Endoplasmic Reticulum Stress and Reactive Oxygen Species in Plants
Abstract
:1. Introduction
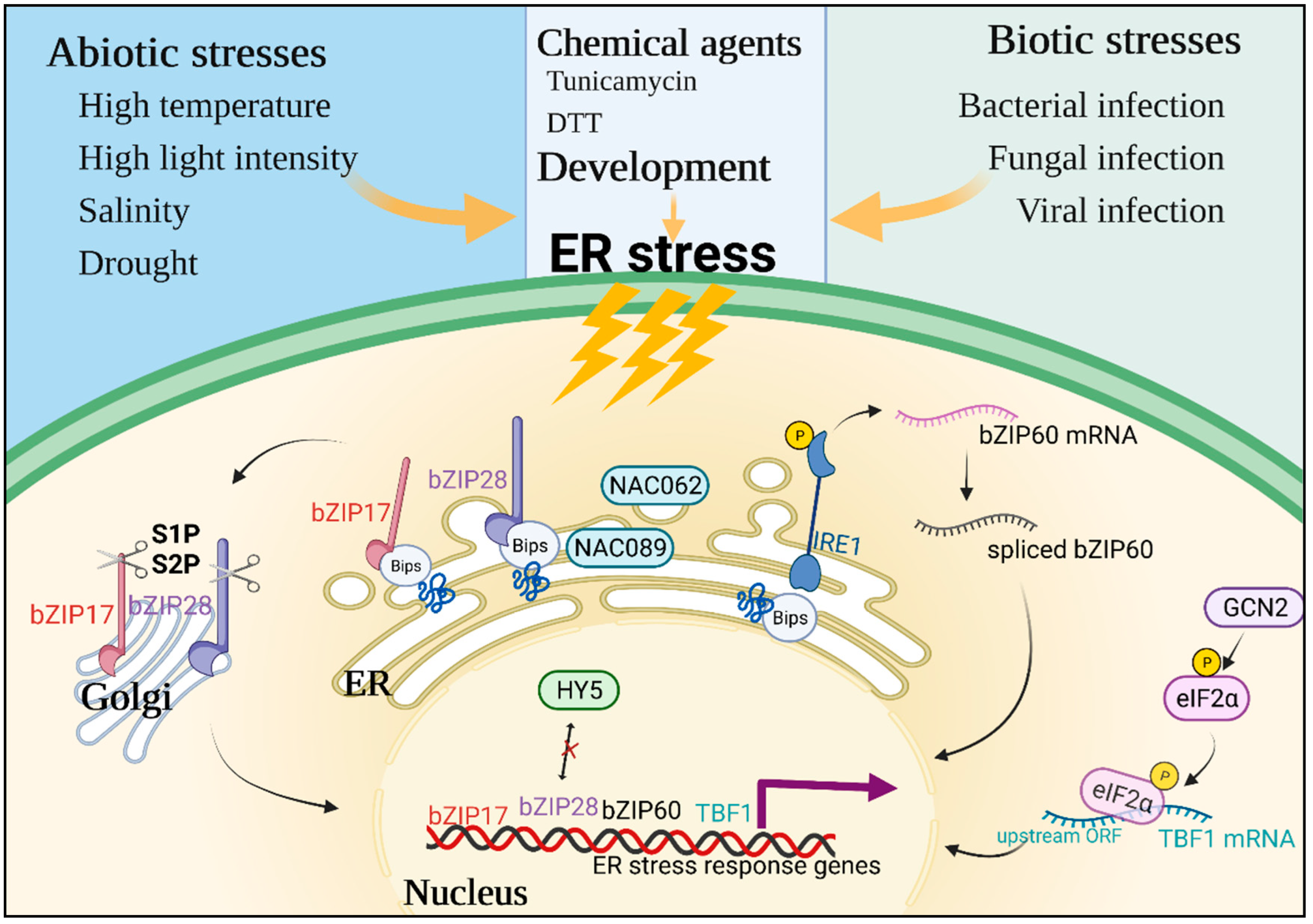
2. Perceptions and Responses under Plant ER Stress
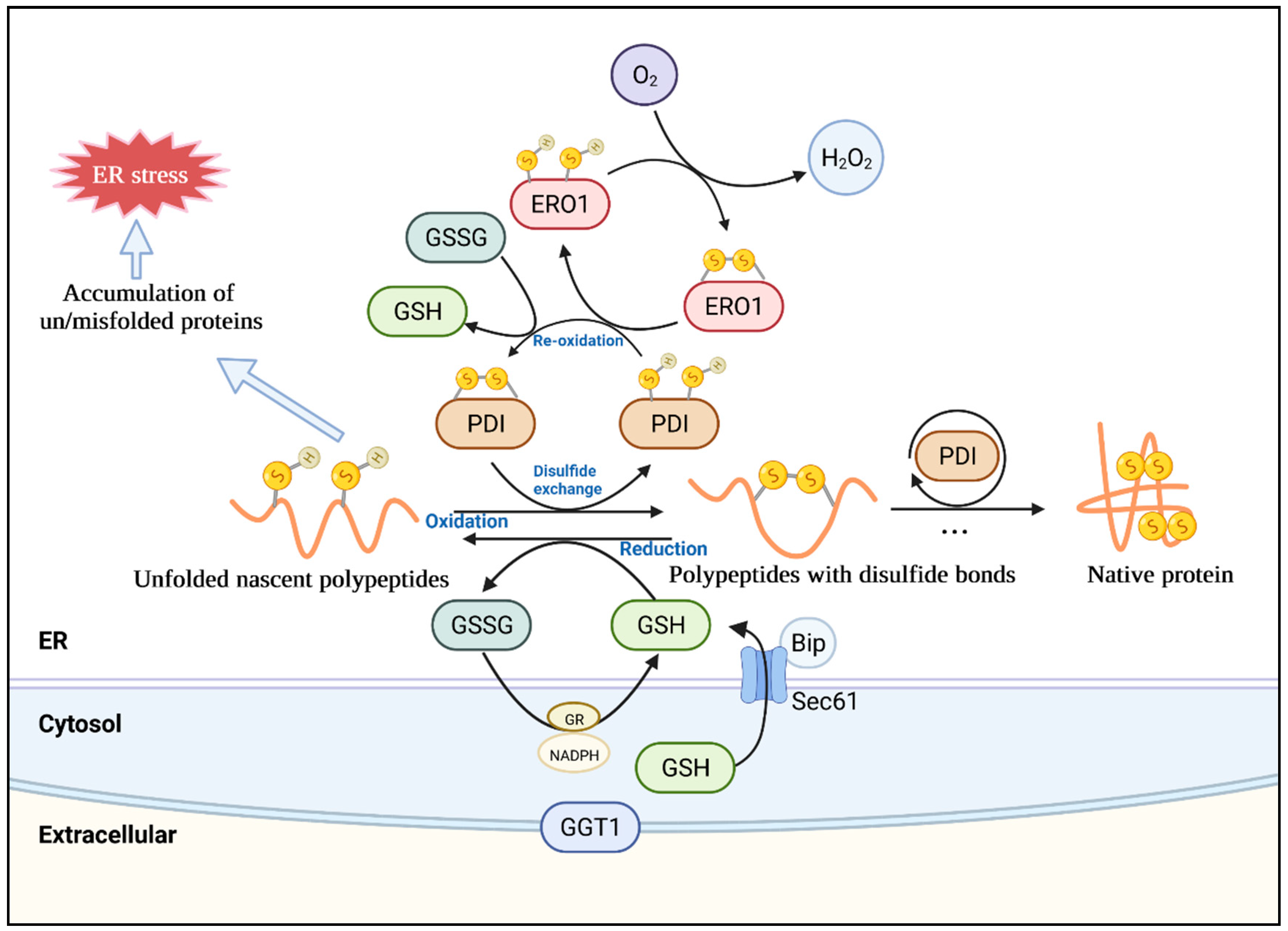
3. Oxidative Protein Folding in the Plant ER
4. Production and Signaling of ROS under ER Stress
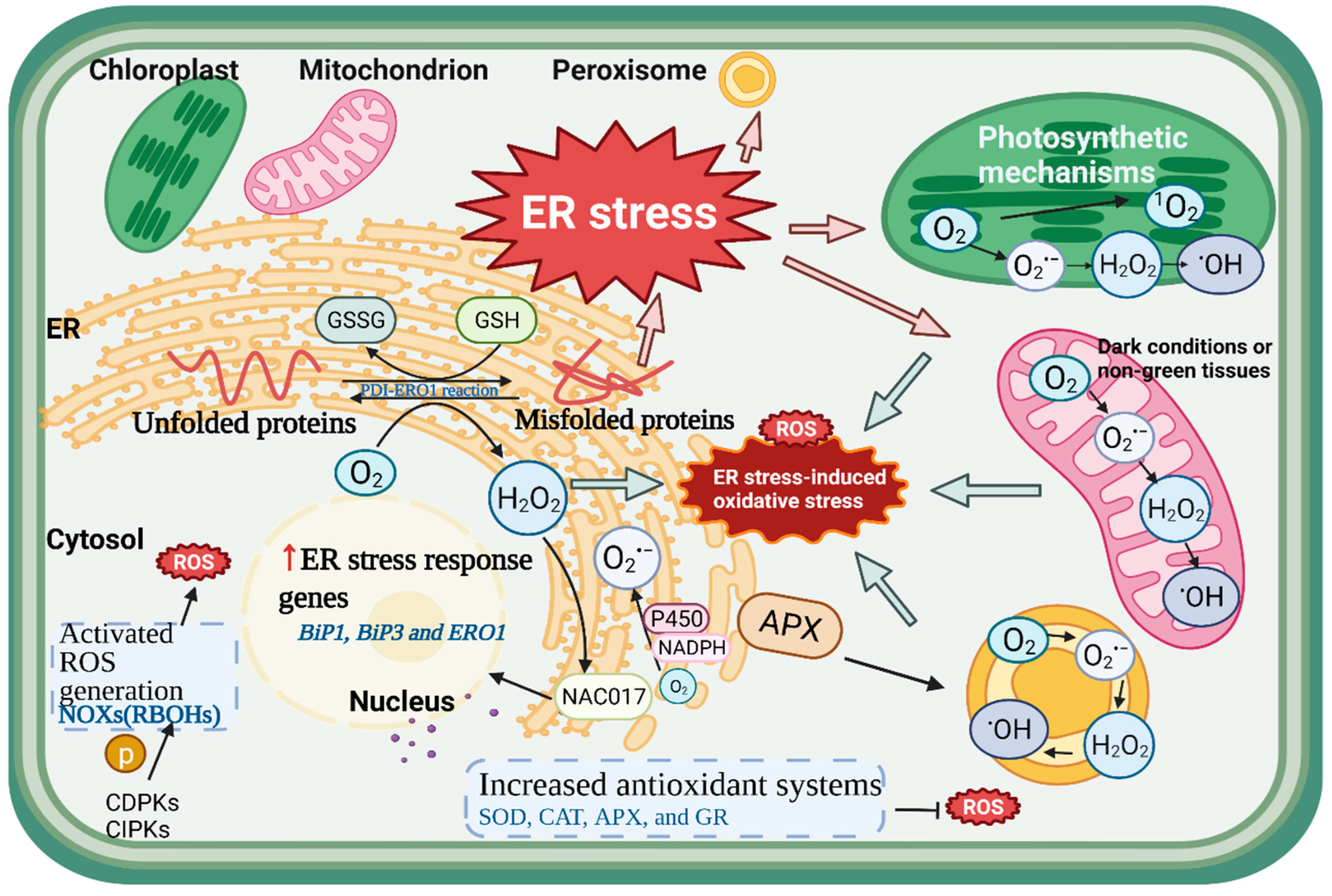
4.1. The Generation of ROS in the ER
4.2. ROS Signaling Triggered by ER Stress
4.3. The Redox Systems in the ER
4.4. ROS as Signaling Molecules between the ER and Other Organelles
4.5. The Interplay between ER Stress and ROS during Biotic/Abiotic Stress
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Stefano, G.; Brandizzi, F. Advances in plant ER architecture and dynamics. Plant Physiol. 2018, 176, 178–186. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelos, E.; Ruberti, C.; Kim, S.J.; Brandizzi, F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 2017, 90, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Ozgur, R.; Uzilday, B.; Iwata, Y.; Koizumi, N.; Turkan, I. Interplay between the unfolded