Aryl Hydrocarbon Receptor-Dependent and -Independent Pathways Mediate Curcumin Anti-Aging Effects
Abstract
1. Introduction
2. Materials and Methods
2.1. C. elegans
2.1.1. C. elegans Strains and Cultivation
2.1.2. Gene Silencing by RNA-Mediated Interference (RNAi)
2.1.3. E. coli Strains and Growth
2.1.4. Lifespan
2.1.5. Movement/Health Span
2.1.6. Curcumin Treatment of C. elegans
2.1.7. Quantification of PolyQ Aggregates
2.1.8. Quantification of α-Synuclein Aggregates
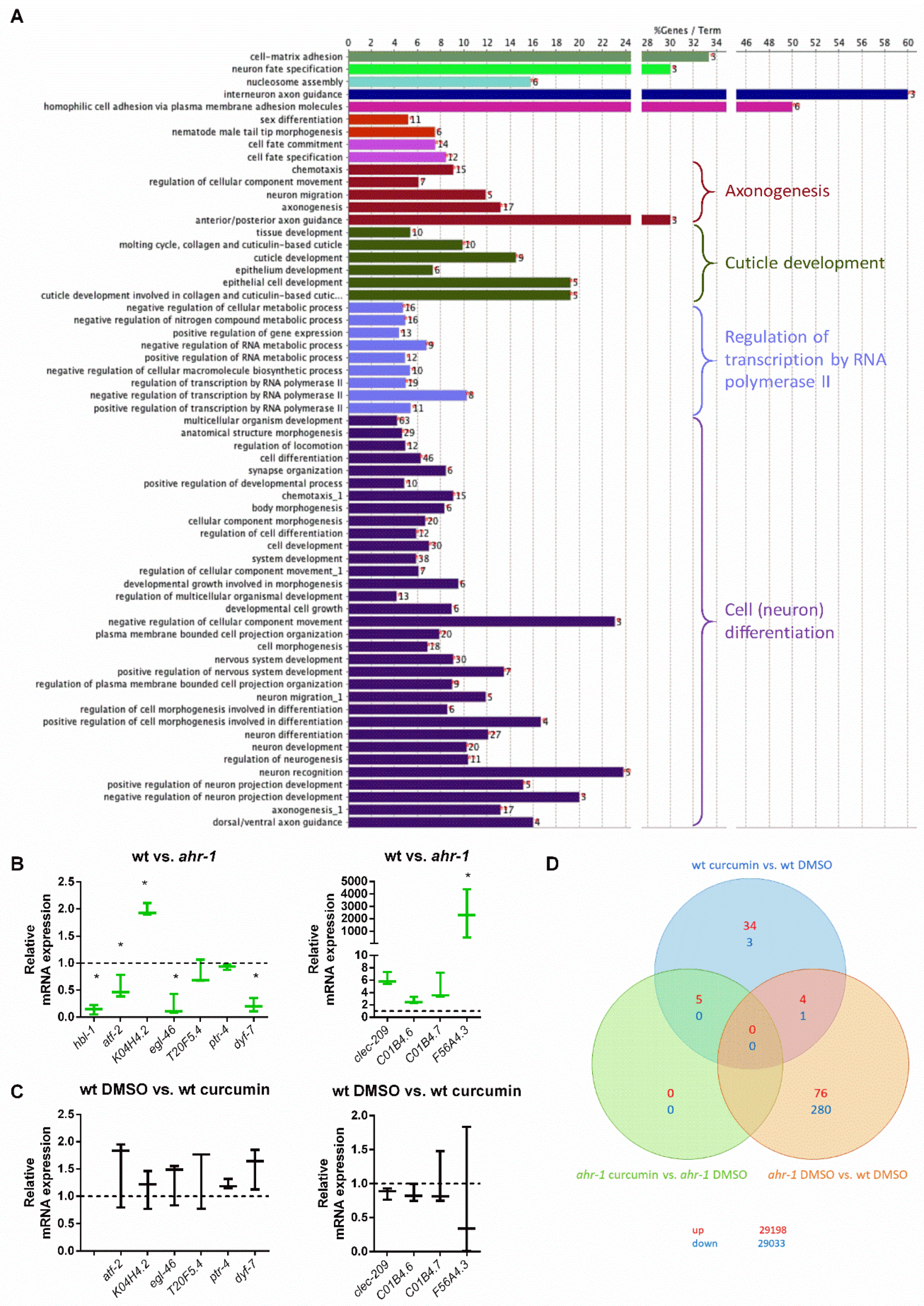
2.1.9. Microarray and GO Term Analysis
2.1.10. ROS Quantification
2.1.11. Tetramethylrhodamine Ethyl Ester (TMRE) Assay
2.1.12. Pharyngeal Pumping Rate and Motility Assay
2.1.13. Acute Juglone Sensitivity Assay
2.1.14. Quantification of the gst-4::GFP Intensity
2.1.15. Semi-Quantitative Real-Time PCR (qPCR) in C. elegans
2.2. Mammalian Cells
2.2.1. Cultivation of Cos7 Cells
2.2.2. Transfection Plasmids
2.2.3. Transient Transfection of Cos7 Cells
2.2.4. Treatment of Cos7 Cells
2.2.5. Luciferase Assay (AhR Activity)
2.2.6. Cultivation of Primary Human EC
2.2.7. Transient Transfection of EC
2.2.8. Scratch Wound Assay of EC
2.2.9. Immunostaining of EC
2.2.10. qPCR in Cells
2.3. Mice
2.3.1. Mouse Lines and Breeding
2.3.2. qPCR in Mice
2.4. In Silico Analyses
Homology Modeling of the CeAhR LBD
2.5. Statistical Analysis
3. Results
3.1. Curcumin Promotes Health Span in an AhR-Dependent and -Independent Manner
3.2. ugt-45 Mediates the Anti-Aging Effects of Curcumin and ahr-1 Depletion
3.3. AHR-1 and Curcumin Independently Protect against Oxidative Stress
3.4. Nrf2/SKN-1 Mediates the AhR-Independent Effects of Curcumin
3.5. Curcumin and Pro-Oxidants Display Opposite Effects on AHR-1 Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poland, A.; Glover, E.; Kende, A.S. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J. Biol. Chem. 1976, 251, 4936–4946. [Google Scholar] [CrossRef]
- Abel, J.; Haarmann-Stemmann, T. An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol. Chem. 2010, 391, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Vazquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, J.; Takeuchi, M.; Usui, Y.; Hattori, T.; Okunuki, Y.; Yamakawa, N.; Kezuka, T.; Kuroda, M.; Goto, H. Suppression of experimental autoimmune uveoretinitis by inducing differentiation of regulatory T cells via activation of aryl hydrocarbon receptor. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Vondracek, J.; Umannova, L.; Machala, M. Interactions of the aryl hydrocarbon receptor with inflammatory mediators: Beyond CYP1A regulation. Curr. Drug Metab. 2011, 12, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Hanieh, H. Toward understanding the role of aryl hydrocarbon receptor in the immune system: Current progress and future trends. BioMed Res. Int. 2014, 2014, 520763. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Nakajima, M.; Fujiki, Y.; Katoh, M.; Gonzalez, F.J.; Yokoi, T. Regulation of insulin-like growth factor binding protein-1 and lipoprotein lipase by the aryl hydrocarbon receptor. J. Toxicol. Sci. 2008, 33, 405–413. [Google Scholar] [CrossRef][Green Version]
- Diani-Moore, S.; Ram, P.; Li, X.; Mondal, P.; Youn, D.Y.; Sauve, A.A.; Rifkind, A.B. Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J. Biol. Chem. 2010, 285, 38801–38810. [Google Scholar] [CrossRef]
- Yi, T.; Wang, J.; Zhu, K.; Tang, Y.; Huang, S.; Shui, X.; Ding, Y.; Chen, C.; Lei, W. Aryl Hydrocarbon Receptor: A New Player of Pathogenesis and Therapy in Cardiovascular Diseases. BioMed. Res. Int. 2018, 2018, 6058784. [Google Scholar] [CrossRef]
- Schmidt, J.V.; Su, G.H.; Reddy, J.K.; Simon, M.C.; Bradfield, C.A. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA 1996, 93, 6731–6736. [Google Scholar] [CrossRef]
- Fernandez-Salguero, P.; Pineau, T.; Hilbert, D.M.; McPhail, T.; Lee, S.S.; Kimura, S.; Nebert, D.W.; Rudikoff, S.; Ward, J.M.; Gonzalez, F.J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 1995, 268, 722–726. [Google Scholar] [CrossRef]
- Fernandez-Salguero, P.M.; Ward, J.M.; Sundberg, J.P.; Gonzalez, F.J. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet. Pathol. 1997, 34, 605–614. [Google Scholar] [CrossRef]
- Brinkmann, V.; Ale-Agha, N.; Haendeler, J.; Ventura, N. The Aryl Hydrocarbon Receptor (AhR) in the Aging Process: Another Puzzling Role for This Highly Conserved Transcription Factor. Front. Physiol. 2020, 10, 1561. [Google Scholar] [CrossRef]
- Eckers, A.; Jakob, S.; Heiss, C.; Haarmann-Stemmann, T.; Goy, C.; Brinkmann, V.; Cortese-Krott, M.M.; Sansone, R.; Esser, C.; Ale-Agha, N.; et al. The aryl hydrocarbon receptor promotes aging phenotypes across species. Sci. Rep. 2016, 6, 19618. [Google Scholar] [CrossRef]
- Williams, E.G.; Mouchiroud, L.; Frochaux, M.; Pandey, A.; Andreux, P.A.; Deplancke, B.; Auwerx, J. An evolutionarily conserved role for the aryl hydrocarbon receptor in the regulation of movement. PLoS Genet. 2014, 10, e1004673. [Google Scholar] [CrossRef]
- Huang, S.; Shui, X.; He, Y.; Xue, Y.; Li, J.; Li, G.; Lei, W.; Chen, C. AhR expression and polymorphisms are associated with risk of coronary arterial disease in Chinese population. Sci. Rep. 2015, 5, 8022. [Google Scholar] [CrossRef]
- Sakakibara, H.; Nakagawa, S.; Wakameda, H.; Nakagiri, Y.; Kamata, K.; Das, S.K.; Tsuji, T.; Kanazawa, K. Effects of Japanese kelp (kombu) on life span of benzo[a]pyrene-fed mice. J. Nutr. Sci. Vitaminol. 2005, 51, 369–373. [Google Scholar] [CrossRef]
- Okey, A.B.; Dube, A.W.; Vella, L.M. Binding of benzo(a)pyrene and dibenz(a,h)anthracene to the Ah receptor in mouse and rat hepatic cytosols. Cancer Res. 1984, 44, 1426–1432. [Google Scholar]
- Gao, D.; Wu, M.; Wang, C.; Wang, Y.; Zuo, Z. Chronic exposure to low benzo[a]pyrene level causes neurodegenerative disease-like syndromes in zebrafish (Danio rerio). Aquat. Toxicol. 2015, 167, 200–208. [Google Scholar] [CrossRef]
- Denison, M.S.; Pandini, A.; Nagy, S.R.; Baldwin, E.P.; Bonati, L. Ligand binding and activation of the Ah receptor. Chem.-Biol. Interact. 2002, 141, 3–24. [Google Scholar] [CrossRef]
- Ashida, H.; Fukuda, I.; Yamashita, T.; Kanazawa, K. Flavones and flavonols at dietary levels inhibit a transformation of aryl hydrocarbon receptor induced by dioxin. FEBS Lett. 2000, 476, 213–217. [Google Scholar] [CrossRef]
- Zhang, S.; Qin, C.; Safe, S.H. Flavonoids as aryl hydrocarbon receptor agonists/antagonists: Effects of structure and cell context. Environ. Health Perspect 2003, 111, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K. Dioxin: A review of its environmental effects and its aryl hydrocarbon receptor biology. J. Comp. Physiol. B 2005, 175, 221–230. [Google Scholar] [CrossRef]
- Marinkovic, N.; Pasalic, D.; Ferencak, G.; Grskovic, B.; Stavljenic Rukavina, A. Dioxins and human toxicity. Arh. Hig. Rada. Toksikol. 2010, 61, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Schiavi, A.; Shaik, A.; Puchta, D.R.; Ventura, N. Dietary and environmental factors have opposite AhR-dependent effects on C. elegans healthspan. Aging 2020, 13, 104–133. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Harikumar, K.B. Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int. J. Biochem. Cell Biol. 2009, 41, 40–59. [Google Scholar] [CrossRef]
- Alavez, S.; Vantipalli, M.C.; Zucker, D.J.; Klang, I.M.; Lithgow, G.J. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature 2011, 472, 226–229. [Google Scholar] [CrossRef]
- Liu, Y.; Samuel, B.S.; Breen, P.C.; Ruvkun, G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 2014, 508, 406–410. [Google Scholar] [CrossRef]
- Caesar, I.; Jonson, M.; Nilsson, K.P.; Thor, S.; Hammarstrom, P. Curcumin promotes A-beta fibrillation and reduces neurotoxicity in transgenic Drosophila. PLoS ONE 2012, 7, e31424. [Google Scholar] [CrossRef]
- Lim, G.P.; Chu, T.; Yang, F.; Beech, W.; Frautschy, S.A.; Cole, G.M. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J. Neurosci. 2001, 21, 8370–8377. [Google Scholar] [CrossRef]
- Fleenor, B.S.; Sindler, A.L.; Marvi, N.K.; Howell, K.L.; Zigler, M.L.; Yoshizawa, M.; Seals, D.R. Curcumin ameliorates arterial dysfunction and oxidative stress with aging. Exp. Gerontol. 2013, 48, 269–276. [Google Scholar] [CrossRef]
- Oliver, J.M.; Stoner, L.; Rowlands, D.S.; Caldwell, A.R.; Sanders, E.; Kreutzer, A.; Mitchell, J.B.; Purpura, M.; Jager, R. Novel Form of Curcumin Improves Endothelial Function in Young, Healthy Individuals: A Double-Blind Placebo Controlled Study. J. Nutr. Metab. 2016, 2016, 1089653. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Li, D.; Yu, W.; Zhang, Q.; Hou, X.; He, Y.; Kou, X. Mechanisms and therapeutic prospects of polyphenols as modulators of the aryl hydrocarbon receptor. Food Funct. 2017, 8, 1414–1437. [Google Scholar] [CrossRef] [PubMed]
- Jeuken, A.; Keser, B.J.; Khan, E.; Brouwer, A.; Koeman, J.; Denison, M.S. Activation of the Ah receptor by extracts of dietary herbal supplements, vegetables, and fruits. J. Agric. Food Chem. 2003, 51, 5478–5487. [Google Scholar] [CrossRef] [PubMed]
- Powell-Coffman, J.A.; Bradfield, C.A.; Wood, W.B. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc. Natl. Acad. Sci. USA 1998, 95, 2844–2849. [Google Scholar] [CrossRef]
- Bell, D.R.; Poland, A. Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein. The role of hsp90. J. Biol. Chem. 2000, 275, 36407–36414. [Google Scholar] [CrossRef]
- Huang, X.; Powell-Coffman, J.A.; Jin, Y. The AHR-1 aryl hydrocarbon receptor and its co-factor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development 2004, 131, 819–828. [Google Scholar] [CrossRef]
- Qin, H.; Powell-Coffman, J.A. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev. Biol. 2004, 270, 64–75. [Google Scholar] [CrossRef]
- Butler, R.A.; Kelley, M.L.; Powell, W.H.; Hahn, M.E.; Van Beneden, R.J. An aryl hydrocarbon receptor (AHR) homologue from the soft-shell clam, Mya arenaria: Evidence that invertebrate AHR homologues lack 2,3,7,8-tetrachlorodibenzo-p-dioxin and beta-naphthoflavone binding. Gene 2001, 278, 223–234. [Google Scholar] [CrossRef]
- Qin, H.; Zhai, Z.; Powell-Coffman, J.A. The Caenorhabditis elegans AHR-1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev. Biol. 2006, 298, 606–615. [Google Scholar] [CrossRef]
- Smith, C.J.; O’Brien, T.; Chatzigeorgiou, M.; Spencer, W.C.; Feingold-Link, E.; Husson, S.J.; Hori, S.; Mitani, S.; Gottschalk, A.; Schafer, W.R.; et al. Sensory Neuron Fates Are Distinguished by a Transcriptional Switch that Regulates Dendrite Branch Stabilization. Neuron 2013, 79, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Shima, Y.; Owaki, A.; Mimura, J.; Oshima, M.; Fujii-Kuriyama, Y.; Morohashi, K.I. Disruption of aryl hydrocarbon receptor (AhR) induces regression of the seminal vesicle in aged male mice. Sex. Dev. Genet. Mol. Biol. Evol. Endocrinol. Embryol. Pathol. Sex Determ. Differ. 2008, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Aarnio, V.; Storvik, M.; Lehtonen, M.; Asikainen, S.; Reisner, K.; Callaway, J.; Rudgalvyte, M.; Lakso, M.; Wong, G. Fatty acid composition and gene expression profiles are altered in aryl hydrocarbon receptor-1 mutant Caenorhabditis elegans. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010, 151, 318–324. [Google Scholar] [CrossRef]
- Hahn, M.E.; Karchner, S.I.; Shapiro, M.A.; Perera, S.A. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc. Natl. Acad. Sci. USA 1997, 94, 13743–13748. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E. Aryl hydrocarbon receptors: Diversity and evolution. Chem.-Biol. Interact. 2002, 141, 131–160. [Google Scholar] [CrossRef]
- Fuse, Y.; Kobayashi, M. Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time. Molecules 2017, 22, 436. [Google Scholar] [CrossRef]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef]
- Morley, J.F.; Brignull, H.R.; Weyers, J.J.; Morimoto, R.I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 10417–10422. [Google Scholar] [CrossRef]
- van Ham, T.J.; Thijssen, K.L.; Breitling, R.; Hofstra, R.M.; Plasterk, R.H.; Nollen, E.A. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008, 4, e1000027. [Google Scholar] [CrossRef]
- Timmons, L.; Fire, A. Specific interference by ingested dsRNA. Nature 1998, 395, 854. [Google Scholar] [CrossRef]
- Kamath, R.S.; Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 2003, 30, 313–321. [Google Scholar] [CrossRef]
- Yang, J.S.; Nam, H.J.; Seo, M.; Han, S.K.; Choi, Y.; Nam, H.G.; Lee, S.J.; Kim, S. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 2011, 6, e23525. [Google Scholar] [CrossRef] [PubMed]
- Han, S.K.; Lee, D.; Lee, H.; Kim, D.; Son, H.G.; Yang, J.S.; Lee, S.V.; Kim, S. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 2016, 7, 56147–56152. [Google Scholar] [CrossRef] [PubMed]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Sommer, C.; Strähle, C.; Köthe, U.; Hamprecht, F.A. lastik: Interactive Learning and Segmentation Toolkit. In Proceedings of the Eighth IEEE International Symposium on Biomedical Imaging (ISBI), Chicago, IL, USA, 30 March–2 April 2011. [Google Scholar]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Kauffmann, A.; Gentleman, R.; Huber, W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Shaham, S. WormBook: Methods in Cell Biology; WormBook: Online, USA, 2006. [Google Scholar]
- Morel, Y.; Barouki, R. Down-regulation of cytochrome P450 1A1 gene promoter by oxidative stress. Critical contribution of nuclear factor 1. J. Biol. Chem. 1998, 273, 26969–26976. [Google Scholar] [CrossRef] [PubMed]
- Haendeler, J.; Hoffmann, J.; Tischler, V.; Berk, B.C.; Zeiher, A.M.; Dimmeler, S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat. Cell Biol. 2002, 4, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Ale-Agha, N.; Goy, C.; Jakobs, P.; Spyridopoulos, I.; Gonnissen, S.; Dyballa-Rukes, N.; Aufenvenne, K.; von Ameln, F.; Zurek, M.; Spannbrucker, T.; et al. CDKN1B/p27 is localized in mitochondria and improves respiration-dependent processes in the cardiovascular system-New mode of action for caffeine. PLoS Biol. 2018, 16, e2004408. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fiser, A.; Do, R.K.; Sali, A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef]
- Marti-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sanchez, R.; Melo, F.; Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.; MacArthur, M.; Moss, D.; Thornton, J. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Andersen, C.A.; Palmer, A.G.; Brunak, S.; Rost, B. Continuum secondary structure captures protein flexibility. Structure 2002, 10, 175–184. [Google Scholar] [CrossRef]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.; Liang, J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. [Google Scholar] [CrossRef]
- Schrödinger, L. The PyMOL Molecular Graphics System; Version 1.3r1; Schödinger, LLC: New York, NY, USA, 2010. [Google Scholar]
- Liao, V.H.; Yu, C.W.; Chu, Y.J.; Li, W.H.; Hsieh, Y.C.; Wang, T.T. Curcumin-mediated lifespan extension in Caenorhabditis elegans. Mech. Ageing Dev. 2011, 132, 480–487. [Google Scholar] [CrossRef]
- Nishiumi, S.; Yoshida, K.; Ashida, H. Curcumin suppresses the transformation of an aryl hydrocarbon receptor through its phosphorylation. Arch. Biochem. Biophys 2007, 466, 267–273. [Google Scholar] [CrossRef]
- Rinaldi, A.L.; Morse, M.A.; Fields, H.W.; Rothas, D.A.; Pei, P.; Rodrigo, K.A.; Renner, R.J.; Mallery, S.R. Curcumin activates the aryl hydrocarbon receptor yet significantly inhibits (-)-benzo(a)pyrene-7R-trans-7,8-dihydrodiol bioactivation in oral squamous cell carcinoma cells and oral mucosa. Cancer Res. 2002, 62, 5451–5456. [Google Scholar]
- Choi, H.; Chun, Y.S.; Shin, Y.J.; Ye, S.K.; Kim, M.S.; Park, J.W. Curcumin attenuates cytochrome P450 induction in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin by ROS-dependently degrading AhR and ARNT. Cancer Sci. 2008, 99, 2518–2524. [Google Scholar] [CrossRef]
- Jones, L.M.; Rayson, S.J.; Flemming, A.J.; Urwin, P.E. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS ONE 2013, 8, e69956. [Google Scholar] [CrossRef]
- Bors, W.; Heller, W.; Michel, C.; Saran, M. Flavonoids as antioxidants: Determination of radical-scavenging efficiencies. Methods Enzymol. 1990, 186, 343–355. [Google Scholar] [CrossRef]
- Sandoval-Acuna, C.; Ferreira, J.; Speisky, H. Polyphenols and mitochondria: An update on their increasingly emerging ROS-scavenging independent actions. Arch. Biochem. Biophys 2014, 559, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, S.; Liu, L.; Jian, Z.; Cui, T.; Yang, Y.; Guo, S.; Yi, X.; Wang, G.; Li, C.; et al. Role of the aryl hydrocarbon receptor signaling pathway in promoting mitochondrial biogenesis against oxidative damage in human melanocytes. J. Dermatol. Sci. 2019, 96, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kubli, S.P.; Bassi, C.; Roux, C.; Wakeham, A.; Gobl, C.; Zhou, W.; Jafari, S.M.; Snow, B.; Jones, L.; Palomero, L.; et al. AhR controls redox homeostasis and shapes the tumor microenvironment in BRCA1-associated breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 3604–3613. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, J.; Gao, H. Benzo(alpha)pyrene induces oxidative stress and inflammation in human vascular endothelial cells through AhR and NF-kappaB pathways. Microvasc Res. 2021, 137, 104179. [Google Scholar] [CrossRef]
- Bazopoulou, D.; Knoefler, D.; Zheng, Y.; Ulrich, K.; Oleson, B.J.; Xie, L.; Kim, M.; Kaufmann, A.; Lee, Y.T.; Dou, Y.; et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 2019, 576, 301–305. [Google Scholar] [CrossRef]
- Lemire, B.D.; Behrendt, M.; DeCorby, A.; Gaskova, D.C. elegans longevity pathways converge to decrease mitochondrial membrane potential. Mech. Ageing Dev. 2009, 130, 461–465. [Google Scholar] [CrossRef]
- Lahteenvuo, J.; Rosenzweig, A. Effects of aging on angiogenesis. Circ. Res. 2012, 110, 1252–1264. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Ahmadi, Z.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Curcumin Activates the Nrf2 Pathway and Induces Cellular Protection Against Oxidative Injury. Curr. Mol. Med. 2020, 20, 116–133. [Google Scholar] [CrossRef]
- Chiu, H.F.; Venkatakrishnan, K.; Wang, C.K. The role of nutraceuticals as a complementary therapy against various neurodegenerative diseases: A mini-review. J. Tradit. Complement. Med. 2020, 10, 434–439. [Google Scholar] [CrossRef]
- Li, W.; Sun, K.; Hu, F.; Chen, L.; Zhang, X.; Wang, F.; Yan, B. Protective effects of natural compounds against oxidative stress in ischemic diseases and cancers via activating the Nrf2 signaling pathway: A mini review. J. Biochem. Mol. Toxicol. 2021, 35, e22658. [Google Scholar] [CrossRef]
- Detienne, G.; Van de Walle, P.; De Haes, W.; Schoofs, L.; Temmerman, L. SKN-1-independent transcriptional activation of glutathione S-transferase 4 (GST-4) by EGF signaling. Worm 2016, 5, e1230585. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abnet, C.C.; Tanguay, R.L.; Heideman, W.; Peterson, R.E. Transactivation activity of human, zebrafish, and rainbow trout aryl hydrocarbon receptors expressed in COS-7 cells: Greater insight into species differences in toxic potency of polychlorinated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners. Toxicol. Appl. Pharmacol. 1999, 159, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Ema, M.; Ohe, N.; Suzuki, M.; Mimura, J.; Sogawa, K.; Ikawa, S.; Fujii-Kuriyama, Y. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J. Biol. Chem. 1994, 269, 27337–27343. [Google Scholar] [CrossRef]
- Maglioni, S.; Arsalan, N.; Hamacher, A.; Afshar, S.; Schiavi, A.; Beller, M.; Ventura, N. High-Content C. elegans Screen Identifies Natural Compounds Impacting Mitochondria-Lipid Homeostasis and Promoting Healthspan. Cells 2022, 11, 100. [Google Scholar] [CrossRef]
- Regitz, C.; Fitzenberger, E.; Mahn, F.L.; Dussling, L.M.; Wenzel, U. Resveratrol reduces amyloid-beta (Abeta(1)(-)(4)(2))-induced paralysis through targeting proteostasis in an Alzheimer model of Caenorhabditis elegans. Eur. J. Nutr. 2016, 55, 741–747. [Google Scholar] [CrossRef]
- Wood, J.G.; Rogina, B.; Lavu, S.; Howitz, K.; Helfand, S.L.; Tatar, M.; Sinclair, D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004, 430, 686–689. [Google Scholar] [CrossRef]
- Motto, I.; Bordogna, A.; Soshilov, A.A.; Denison, M.S.; Bonati, L. New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J. Chem. Inf. Model. 2011, 51, 2868–2881. [Google Scholar] [CrossRef]
- Fraccalvieri, D.; Soshilov, A.A.; Karchner, S.I.; Franks, D.G.; Pandini, A.; Bonati, L.; Hahn, M.E.; Denison, M.S. Comparative analysis of homology models of the AH receptor ligand binding domain: Verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochemistry 2013, 52, 714–725. [Google Scholar] [CrossRef]
- Poland, A.; Glover, E. Genetic Expression of Aryl Hydrocarbon Hydroxylase by 2,3,7,8-Tetrachlorodibenzo-p-dioxin: Evidence for a Receptor Mutation in Genetically Non-responsive Mice. Mol. Pharmacol. 1975, 11, 389–398. [Google Scholar]
- Kahn, N.W.; Rea, S.L.; Moyle, S.; Kell, A.; Johnson, T.E. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 2008, 409, 205–213. [Google Scholar] [CrossRef]
- Pecker, M.S.; Im, W.B.; Sonn, J.K.; Lee, C.O. Effect of norepinephrine and cyclic AMP on intracellular sodium ion activity and contractile force in canine cardiac Purkinje fibers. Circ. Res. 1986, 59, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, E.; Schafer, C.; Calles, C.; Bernsmann, T.; Bernshausen, T.; Wurm, M.; Hubenthal, U.; Cline, J.E.; Hajimiragha, H.; Schroeder, P.; et al. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc. Natl. Acad. Sci. USA 2007, 104, 8851–8856. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Borana, M.S.; Chaudhary, A.P. Understanding curcumin-induced modulation of protein aggregation. Int. J. Biol. Macromol. 2017, 100, 89–96. [Google Scholar] [CrossRef]
- Spyridopoulos, I.; Fichtlscherer, S.; Popp, R.; Toennes, S.W.; Fisslthaler, B.; Trepels, T.; Zernecke, A.; Liehn, E.A.; Weber, C.; Zeiher, A.M.; et al. Caffeine enhances endothelial repair by an AMPK-dependent mechanism. Arter. Thromb. Vasc. Biol. 2008, 28, 1967–1974. [Google Scholar] [CrossRef] [PubMed]
- Dostal, V.; Roberts, C.M.; Link, C.D. Genetic mechanisms of coffee extract protection in a Caenorhabditis elegans model of beta-amyloid peptide toxicity. Genetics 2010, 186, 857–866. [Google Scholar] [CrossRef]
- Kohle, C.; Bock, K.W. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem. Pharm. 2007, 73, 1853–1862. [Google Scholar] [CrossRef]
- Ciolino, H.P.; Daschner, P.J.; Wang, T.T.; Yeh, G.C. Effect of curcumin on the aryl hydrocarbon receptor and cytochrome P450 1A1 in MCF-7 human breast carcinoma cells. Biochem. Pharm. 1998, 56, 197–206. [Google Scholar] [CrossRef]
- Mohammadi-Bardbori, A.; Akbarizadeh, A.R.; Delju, F.; Rannug, A. Chromatin remodeling by curcumin alters endogenous aryl hydrocarbon receptor signaling. Chem.-Biol. Interact. 2016, 252, 19–27. [Google Scholar] [CrossRef]
- Gouedard, C.; Barouki, R.; Morel, Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol. Cell. Biol. 2004, 24, 5209–5222. [Google Scholar] [CrossRef]
- Guyot, E.; Chevallier, A.; Barouki, R.; Coumoul, X. The AhR twist: Ligand-dependent AhR signaling and pharmaco-toxicological implications. Drug Discov. Today 2013, 18, 479–486. [Google Scholar] [CrossRef]
- Smirnova, A.; Wincent, E.; Vikstrom Bergander, L.; Alsberg, T.; Bergman, J.; Rannug, A.; Rannug, U. Evidence for New Light-Independent Pathways for Generation of the Endogenous Aryl Hydrocarbon Receptor Agonist FICZ. Chem. Res. Toxicol. 2016, 29, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Heck, D.E.; Vetrano, A.M.; Mariano, T.M.; Laskin, J.D. UVB light stimulates production of reactive oxygen species: Unexpected role for catalase. J. Biol. Chem. 2003, 278, 22432–22436. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Huang, C.; Taki, F.A.; Zhang, Y.; Dobbins, D.L.; Li, L.; Yan, H.; Pan, X. Benzo-alpha-pyrene induced oxidative stress in Caenorhabditis elegans and the potential involvements of microRNA. Chemosphere 2015, 139, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Ristow, M.; Schmeisser, K. Mitohormesis: Promoting Health and Lifespan by Increased Levels of Reactive Oxygen Species (ROS). Dose Response 2014, 12, 288–341. [Google Scholar] [CrossRef] [PubMed]
- Herholz, M.; Cepeda, E.; Baumann, L.; Kukat, A.; Hermeling, J.; Maciej, S.; Szczepanowska, K.; Pavlenko, V.; Frommolt, P.; Trifunovic, A. KLF-1 orchestrates a xenobiotic detoxification program essential for longevity of mitochondrial mutants. Nat. Commun. 2019, 10, 3323. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Ji, F.; Breen, P.; Sewell, A.; Han, M.; Sadreyev, R.; Ruvkun, G. Mitochondrial Dysfunction in C. elegans Activates Mitochondrial Relocalization and Nuclear Hormone Receptor-Dependent Detoxification Genes. Cell Metab. 2019, 29, 1182–1191 e1184. [Google Scholar] [CrossRef]






| Strongest Over-/Under-Expressed Genes by Curcumin in an ahr-1-Dependent Manner | |||||
|---|---|---|---|---|---|
| Gene/Sequence Name | Gene Class a | Molecular Function a | logFC b | adj. p-Value c | Selected Modulators a |
| H43E16.1 | unknown | unknown | 1.53 | 0.021 | bacterial infection, quercetin, rotenone, aging, nuo-6(qm200) |
| numr-1 | Nuclear localized metal responsive | unknown | 1.42 | 0.034 | bacterial infection, quercetin, spg-7 RNAi, isp-1(qm150), nuo-6(qm200), aging |
| mul-1 | Mucin-like | unknown | 1.34 | 0.029 | resveratrol, bacterial infection, spg-7 RNAi, rotenone, paraquat, indole, isp-1(qm150), nuo-6(qm200) |
| oac-14 | O-acyltransferase homolog | transferase activity, transferring acyl groups other than amino-acyl groups | 1.24 | 0.085 | bacterial infection, quercetin, tryptophan, rotenone, paraquat, indole, nuo-6(qm200) |
| F58B4.5 | unknown | unknown | 1.21 | 0.030 | resveratrol, quercetin, spg-7 RNAi, tryptophan, isp-1(qm150), nuo-6(qm200), paraquat indole, |
| comt-4 | Catechol-O-methyl-transferase | O-methyltransferase activity | 1.17 | 0.030 | pathogenic bacteria, ahr-1(ju145), quercetin, rotenone, paraquat, isp-1(qm150), nuo-6(qm200), indole, aging |
| F09C8.1 | Ortholog of human phospholipase B1 | Phospholipase activity; hydrolase activity, acting on ester bonds | 1.08 | 0.030 | ahr-1(ju145), bacterial infection, quercetin, rotenone, paraquat, isp-1(qm150), nuo-6(qm200), aging |
| ugt-48 | UDP-glucuronosyl-transferase | calmodulin binding, glucuronosyltransferase activity, UDP-glycosyltransferase activity, transferase activity, transferring hexosyl and glycosyl groups | 1.07 | 0.034 | bacterial infection, rotenone, aging |
| cyp-13A5 | Cytochrome P450 | monooxygenase activity, metal ion binding, heme binding, oxidoreductase activity | 1.05 | 0.057 | bacterial infection, quercetin, spg-7 RNAi, tryptophan, isp-1(qm150), nuo-6(qm200), indole |
| T19C9.8 | unknown | unknown | 0.96 | 0.084 | bacterial infection, quercetin, spg-7 RNAi, tryptophan, paraquat, isp-1(qm150), nuo-6(qm200) |
| lys-7 | Lysozyme | unknown | −1.54 | 0.084 | bacterial infection, aging, rotenone, isp-1(qm150), nuo-6(qm200) |
| cyp-35A5 | Cytochrome P450 | monooxygenase activity, metal ion binding, heme binding, oxidoreductase activity, steroid hydroxylase activity | −1.08 | 0.087 | bacterial infection, ahr-1(ju145), aging, tryptophan, rotenone, isp-1(qm150), nuo-6(qm200) |
| C14A4.9 | unknown | unknown | −0.55 | 0.079 | bacterial infection, quercetin, rotenone, indole |
| Genes Regulated in the Same Way by Curcumin or ahr-1 Depletion | |||||
| Gene/Sequence Name | Gene Class a | Molecular Function a | LogFC d | LogFC e | |
| slc-17.5 | Solute carrier homolog | transmembrane transporter activity | 0.64 | 0.46 | |
| ugt-9 | UDP-glucuronosyl-transferase | glucuronosyltransferase activity, transferase activity, transferring hexosyl groups | 0.57 | 0.41 | |
| nhr-239 | Nuclear hormone receptor | metal ion binding, zinc ion binding, transcription factor activity, sequence-specific DNA binding | 0.43 | 0.39 | |
| ugt-29 | UDP-glucuronosyl-transferase | glucuronosyltransferase activity, transferase activity, transferring hexosyl and glycosyl groups | 0.36 | 0.34 | |
| C14A4.9 | unknown | unknown | −0.55 | −0.63 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brinkmann, V.; Romeo, M.; Larigot, L.; Hemmers, A.; Tschage, L.; Kleinjohann, J.; Schiavi, A.; Steinwachs, S.; Esser, C.; Menzel, R.; et al. Aryl Hydrocarbon Receptor-Dependent and -Independent Pathways Mediate Curcumin Anti-Aging Effects. Antioxidants 2022, 11, 613. https://doi.org/10.3390/antiox11040613
Brinkmann V, Romeo M, Larigot L, Hemmers A, Tschage L, Kleinjohann J, Schiavi A, Steinwachs S, Esser C, Menzel R, et al. Aryl Hydrocarbon Receptor-Dependent and -Independent Pathways Mediate Curcumin Anti-Aging Effects. Antioxidants. 2022; 11(4):613. https://doi.org/10.3390/antiox11040613
Chicago/Turabian StyleBrinkmann, Vanessa, Margherita Romeo, Lucie Larigot, Anne Hemmers, Lisa Tschage, Jennifer Kleinjohann, Alfonso Schiavi, Swantje Steinwachs, Charlotte Esser, Ralph Menzel, and et al. 2022. "Aryl Hydrocarbon Receptor-Dependent and -Independent Pathways Mediate Curcumin Anti-Aging Effects" Antioxidants 11, no. 4: 613. https://doi.org/10.3390/antiox11040613
APA StyleBrinkmann, V., Romeo, M., Larigot, L., Hemmers, A., Tschage, L., Kleinjohann, J., Schiavi, A., Steinwachs, S., Esser, C., Menzel, R., Giani Tagliabue, S., Bonati, L., Cox, F., Ale-Agha, N., Jakobs, P., Altschmied, J., Haendeler, J., Coumoul, X., & Ventura, N. (2022). Aryl Hydrocarbon Receptor-Dependent and -Independent Pathways Mediate Curcumin Anti-Aging Effects. Antioxidants, 11(4), 613. https://doi.org/10.3390/antiox11040613







_Haendeler.png)


