Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients
Abstract
:1. Introduction
2. Methods
2.1. Subjects
2.2. Fibroblasts Cell Culture
2.3. C9ORF72 and SUMO2/3 Protein Quantification
2.4. Oxidative Stress Assessment
2.5. Mitochondrial Metabolism Assay
2.6. Autophagic Flux Analysis
2.7. Functional Enrichment Analysis
2.8. Accession of Data
2.9. Statistical Analysis
3. Results
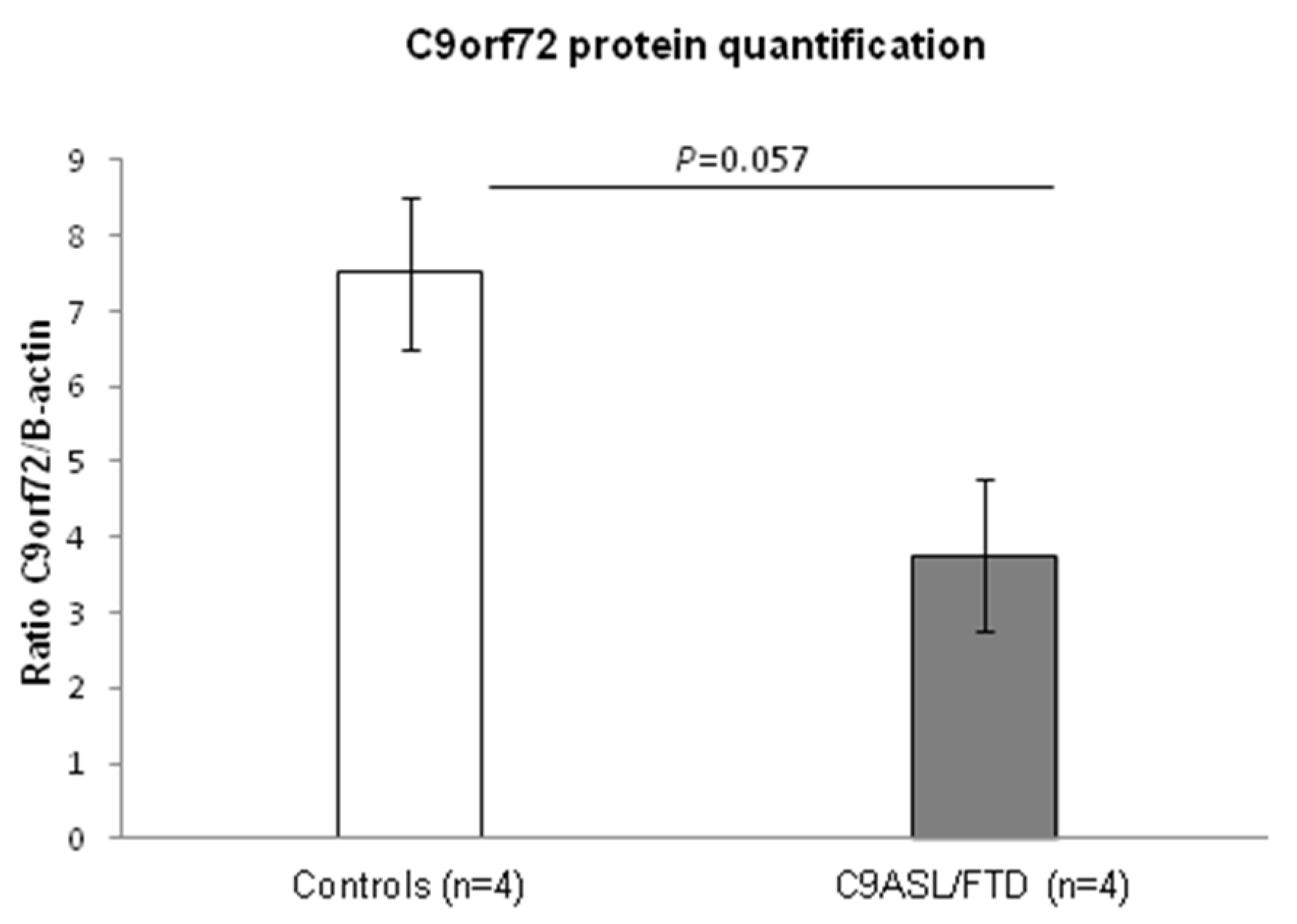
3.1. Fibroblasts from C9ALS/FTD Patients Show Reduced C9orf72 Protein Levels
3.2. Fibroblasts from C9ALS/FTD Patients Evidence Increased Levels of Oxidative Stress
3.3. Fibroblasts from C9ALS/FTD Patients Evidence Mitochondrial Dysfunction and Altered Metabolism
3.4. Fibroblasts from C9ALS/FTD Patients Show Increased SUMO2/3 Protein Quantification
3.5. Autophagy- and SUMOylation-Related Pathways Are Significantly Altered in Fibroblasts from C9ALS/FTD Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Saánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balendra, R.; Isaacs, A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018, 14, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.D.; Pattamatta, A.; Ranum, L.P.W. Repeat-associated non-ATG (RAN) translation. J. Biol. Chem. 2018, 293, 16127–16141. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-B.; Chen, H.-J.; Peres, J.N.; Gomez-Deza, J.; Attig, J.; Štalekar, M.; Troakes, C.; Nishimura, A.L.; Scotter, E.L.; Vance, C.; et al. Hexanucleotide Repeats in ALS/FTD Form Length-Dependent RNA Foci, Sequester RNA Binding Proteins, and Are Neurotoxic. Cell Rep. 2013, 5, 1178–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, K.M.; Linsalata, A.E.; Todd, P.K. RAN translation-What makes it run? Brain Res. 2016, 1647, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Campoy, O.; Ávila-Polo, R.; Grau-Rivera, O.; Antonell, A.; Clarimón, J.; Rojas-Garcia, R.; Charif, S.; Santiago-Valera, V.; Hernández, I.; Aguilar, M.; et al. Systematic Screening of Ubiquitin/p62 Aggregates in Cerebellar Cortex Expands the Neuropathological Phenotype of the C9orf72 Expansion Mutation. J. Neuropathol. Exp. Neurol. 2018, 77, 703–709. [Google Scholar] [CrossRef]
- MacKenzie, I.R.A.; Frick, P.; Neumann, M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol. 2014, 127, 347–357. [Google Scholar] [CrossRef]
- Monaco, A.; Fraldi, A. Protein Aggregation and Dysfunction of Autophagy-Lysosomal Pathway: A Vicious Cycle in Lysosomal Storage Diseases. Front. Mol. Neurosci. 2020, 13, 37. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Princz, A.; Tavernarakis, N. SUMOylation in Neurodegenerative Diseases. Gerontology 2020, 66, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Marmor-Kollet, H.; Siany, A.; Kedersha, N.; Knafo, N.; Rivkin, N.; Danino, Y.M.; Moens, T.G.; Olender, T.; Sheban, D.; Cohen, N.; et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol. Cell 2020, 80, 876–891.e6. [Google Scholar] [CrossRef]