protein response and reactive oxygen species: A dynamic duo. J. Exp. Bot. 2018, 69, 3333–3345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.X.; Howell, S.H. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 2010, 22, 2930–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Yu, F.; Xie, Q. Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 2020, 226, 345–350. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef] [Green Version]
- Ozgur, R.; Turkan, I.; Uzilday, B.; Sekmen, A.H. Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1377–1390. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Aller, I.; Meyer, A.J. The oxidative protein folding machinery in plant cells. Protoplasma 2013, 250, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Hajnoczky, G. SR/ER-mitochondrial local communication: Calcium and ROS. Biochim. Biophys. Acta 2009, 1787, 1352–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drerup, M.M.; Schlucking, K.; Hashimoto, K.; Manishankar, P.; Steinhorst, L.; Kuchitsu, K.; Kudla, J. The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 2013, 6, 559–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilroy, S.; Bialasek, M.; Suzuki, N.; Gorecka, M.; Devireddy, A.R.; Karpinski, S.; Mittler, R. ROS, Calcium, and electric signals: Key mediators of rapid systemic signaling in plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef] [Green Version]
- Chong, W.C.; Shastri, M.D.; Eri, R. Endoplasmic reticulum stress and oxidative stress: A vicious nexus implicated in bowel disease pathophysiology. Int. J. Mol. Sci. 2017, 18, 771. [Google Scholar] [CrossRef]
- Laurindo, F.R.M.; Araujo, T.L.S.; Abrahão, T.B. Nox NADPH oxidases and the endoplasmic reticulum. Antioxid. Redox Signal. 2014, 20, 2755–2775. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, C.D.; Wu, R.F.; Terada, L.S. ROS signaling and ER stress in cardiovascular disease. Mol. Asp. Med. 2018, 63, 18–29. [Google Scholar] [CrossRef]
- Zeeshan, H.M.; Lee, G.H.; Kim, H.R.; Chae, H.J. Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 2016, 17, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Mhamdi, A.; van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braakman, I.; Bulleid, N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef] [PubMed]
- Urade, R. Oxidative protein folding in the plant endoplasmic reticulum. Biosci. Biotechnol. Biochem. 2019, 83, 781–793. [Google Scholar] [CrossRef]
- Afrin, T.; Diwan, D.; Sahawneh, K.; Pajerowska-Mukhtar, K. Multilevel regulation of endoplasmic reticulum stress responses in plants: Where old roads and new paths meet. J. Exp. Bot. 2020, 71, 1659–1667. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, J. Communications between the endoplasmic reticulum and other organelles during abiotic stress response in plants. Front. Plant Sci. 2019, 10, 749. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Howell, S.H. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 2008, 31, 1735–1743. [Google Scholar] [CrossRef]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a Signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef] [Green Version]
- Nawkar, G.M.; Kang, C.H.; Maibam, P.; Park, J.H.; Jung, Y.J.; Chae, H.B.; Chi, Y.H.; Jung, I.J.; