; conjugation; cellular toxic physical and chemical factors, as well as spontaneous transposition [16]. Here, we summarised several factors, including host factors, environmental factors, such as temperature and nutrition, ROS-producing radiations, redox imbalance and some known redox-signalling events by oxidative stressors.
4.1. Host Factors
Nucleoid-associated proteins that induce changes in DNA structural topology can affect the behaviour of IS elements. For example, H-NS (histone-like nucleoid structuring protein) may explain the targeting preferences of H-NS mutants for IS903 and IS10 [45], IS1 [46] and IS5 [47]. H-NS is necessary for efficient IS transposition [47,48]. In addition, IS transposition can also be modulated by host DNA stabilisers, such as IHF (integration host factor), HU (heat-unstable protein) and FIS (factor for inversion stimulation), the replication initiator DnaA, the protein chaperone/protease ClpX/P/A, the SOS control protein LexA, the DNA methylase Dam and GTP levels [44,48,49,50]. Nevertheless, the in vitro detection method of some host factors of active transposition is limited because the screenings always have a positive effect. General host factors have affected chromosome stabilising, resulting in the need for multiple-level signalling for IS transposition.
Bacterial IS transposition reactions have also been shown to be regulated by Hfq. This well-studied RNA chaperone binds small regulatory RNA (sRNA) and plays a central role in complex post-transcriptional regulatory networks for target genes in many bacteria [51,52,53]. For example, it interacts directly with the ribosome-binding site of IS10 Tpase mRNA, resulting in translation inhibition [54].
Dps works in concert with other host factors to organise the chromosome. It is abundant in starved E. coli cells and is involved in stress resistance. Dps is dominantly expressed in the stationary phase of E. coli and affected IS transposition, although the exact mechanism is not yet known. In the case of D. geothermalis, dps-deficient mutants exhibited active transposition of specific IS family members [24].
The transpositional activity of IS elements is under strict regulation, presumably to limit their effects on the host cell. Vandecraen et al. aptly summarised the regulatory factors controlling IS transposition [16]: Transcriptional repressors [55,56], translational inhibitors [57], ribosome frameshifting [58], impinging of transcription by secondary mRNA structures [59], methylation sites [60], Tpase instability [61] and target site preference [62,63].
4.2. Nutrition and Temperature
The nutrient-rich environment (e.g., high-glucose level) of the host reduces the requirement for many genes that are essential for free-living bacteria. This allows the fixation of slightly deleterious mutations in the population by random stochastic IS transposition and the concomitant increase in the IS copy number in the genome [2,64].
In E. coli, the IS1, IS30 and IS911 families show temperature-sensitive transposition [46,65]. Specifically, high-temperature conditions decreased the transposition activity. In contrast to E. coli, the transposition of novel IS elements of IS5 and IS21 family members from a soil-derived Burkholderia multivorans strain was enhanced 7-fold under a high temperature at 42 °C but not under oxidative stress and starvation conditions [34]. Genome-wide sequencing and Southern blot analysis of IS elements revealed that the G. kaustophilus wild-type genome contained 19 IS families and 118 copies of full-length IS elements. Several IS families showed growth inhibition-, sigma factor- and heat shock-dependent active transposition [37].
Using the papillation screen method in E. coli, various stressors positively affected IS transposition by different host factors, such as regulators, metabolism, protein stability and folding and DNA metabolism [51]. In conclusion, the IS transposition induction in prokaryotes is a complex and orchestrated phenomenon that is affected by a large number of host factors and environmental factors.
4.3. Gamma-Irradiation and Dielectric Bilayer Discharge Plasma
Gamma-irradiation is a strong DNA-damaging agent that results in genome DNA fragmentation. Interestingly, in this extreme stress state of D. radiodurans, five IS elements (IS2621, ISDra2, ISDra3, ISDra4 and ISDra5) are actively transposed to the thyA gene by gamma-irradiation [19,30,66]. Under ultraviolet radiation, the transposition of IS2621 into the uvrA gene encoding for replication, repair and recombination factors was detected [67]. ISDra2 of the IS200/IS605 family is induced 100-fold by gamma rays [66].
We recently performed gamma-irradiation treatment of D. geothermalis WT and several targeted gene-disrupted mutants. Our carotenoid-deficient screening test successfully obtained non-pigment mutants with the disruption of the two genes encoding phytoene cyclase and phytoene desaturase, respectively, caused by the IS integration. Gamma-irradiation led to additional transpositions of IS families compared to the oxidative stress response to H2O2. However, each IS family still followed a specific selectivity (Ye et al., submitted). For example, ISDge2 and ISDge3 of the IS1 family are limitedly transposed by gamma-irradiation. In addition, the dielectric bilayer discharge (DBD) plasma-radiation was also performed to detect active IS transposition. This approach successfully increases the frequency of active IS transposition to the target genes. Interestingly, these IS transposition events of DBD plasma-radiation are the same IS types and transposition loci as those under H2O2-induced oxidative stress.
4.4. Redox Imbalance
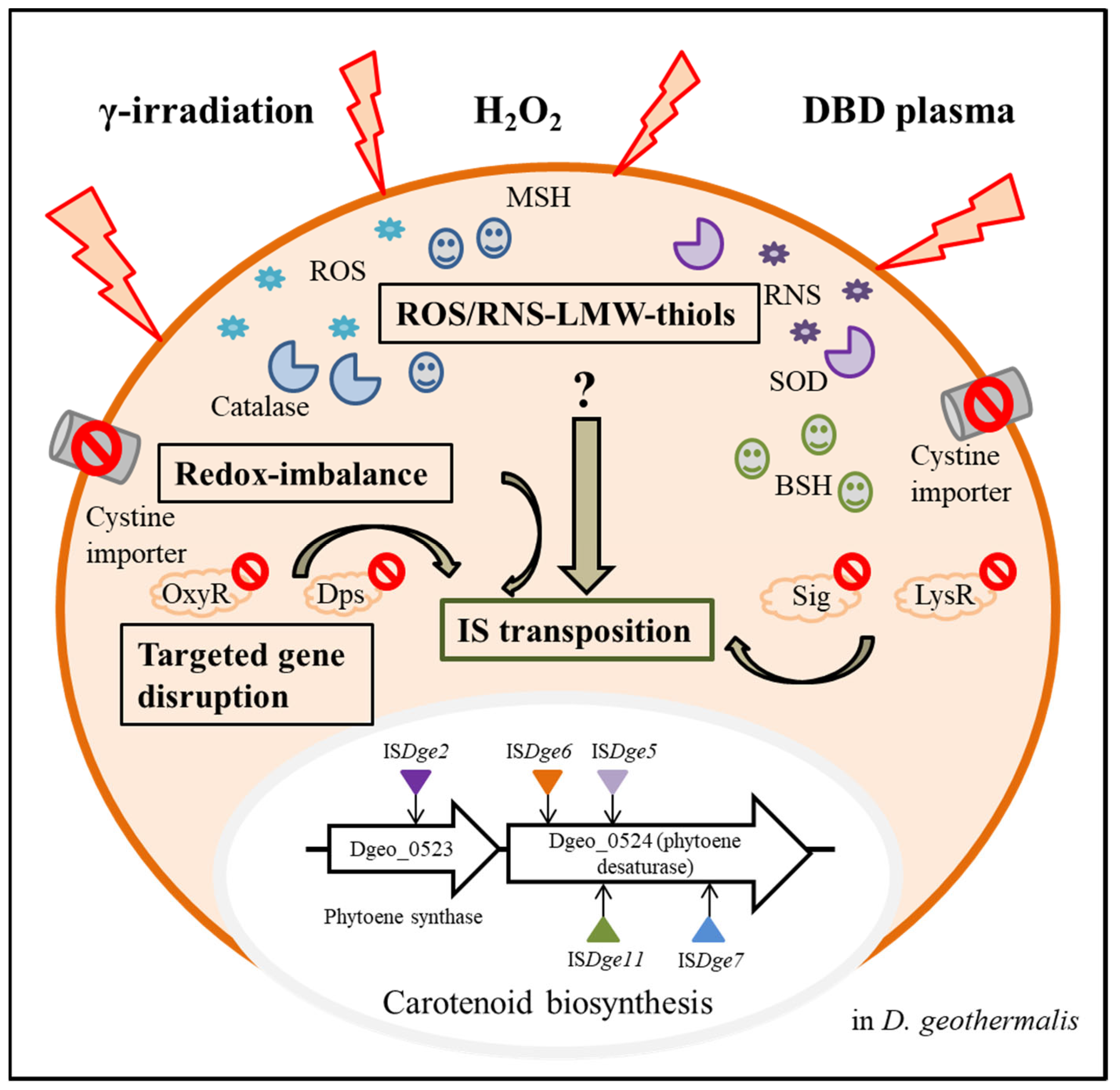
Furthermore, we additionally demonstrated specialised IS transposition in the redox-imbalance condition using a cystine importer-disrupted mutant and its complementary strain [26]. Interestingly, both ISDge5 of the IS701 family and ISDge7 of the IS5 family transposed to carotenoid biosynthesis genes, resulting in the non-pigmented phenotype (Figure 3). This selectable IS transposition mirrors the phenomenon of the dgeo_0257 (DgDps3)-disrupted mutant [24]. In the redox-imbalanced state, the expression level of DgDps3 was enhanced, but DgDps1 was strictly downregulated. DgDps1 and DgDps3 showed complementary effects on their expression levels depending on the growth phases. DgDps1 expression was enhanced during growth in the absence of DgDps3. Similarly, DgDps3 was strongly expressed at the early and late exponential growth phases in the DgDps1-deficient condition under oxidative stress (Bae et al., manuscript prepared). DgDps3-deficient conditions revealed active transposition of ISDge5 of the IS701 family. Both redox-imbalance and dps-deficient conditions often revealed a common transposition phenomenon of the IS element. This leaves a pertinent question regarding Tpase induction signalling and its network regulation between redox-imbalance and dps-deficient conditions under oxidative stress. Interestingly, dps gene-deficient mutants no longer expressed bshA, a gene required for bacillithiol biosynthesis [31]. However, WT and redox-imbalanced strains revealed similar expression levels of bshA [26]. Under oxidative stress, dps-deficient mutants revealed dramatically enhanced expression of mycothiol biosynthesis enzymes, such as MshBCD, specifically at the early exponential growth phase. Therefore, specific redox-sensing signal transduction for LMW-thiols-mediated induction could modulate the redox-sensing regulators involved in the oxidative stress response. However, deconvoluting the signalling pathways from ROS sensing to activating IS transposition remains a challenge.
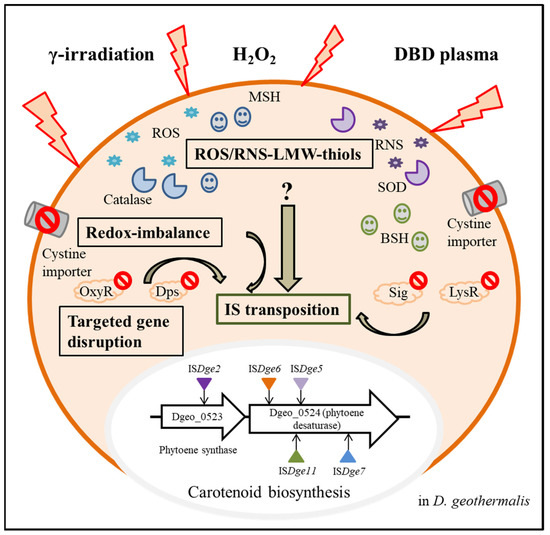
Figure 3.
Overview of active transposition of ISs through various intracellular factors in Deinococcus geothermalis from three extracellular stressors. Three extracellular stressors induced ROS and RNS, and a cystine importer-disrupted mutant induced the intracellular redox imbalance state. LMW-thiols, such as bacillithiol and mycothiol, and antioxidant enzymes were produced. As a result of oxidation stress in WT and mutants with targeted gene disruption of oxyR, lysR, dps and sig, particular IS elements were transposed to the carotenoid biosynthesis genes, and phenotypic non-pigment colonies were isolated.
4.5. Antibiotics
Recently, we applied oxidative stress-induced IS transposition to streptomycin-resistant phenotypic selection because antibiotic resistance is a powerful selectable biomarker, and the genetic network of the target genes and biochemical functions are normally well defined. Generally, peptide translocation-blocking aminoglycoside streptomycin was affected on the ribosomal structure via direct interaction with its component’s ribosomal proteins and 16S rRNA [68]. The streptomycin-resistant phenotype is easily obtained through low-level streptomycin induction, resulting in spontaneous mutations in ribosomal component-related genes, such as rpsL encoding a S12 ribosomal protein, rsmG which is a chemical modifier of m7G527 on the 16S rRNA and mthA encoding an enzyme involved in the S-adenosylmethionine (SAM) recycling pathway (see Section 3.1) [69,70]. These spontaneous mutations revealed MIC (minimum inhibitory concentration) level-dependent resistance [71]. Interestingly, when we induced oxidative stress by H2O2 treatment in a LysR family regulator-deficient mutant, the IS5 family element was actively transposed to rsmG, resulting in strains with high streptomycin resistance (MIC > 10,000 µg/mL; Lee et al., submitted).
This observation is similar to particular IS transposition in the thermophilic bacterium T. thermophilus. Researchers performed serial cultivation with gradual streptomycin treatment and selected the streptomycin-resistant phenotype, which originated from many spontaneous mutations and the transposition of the ISTth7 to rsmG [29,72]. Surprisingly, transposition of the IS by oxidative stress (H2O2 treatment) for streptomycin resistance was limited in a LysR family regulator-deficient mutant.
4.6. Metals
The genome of the famous metal-resistant bacterium Cupriavidus metallidurans CH34 contains 21 distinct IS elements (a total of 57 full-length ISs) belonging to 10 IS families [73]. When zinc ions (Zn2+) at an 0.8 mM concentration were present in the culture plates, zinc-resistant mutants from C. metallidurans AE126 (a strain cured for pMOL30 mega-plasmid) with a compromised czc (cadmium/zinc/cobalt resistance) operon were isolated. Vandercraen et al. performed colony PCR experiments to determine the active transposition of IS elements affecting the cnr (cobalt-nickel resistance) operon and found the transposition of mainly ISRme5 and another six IS elements in the first population [74]. Moreover, the second population enhanced the contribution of IS elements with the identification of IS1087B and IS1088, in addition to ISRme5. Interestingly, these major three IS elements increased the endogenous promoter activity of Tpase via Zn2+ and Cd2+ (cadmium ion) induction. Further research is needed to understand the relationship between metal ion stress and the transposition of IS elements.
5. Redox-Switched Regulators and Redox Signalling
Currently, there is no clear evidence that the multiple stress-sensing regulators directly cause IS induction. Is this because of what happens in various stressed cells, including oxidative stress, and which triggering factors selectively activate particular types of IS element silencing in the genomes?
Very recently, chromatin immunoprecipitation sequencing (ChIP-Seq) analysis and RNA sequencing (RNA-Seq) analysis showed that the radiation/desiccation response (RDR) regulon operated through the cis-acting sequence RDRM (Radiation Desiccation Response Motif), the trans-acting repressor DdrO and the protease IrrE possibly controlled a unique IS family [75]. The authors performed a systematic and comprehensive sequence analysis for the RDR motif and newly found the DdrO target genes, which involved two Tpase that included the IS5 and Tn3 families. The DdrO-IrrE regulatory system is a recently well-defined regulatory system in the extremely radiation-resistant bacterium D. radiodurans [76,77,78]. When cells were exposed to oxidative stress, most of the ROS produced was directly detoxified by the enzymatic cooperation of catalase, peroxidase and superoxide dismutase and the scavenging of LMW thiols. Nevertheless, ROS triggered the chemical modification of several transcriptional regulators, resulting in the activation of proteins, such as the redox-sensitive transcriptional regulator OxyR. When cells were exposed to radiation and desiccation stresses, the intracellular redox balance was destroyed and the Zn2+-chelating proteins were distorted, releasing Zn2+. Zinc is a cofactor element for the IrrE protein. IrrE is a metalloprotease that cleaves the dimeric form of DdrO bound to the RDRM motif of the RDR regulon. By inactivating the repressor DdrO, gene expression mediated by the RDR regulon is activated [75,78]. We expect this is an example of a defined redox-sensing pathway. Perhaps multiple signalling pathways for sensing a redox imbalance are connected to different environmental stressors. Logically, many transcriptional regulators recognise redox changes in the cytoplasm, and particular regulators might control the expression of Tpase as a key player in IS transposition [79,8081]. Recently, the proteome data of Deinococcus were updated, thus enabling the use of this model organism for investigating multi-scale proteomic questions, including how response mechanisms cope with physicochemical stresses [82,83]. This information provides research opportunities to identify the redox-signalling pathways and regulators of active IS transposition.
6. The Evolution of Prokaryotic Genomes via IS Elements
There are many hypothetical backgrounds for bacterial genomic evolution. The proposed extracellular origin is based on horizontal gene transfer mechanisms. Prokaryotic cells acquire foreign DNA through several gene transfer systems. The lateral gene transfer of genes can occur through transformation, transduction and conjugation. The intracellular mechanisms for genome plasticity are gene duplication by recombination, deletion, homopolymeric tracts by the slippage of DNA polymerase [84] and the moving of transposable elements. The impact of IS transposition on the bacterial genomes can have positive, neutral or negative effects on cell fitness. Under selective conditions (e.g., antibiotic pressure), IS-mediated beneficial mutations are favoured, and fixation of the IS at that particular target site depends on its exerted effect (e.g., gene inactivation or modulation of gene expression).
Perhaps the most common effect of IS transposition is gene inactivation. Many cases have been described illustrating the modulation of antibiotic and xenobiotic resistance, virulence and metabolic activity modulation by IS-mediated gene inactivation. IS transposition to non-coding regions can lead to the altered expression of neighbouring genes by IS elements carrying a complete outward-directed promoter [16].
Vandecraen et al. [16] posited four hypotheses as an interesting question regarding how ISs are maintained in a bacterial genome on an evolutionary timescale: The parasitic nature with self-replicating ability, generating occasional beneficial mutations in promoting genetic variation, selectively neutral and coexistence in a dynamic equilibrium.
IS elements can have an important impact on the evolution of their hosts. In the case of D. geothermalis, chronic H2O2-exposed cells exhibited the streptomycin resistance phenotype due to IS transposition and several point mutations in streptomycin resistance-related genes, such as rpsL and rsmG (Lee et al., submitted). In particular, streptomycin-dependent phenotypic strains grew better in the presence of streptomycin and under the oxidative stress condition compared to the wild-type strain. Thus, whether or not IS elements are selfish and parasitic, their impact on the architecture of microbial genomes regarding adaptation and survival is undeniable.
The insertion of specific IS elements, such as IS1, IS3 and IS5, can result in topological changes in the stress-induced DNA destabilisation (SIDD) region near the promoter as a “hotspot” and transcriptional activation of the flhDC-encoded master regulator in the flagella system [85,86]. In E. coli, the normally silent β-glucoside (bgl) catabolic operon, the glpFK for the glycerol utilisation operon and a cryptic anabolic functional operon are activated by the insertion of IS5 upstream of the promoter under starvation in the presence of glycerol, and specific genes encoding nitroreductases are inactivated under antibiotic stress [16,47]. IS5 can lead to gene activation via integration into a specific recognition sequence in the SIDD region, and precise excision of IS5 from the fucPIK operon can reverse the activation [87]. Humayun et al. reported that environmental stress conditions appeared to influence IS5 insertion through the relatively non-specific nucleoid protein H-NS and locus-specific DNA-binding proteins, such as GlpR and Crp [47]. These findings provide supporting evidence for a highly evolved and mutually beneficial relationship between IS and the host genome.
We expect that studies of the active transposition of ISs in Deinococcus as a model system will explain the molecular evolution of genome plasticity in the imminent future. In particular, the classification of active transposition of IS types in WT and targeted gene disruption mutants offers a clue to define specific IS selectivity and functional regulatory networks between intracellular redox-imbalance and redox-sensitive regulators (e.g., a LysR family member, a sigma factor and a putative Dps protein) under differential oxidative stress-inducing treatments.
7. Conclusions
In this review, we covered the active transposition of IS elements in prokaryotic genomes. The activation of IS transposition was triggered by various extracellular and intracellular redox imbalances, including oxidative stress and physicochemical stress. Currently, there is a gap linking the redox-sensing regulations with the active IS transposition of prokaryotes. In the near future, it is expected that further mobilome analysis and full genome sequence analysis by deep sequencing and IS-sequence technology will assist in understanding the IS distribution and stressor-dependent IS transposition patterns in several natural isolates of the same bacterial species. Therefore, D. geothermalis is a good model organism for explaining the network regulations, including the active transposition of IS elements, redox-sensitive regulators and intracellular stress responses.
Author Contributions
E.S., Q.Y. and S.-J.L. participated in conceptual design and data collection and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2020R1F1A1070779).
Acknowledgments
The authors thank K. Choo, M. Kim and C. Lee for the construction of mutant strains, RNA-Seq analysis and project initiative.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviation
| IS | insertion sequence |
| Tpase (Tnp) | transposase |
| InsA; InsB’ | two ORFs in IS element and in some cases both genes were produced a fused protein (InsAB’) or InsA regulated InsB’ expression |
| TIR | terminal inverted repeat |
| DR | direct repeat |
| Dps | DNA-protection protein from starved cell |
| H-NS | histone-like nucleoid structuring protein |
| LysR | a broad transcriptional regulator family |
| DBD | dielectric bilayer discharge |
| DdrO-IrrE | Deinococcus unique regulatory system for RDR regulon |
| RDR | radiation-desiccation responded regulon |
| ROS | reactive oxygen species |
| SIDD | stress-induced DNA destabilisation region |
References
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s guide to bacterial insertion sequences. Microbiol. Spectrum 2015, 3, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Sanchez, M.; Sacristan-Horcajada, E.; Fuente, S.G.; Peiro, R.; Berenguer, J. Into the Thermus mobilome: Presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Fayad, N.; Awad, M.K.; Mahillon, J. Diversity of Bacillus cereus sensu lato mobilome. BMC Genom. 2019, 20, 436. [Google Scholar] [CrossRef]
- Durrant, M.G.; Li, M.M.; Siranosian, B.A.; Montgomery, S.B.; Bhatt, A.S. A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 2020, 27, 140–153. [Google Scholar] [CrossRef]
- Lee, C.; Bae, M.K.; Choi, N.; Lee, S.J.; Lee, S.-J. Genome plasticity by insertion sequences learned from a case of radiation-resistant bacterium Deinococcus geothermalis. Bioinformat. Biol. Insights 2021, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; Hamidian, M.; Wick, R.R.; Edwards, D.J.; Billman-Jacobe, H.; Hall, R.M.; Holt, K.E. ISMapper: Identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genom. 2015, 16, 667. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Gauthier, D.T.; Ranjan, D.; Zubair, M. ISQuest: Finding insertion sequences in prokaryotics sequence fragment data. Bioinformatics 2015, 31, 3406–3412. [Google Scholar] [CrossRef]
- Robinson, D.G.; Lee, M.-C.; Marx, C.J. Oasis: An automatic program for global investigation of bacterial and archaeal insertion sequences. Nucleic Acid Res. 2012, 40, e174. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Dyda, F. Mechanisms of DNA transposition. Microbiol. Spectrum 2015, 3, 3.2.12. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Dyda, F. DNA transposition at work. Chem. Rev. 2016, 116, 12758–12784. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Machida, Y. Regulation of transposition by the insA gene product. J. Mol. Biol. 1989, 208, 567–574. [Google Scholar] [CrossRef]
- Ton-Hoang, B.; Turlan, C.; Chandler, M. Functional domains of the IS1 transposase: Analysis in vivo and in vitro. Mol. Microbiol. 2004, 53, 1529–1543. [Google Scholar] [CrossRef] [PubMed]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Barabas, O.; Ronning, D.R.; Guynet, C.; Hickman, A.B.; Ton-Hoang, B.; Chandler, M.; Dyda, F. Mechanism of IS200/IS605 family DNA transposases: Activation and transposon-directed target site selection. Cell 2008, 132, 208–220. [Google Scholar] [CrossRef]
- Hickman, A.B.; James, J.A.; Barabas, O.; Pasternak, C.; Ton-Hoang, B.; Chandler, M.; Sommer, S.; Dyda, F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J. 2010, 29, 3840–3852. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Corneloup, A.; Guynet, C.; Lavatine, L.; Caumont-Sarcos, A.; Siguier, P.; Marty, B.; Dyda, F.; Chandler, M.; Ton Hoang, B. The IS200/IS605 family and “peel and paste” single-strand transposition mechanism. Microbiol. Spectrum 2015, 3, 3.4.02. [Google Scholar] [CrossRef] [PubMed]
- Lavatine, L.; He, S.; Caumont-Sarcos, A.; Guynet, C.; Marty, B.; Chandler, M.; Ton-Hoang, B. Single strand transposition at the host replication fork. Nucleic Acids Res. 2016, 44, 7866–7883. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Known knowns, known unknowns and unknown unknowns in prokaryotic transposition. Curr. Opin. Microbiol. 2017, 38, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hall, R.M. An analysis of the IS6/IS26 family of insertion sequences: Is it a single family? Microb. Genom. 2019, 5, e000291. [Google Scholar] [CrossRef] [PubMed]
- Varani, A.; He, S.; Siguier, P.; Ross, K.; Chandler, M. The IS6 family, a clinically important group of insertion sequences including IS26. Mob. DNA 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choi, N.; Bae, M.K.; Choo, K.; Lee, S.-J. Transposition of insertion sequences was triggered by oxidative stress in radiation-resistant bacterium Deinococcus geothermalis. Microorganisms 2019, 7, 446. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choo, K.; Lee, S.-J. Active transposition of insertion sequences by oxidative stress in Deinococcus geothermalis. Front. Microbiol. 2020, 11, 558747. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Lee, C.; Shin, E.; Lee, S.-J. Influence of redox imbalances on the transposition of insertion sequences in Deinococcus geothermalis. Antioxidants 2021, 10, 1623. [Google Scholar] [CrossRef]
- Pasternak, C.; Dulermo, R.; Ton-Hoang, B.; Debuchy, R.; Siguier, P.; Coste, G.; Chandler, M.; Sommer, S. ISDra2 transpositionin Deinococcus radiodurans is downregulated by TnpB. Mol. Microbiol. 2013, 88, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Quentin, Y.; Siguier, P.; Chandler, M.; Fichant, G. Single-strand DNA processing: Phylogenomics and sequence diversity of a superfamily of potential prokaryotic HuH endonuclease. BMC Genom. 2018, 19, 475. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.T.; Dahlberg, A.E. Transposition of an insertion sequence, ISTth7, in the genome of the extreme thermophile Thermus thermophilus HB8. FEMS Lett. 2008, 289, 187–192. [Google Scholar] [CrossRef]
- Mennecier, S.; Servant, P.; Coste, G.; Bailone, A.; Sommer, S. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 2006, 59, 317–325. [Google Scholar] [CrossRef]
- Lee, C.; Bae, M.K.; Lee, S.-J. An antioxidant defense system in radiation-resistant bacterium Deinococcus geothermalis against oxidative stress. In Antioxidants: Benefits, Sources, Mechanisms of Action; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Chandler, M.; Fayet, O.; Rousseau, P.; Ton-Hoang, B.; Duval-Valentin, G. Copy-out-paste-in transposition of IS911: A major transposition pathway. Miocrobiol. Spectrum 2015, 3, 3.4.01. [Google Scholar]
- Haren, L.; Betermier, M.; Polard, P.; Chandler, M. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol. Microbiol. 1997, 25, 531–540. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Genka, H.; Komatsu, H.; Nagata, Y.; Tsuda, M. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC17616. Appl. Environ. Microbiol. 2005, 71, 1822–1828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Christie-Oleza, J.A.; Lanfranconi, M.P.; Nogales, B.; Lalucat, J.; Bosch, R. Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J. Bacteriol. 2009, 191, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Baldwin, A.; Lindenburg, L.; Joshi, L.T.; Marchbank, A.; Vosahlikova, S.; Dowson, C.G.; Mahenthiralingam, E. Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J. Clin. Microbiol. 2010, 48, 34–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suzuki, H.; Taketani, T.; Tanabiki, M.; Ohara, M.; Kobayashi, J.; Ohshiro, T. Frequent transposition of multiple insertion sequences in Geobacillus kaustophilus HTA426. Front. Microbiol. 2021, 12, 650461. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Sekine, Y.; Chibazakura, T.; Yoshikawa, H. Development of an intermolecular transposition assay system in Bacillus subtilis 168 using IS4Bsu1 from Bacillus subtilis (natto). Microbiology 2007, 153, 2553–2559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Filée, J.; Siguier, P.; Chandler, M. Insertion sequence diversity in archaea. Micribiol. Mol. Biol. Rev. 2007, 71, 121–157. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, F.; Blaseio, U. Transposition burst of the ISH27 insertion element family in Halobacterium halobium. Nucleic Acids Res. 1990, 18, 6921–6925. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic, Y.; Lopez, P.; Philippe, H.; Forterre, P. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 2002, 30, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Fukiya, S.; Kobayashi, R.; Abe, A.; Hirayama, Y.; Kano, Y.; Yokota, A. Isolation and transposition properties of ISBlo11, an active insertion sequence belonging to the IS3 family, from Bifidobacterium longum 105-A. FEMS Lett. 2015, 362, fnv032. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Nakakawaji, S.; Nakajima, S.; Fukiya, S.; Abe, A.; Saburi, W.; Mori, H.; Yokota, A. A transposon mutagenesis system for Bifidobacterium longum subsp. longum based on an IS3 family insertion sequence, ISBlo11. Appl. Environ. Microbiol. 2018, 84, e00824-18. [Google Scholar]
- Nagy, Z.; Chandler, M. Regulation of transposition in bacteria. Res. Microbiol. 2004, 155, 387–398. [Google Scholar] [CrossRef]
- Swingle, B.; O’Carroll, M.; Haniford, D.; Derbyshire, K.M. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 2004, 52, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- Shiga, Y.; Sekina, Y.; Kano, Y.; Ohtsubo, E. Involvment of H-NS in transpositional recombination mediated by IS1. J. Bacteriol. 2001, 183, 2476–2484. [Google Scholar] [CrossRef]
- Humayun, M.Z.; Zhang, Z.; Butcher, A.M.; Moshayedi, A.; Saier, M.H., Jr. Hopping into a host seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress. PLoS ONE 2017, 12, e0180156. [Google Scholar] [CrossRef]
- Rouquette, C.; Serre, M.-C.; Lane, D. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 2004, 186, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; Mahillon, J. Insertion sequences revisited. In Mobile DNA; ASM Press: Washington, DC, USA, 2002; Volume II, pp. 305–366. [Google Scholar]
- Coros, A.M.; Twiss, E.; Tavakoli, N.P.; Derbyshire, K.M. Genetic Evidence that GTP Is Required for Transposition of IS903 and Tn552 in Escherichia coli. J. Bacteriol. 2005, 187, 4598–4606. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Twiss, E.; Coros, A.M.; Tavakoli, N.P.; Derbyshire, K.M. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 2005, 57, 1593–1607. [Google Scholar] [CrossRef]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Sobrero, P.; Valverde, C. The bacterial protein Hfq: Much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012, 38, 276–299. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Trussler, R.S.; Haniford, D.B. Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 2015, 96, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Zerbib, D.; Polard, P.; Escoubas, J.M.; Galas, D.; Chandler, M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol. Microbiol. 1990, 4, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.T.; Hwang, J.H.; Lee, L.C.; Lee, C.H.; Li, P.L.; Hsieh, Y.C. Functional analysis of the 14 kDa protein of insertion sequence 2. J. Mol. Biol. 1994, 236, 503–513. [Google Scholar] [CrossRef]
- Kleckner, N.; Chalmers, R.M.; Kwon, D.; Sakai, J.; Bolland, S. Tn10 and IS10 transposition and chromosome rearrangements: Mechanism and regulation in vivo and in vitro. In Transposable Elements; Springer: Berlin/Heidelberg, Germany, 1996; pp. 49–82. [Google Scholar]
- Escoubas, J.M.; Prere, M.F.; Fayet, O.; Salvignol, I.; Galas, D.; Zerbib, D.; Chandler, M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991, 10, 705–712. [Google Scholar] [CrossRef]
- Beuzon, C.R.; Marques, S.; Casadesus, J. Repression of IS200 transposase synthesis by RNA secondary structures. Nucleic Acids Res. 1999, 27, 3690–3695. [Google Scholar] [CrossRef][Green Version]
- Roberts, D.; Hoopes, B.C.; McClure, W.R.; Kleckner, N. IS10 transposition is regulated by DNA adenine methylation. Cell 1990, 43, 117–130. [Google Scholar] [CrossRef]
- Derbyshire, K.M.; Kramer, M.; Grindley, N.D. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc. Natl. Acad. Sci. USA 1990, 87, 4048–4052. [Google Scholar] [CrossRef]
- Olasz, F.; Kiss, J.; Konig, P.; Buzas, Z.; Stalder, R.; Arber, W. Target specificity of insertion element IS30. Mol. Microbiol. 1998, 28, 691–704. [Google Scholar] [CrossRef]
- Kiss, J.; Nagy, Z.; Toth, G.; Kiss, G.B.; Jakab, J.; Chandler, M.; Olasz, F. Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol. Microbiol. 2007, 63, 1731–1747. [Google Scholar] [CrossRef]
- Kharat, A.; Coursange, E.; Noirclerc-Savoye, M.; Lacoste, J.; Blot, M. IS1 transposition is enhanced by translation errors and by bacterial growth at extreme glucose levels. Acta Biochim. Pol. 2006, 53, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Reif, H.J.; Saedler, H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol. Gen. Genet. 1975, 137, 17–28. [Google Scholar] [CrossRef]
- Pasternak, C.; Ton-Hoang, B.; Coste, G.; Bailone, A.; Chandler, M.; Sommer, S. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 2010, 6, e1000799. [Google Scholar] [CrossRef] [PubMed]
- Narumi, I.; Cherdchu, K.; Kitayama, S.; Watanabe, H. The Deinococcus radiodurans uvrA gene: Identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 1997, 198, 115–126. [Google Scholar] [CrossRef]
- Demirci, H.; Murphy, I.V.F.; Murphy, E.; Gregory, S.T.; Dahlberg, A.E.; Jogl, G. A structural basis for Sm-induced misreading of the genetic code. Nat. Commun. 2013, 4, 1355. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.F.; Hamburg, D.-M.; Gregory, S.T.; Limbach, P.A.; Dahlberg, A.E. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. J. Bacteriol. 2006, 188, 2020–2023. [Google Scholar] [CrossRef]
- Okamoto, S.; Tamaru, A.; Nakajima, C.; Nishimura, K.; Tanaka, Y.; Tokuyama, S.; Suzuki, Y.; Ochi, K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007, 63, 1096–1106. [Google Scholar] [CrossRef]
- Paine, T.F.; Finland, M. Streptomycin-sensitive, -dependent, and -resistant bacteria. Science 1948, 107, 143–144. [Google Scholar] [CrossRef]
- Gregory, S.T.; Cate, J.H.; Dahlberg, A.E. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 2001, 309, 333–338. [Google Scholar] [CrossRef]
- Mijnendonckx, K.; Provoost, A.; Monsieurs, P.; Leys, N.; Mergeay, M.; Mahillon, J.; Van Houdt, R. Insertion sequence elements in Cupriavidus metallidurans CH34: Distribution and role in adaptation. Plasmid 2011, 65, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Monsieurs, P.; Mergeay, M.; Leys, N.; Aertsen, A.; Van Houdt, R. Zinc-induced transposition of insertion sequence elements contributes to increased adaptability of Cupriavidus metallidurans. Front. Microbiol. 2016, 7, 359. [Google Scholar] [CrossRef]
- Eugénie, N.; Zivanovic, Y.; Lelandais, G.; Coste, G.; Bouthier de la Tour, C.; Bentchikou, E.; Servant, P.; Confalonieri, F. Characterization of the radiation desiccation response regulon of the radioresistant bacterium Deinococus radiodurans by integrative genomic analyses. Cells 2021, 10, 2536. [Google Scholar] [CrossRef] [PubMed]
- Ludanyi, M.; Blanchard, L.; Dulermo, R.; Brandelet, G.; Bellanger, L.; Pignol, D.; Lemaire, D.; de Groot, A. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol. Microbiol. 2014, 94, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, L.; Guerin, P.; Roche, D.; Cruveiller, S.; Pignol, D.; Vallenet, D.; Armengaud, J.; de Groot, A. Conservation and diversity of the IrrE/DdrO-controlled radiation response in radiation-resistant Deinococcus bacteria. MicrobiologyOpen 2017, 6, e477. [Google Scholar] [CrossRef] [PubMed]
- Magerand, R.; Rey, P.; Blanchard, L.; de Groot, A. Redox signaling through zinc activates the radiation response in Deinococcus bacteria. Sci. Rep. 2021, 11, 4528. [Google Scholar] [CrossRef] [PubMed]
- Antelmann, H.; Helmann, J.D. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011, 14, 1049–1063. [Google Scholar] [CrossRef]
- Hillion, M.; Antelmann, H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015, 396, 415–444. [Google Scholar] [CrossRef]
- Sevilla, E.; Bes, M.T.; González, A.; Peleato, M.L.; Fillat, M.F. Redox-based transcriptional regulation in prokaryotes: Revisiting model mechanisms. Antioxid. Redox Signal. 2019, 30, 1651–1696. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jung, J.-H.; Blanchard, L.; de Groot, A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, Z.; Chen, X.; Zhang, W.; Lin, M.; Chen, M. Comparative proteomics analysis reveals new features of the oxidative stress response in the polyextremophilic bacterium Deinococcus radiodurans. Microorganisms 2020, 8, 451. [Google Scholar] [CrossRef]
- Orsi, R.H.; Bowen, B.M.; Wiedmann, M. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genom. 2010, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, K.A.; Berg, H.C. Mutations that stimulate flhDC expression in Escherichia coli K-12. J. Bacteriol. 2015, 197, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kukita, C.; Humayun, M.Z.; Saier, M.H., Jr. Environmental-directed activation of the Escherichia coli flhDC operon by transposons. Microbiology 2017, 163, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yen, M.R.; Saier, M.H., Jr. Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J. Bacteriol. 2010, 192, 2013–2019. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).