- Ma, L.; Herren, A.W.; Espinal, G.; Randol, J.; McLaughlin, B.; Martinez-Cerdeño, V.; Pessah, I.N.; Hagerman, R.J.; Hagerman, P.J. Composition of the Intranuclear Inclusions of Fragile X-associated Tremor/Ataxia Syndrome. Acta Neuropathol. Commun. 2019, 7, 143. [Google Scholar] [CrossRef] [Green Version]
- Marinello, M.; Werner, A.; Giannone, M.; Tahiri, K.; Alves, S.; Tesson, C.; Dunnen, W.D.; Seeler, J.-S.; Brice, A.; Sittler, A. SUMOylation by SUMO2 is implicated in the degradation of misfolded ataxin-7 via RNF4 in SCA7 models. Dis. Model. Mech. 2019, 12, dmm036145. [Google Scholar] [CrossRef] [Green Version]
- Maraschi, A.; Gumina, V.; Dragotto, J.; Colombrita, C.; Mompeán, M.; Buratti, E.; Silani, V.; Feligioni, M.; Ratti, A. SUMOylation Regulates TDP-43 Splicing Activity and Nucleocytoplasmic Distribution. Mol. Neurobiol. 2021, 58, 5682–5702. [Google Scholar] [CrossRef]
- González-Casacuberta, I.; Juárez-Flores, D.-L.; Ezquerra, M.; Fucho, R.; García, M.C.; Guitart-Mampel, M.; Tobías, E.; García-Ruiz, C.; Fernández-Checa, J.C.; Tolosa, E.; et al. Mitochondrial and autophagic alterations in skin fibroblasts from Parkinson disease patients with Parkin mutations. Aging 2019, 11, 3750–3767. [Google Scholar] [CrossRef]
- Garrabou, G.; Hernàndez, A.-S.; García, M.C.; Morén, C.; Tobías, E.; Córdoba, S.; López, M.; Figueras, F.; Grau, J.M.; Cardellach, F. Molecular basis of reduced birth weight in smoking pregnant women: Mitochondrial dysfunction and apoptosis. Addict. Biol. 2016, 21, 159–170. [Google Scholar] [CrossRef]
- Medja, F.; Allouche, S.; Frachon, P.; Jardel, C.; Malgat, M.; Mousson de Camaret, B.; Slama, A.; Lunardi, J.; Mazat, J.P.; Lombès, A. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion 2009, 9, 331–339. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Cuervo, A.M.; Ravikumar, B.; Sarkar, S.; Korolchuk, V.; Kaushik, S.; Klionsky, D.J. In search of an “autophagomometer”. Autophagy 2009, 5, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Glick, D.; Macleod, K.F. Autophagy: Assays and artifacts. J. Pathol. 2010, 221, 117–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miró, O.; López, S.; De La Concepción, M.R.; Martínez, E.; Pedrol, E.; Garrabou, G.; Giralt, M.; Cardellach, F.; Gatell, J.M.; Vilarroya, F.; et al. Upregulatory Mechanisms Compensate for Mitochondrial DNA Depletion in Asymptomatic Individuals Receiving Stavudine Plus Didanosine. JAIDS J. Acquir. Immune Defic. Syndr. 2004, 37, 1550–1555. [Google Scholar] [CrossRef]
- Montaner, D.; Dopazo, J. Multidimensional Gene Set Analysis of Genomic Data. PLoS ONE 2010, 5, e10348.25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar]
- Gijselinck, I.; Van Langenhove, T.; van der Zee, J.; Sleegers, K.; Philtjens, S.; Kleinberger, G.; Janssens, J.; Bettens, K.; Van Cauwenberghe, C.; Pereson, S.; et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol. 2012, 11, 54–65. [Google Scholar] [CrossRef]
- Popelka, H.; Klionsky, D.J. Post-translationally-modified structures in the autophagy machinery: An integrative perspective. FEBS J. 2015, 282, 3474–3488. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Kang, J.-H.; Lee, S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. [Google Scholar] [CrossRef]
- Lualdi, M.; Shafique, A.; Pedrini, E.; Pieroni, L.; Greco, V.; Castagnola, M.; Cucina, G.; Corrado, L.; Di Pierro, A.; De Marchi, F.; et al. C9ORF72 Repeat Expansion Affects the Proteome of Primary Skin Fibroblasts in ALS. Int. J. Mol. Sci. 2021, 22, 10385. [Google Scholar] [CrossRef]
- Waite, A.J.; Bäumer, D.; East, S.; Neal, J.; Morris, H.; Ansorge, O.; Blake, D.J. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol. Aging 2014, 35, 1779.e5–1779.e13. [Google Scholar] [CrossRef] [Green Version]
- Leskelä, S.; Hoffmann, D.; Rostalski, H.; Huber, N.; Wittrahm, R.; Hartikainen, P.; Korhonen, V.; Leinonen, V.; Hiltunen, M.; Solje, E.; et al. FTLD Patient–Derived Fibroblasts Show Defective Mitochondrial Function and Accumulation of p62. Mol. Neurobiol. 2021, 58, 5438–5458. [Google Scholar] [CrossRef] [PubMed]
- Marrocco, I.; Altieri, F.; Peluso, I. Measurement and Clinical Significance of Biomarkers of Oxidative Stress in Humans. Oxidative Med. Cell. Longev. 2017, 2017, 6501046. [Google Scholar] [CrossRef]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Farg, M.A.; Sundaramoorthy, V.; Sultana, J.M.; Yang, S.; Atkinson, R.; Levina, V.; Halloran, M.A.; Gleeson, P.; Blair, I.; Soo, K.Y.; et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014, 23, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Sellier, C.; Campanari, M.L.; Julie Corbier, C.; Gaucherot, A.; Kolb-Cheynel, I.; Oulad-Abdelghani, M.; Ruffenach, F.; Page, A.; Ciura, S.; Kabashi, E.; et al. Loss of C9 ORF 72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016, 35, 1276–1297. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.P.; Smith, E.F.; Bauer, C.S.; Moller, A.; Hautbergue, G.M.; Ferraiuolo, L.; Myszczynska, M.; Higginbottom, A.; Walsh, M.J.; Whitworth, A.J.; et al. The C9orf72 protein interacts with Rab1a and the ULK 1 complex to regulate initiation of autophagy. EMBO J. 2016, 35, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liang, C.; Swaminathan, K.; Herrlinger, S.; Lai, F.; Shiekhattar, R.; Chen, J.F. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv. 2016, 2, e1601167. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Burberry, A.; Wang, J.-Y.; Sandoe, J.; Ghosh, S.; Udeshi, N.D.; Svinkina, T.; Mordes, D.A.; Mok, J.; Charlton, M.; et al. The C9orf72-interacting protein Smcr8 is a negative regulator of autoimmunity and lysosomal exocytosis. Genes Dev. 2018, 32, 929–943. [Google Scholar] [CrossRef] [Green Version]
- Chitiprolu, M.; Jagow, C.; Tremblay, V.; Bondy-Chorney, E.; Paris, G.; Savard, A.; Palidwor, G.; Barry, F.A.; Zinman, L.; Keith, J.; et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat. Commun. 2018, 9, 2794. [Google Scholar] [CrossRef] [Green Version]
- Aoki, Y.; Manzano, R.; Lee, Y.; Dafinca, R.; Aoki, M.; Douglas, A.G.L.; Varela, M.A.; Sathyaprakash, C.; Scaber, J.; Barbagallo, P.; et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain 2017, 140, 887–897. [Google Scholar] [CrossRef]
- Sullivan, P.M.; Zhou, X.; Robins, A.M.; Paushter, D.H.; Kim, D.; Smolka, M.B.; Hu, F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol. Commun. 2016, 4, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amick, J.; Ferguson, S.M. C9orf72: At the intersection of lysosome cell biology and neurodegenerative disease. Traffic 2017, 18, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Abo-Rady, M.; Kalmbach, N.; Pal, A.; Schludi, C.; Janosch, A.; Richter, T.; Freitag, P.; Bickle, M.; Kahlert, A.-K.; Petri, S.; et al. Knocking out C9ORF72 Exacerbates Axonal Trafficking Defects Associated with Hexanucleotide Repeat Expansion and Reduces Levels of Heat Shock Proteins. Stem Cell Rep. 2020, 14, 390–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boivin, M.; Pfister, V.; Gaucherot, A.; Ruffenach, F.; Negroni, L.; Sellier, C.; Charlet-Berguerand, N. Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders. EMBO J. 2020, 39, e100574. [Google Scholar] [CrossRef]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef] [Green Version]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Gump, J.M.; Staskiewicz, L.; Morgan, M.J.; Bamberg, A.; Riches, D.W.H.; Thorburn, A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat. Cell Biol. 2014, 16, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Lin, S.; Staats, K.; Li, Y.; Chang, W.-H.; Hung, S.-T.; Hendricks, E.; Linares, G.R.; Wang, Y.; Son, E.Y.; et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 2018, 24, 313–325. [Google Scholar] [CrossRef]
- Janer, A.; Werner, A.; Takahashi-Fujigasaki, J.; Daret, A.; Fujigasaki, H.; Takada, K.; Duyckaerts, C.; Brice, A.; Dejean, A.; Sittler, A. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded ataxin-7. Hum. Mol. Genet. 2010, 19, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Thomas, M.; Dadgar, N.; Lieberman, A.P.; Iñiguez-Lluhí, J.A. Small Ubiquitin-like Modifier (SUMO) Modification of the Androgen Receptor Attenuates Polyglutamine-mediated Aggregation. J. Biol. Chem. 2009, 284, 21296–21306. [Google Scholar] [CrossRef] [Green Version]




| Sex | Age (Years) | G4C2 | Clinical Diagnosis | |
|---|---|---|---|---|
| C9ORF72_1 | Female | 70 | 2, >145 | FTD |
| C9ORF72_2 | Male | 76 | 2, >145 | ALS/FTD |
| C9ORF72_3 | Female | 65 | 2, >145 | ALS/FTD |
| C9ORF72_4 | Female | 69 | 2, >145 | FTD |
| Control_1 | Female | 82 | 2, 2 | Control |
| Control_2 | Male | 57 | 2, 6 | Control |
| Control_3 | Male | 86 | 2, 4 | Control |
| Control_4 | Female | 68 | 2, 2 | Control |
| Enzyme Activities 1 | Fibroblasts | ||||
| C9ORF72_1 | C9ORF72_2 | C9ORF72_3 | C9ORF72_4 | Control Range | |
| Complex I+III | 17.4 | 9.6 * | 0 * | 25.6 | 15–42 |
| Complex II | 19.2 * | 34 | 18.1 * | 40.6 | 22–35 |
| Complex II+III | 3.4 * | 9.8 * | 1.8 * | 15.8 | 11–20 |
| Complex III | 4.6 * | 8.3 * | 0 * | 14.2 * | 25–48 |
| Complex IV | 67 | 42.2 | 18.1 * | 68.5 | 33–57 |
| Citrate synthetase (CS) | 18.7 * | 54 | 15.8 * | 61.8 | 41–62 |
| Enzyme Activities 2 | C9ORF72_1 | C9ORF72_2 | C9ORF72_3 | C9ORF72_4 | Control Range |
| Complex I+III/CS | 0.930 | 0.178 * | 0 * | 0.414 | 0.24–0.85 |
| Complex II/CS | 1.027 | 0.630 | 1.146 | 0.657 | 0.53–0.75 |
| Complex II+III/CS | 0.182 * | 0.181 * | 0.114 * | 0.256 | 0.25–0.42 |
| Complex III/CS | 0.246 * | 0.154 * | 0 * | 0.230 * | 0.49–1.06 |
| Complex IV/CS | 3.583 | 0.781 | 1.146 | 1.108 | 0.84–1.15 |
| PATH_ID | PATH_NAME | Size | padj | LOR |
|---|---|---|---|---|
| R-HSA-1632852 | Macroautophagy | 120 | 0.00004267 | −0.4304 |
| R-HSA-9612973 | Autophagy | 132 | 0.0002208 | −0.3759 |
| R-HSA-9663891 | Selective autophagy | 67 | 0.0005462 | −0.4965 |
| R-HSA-3108232 | SUMO E3 ligases SUMOylate target proteins | 151 | 0.000003633 | 0.4239 |
| R-HSA-2990846 | SUMOylation | 157 | 0.000006105 | 0.4079 |
| R-HSA-983168 | Antigen processing: Ubiquitination and Proteasome degradation | 284 | 0.004528 | −0.2058 |
| R-HSA-8852135 | Protein ubiquitination | 63 | 0.005484 | −0.4211 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Mora, M.I.; Garrabou, G.; Barcos, T.; Garcia-Garcia, F.; Grillo-Risco, R.; Peruga, E.; Gort, L.; Borrego-Écija, S.; Sanchez-Valle, R.; Canto-Santos, J.; et al. Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients. Antioxidants 2022, 11, 1129. https://doi.org/10.3390/antiox11061129
Alvarez-Mora MI, Garrabou G, Barcos T, Garcia-Garcia F, Grillo-Risco R, Peruga E, Gort L, Borrego-Écija S, Sanchez-Valle R, Canto-Santos J, et al. Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients. Antioxidants. 2022; 11(6):1129. https://doi.org/10.3390/antiox11061129
Chicago/Turabian StyleAlvarez-Mora, Maria Isabel, Gloria Garrabou, Tamara Barcos, Francisco Garcia-Garcia, Ruben Grillo-Risco, Emma Peruga, Laura Gort, Sergi Borrego-Écija, Raquel Sanchez-Valle, Judith Canto-Santos, and et al. 2022. "Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients" Antioxidants 11, no. 6: 1129. https://doi.org/10.3390/antiox11061129
APA StyleAlvarez-Mora, M. I., Garrabou, G., Barcos, T., Garcia-Garcia, F., Grillo-Risco, R., Peruga, E., Gort, L., Borrego-Écija, S., Sanchez-Valle, R., Canto-Santos, J., Navarro-Navarro, P., & Rodriguez-Revenga, L. (2022). Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients. Antioxidants, 11(6), 1129. https://doi.org/10.3390/antiox11061129






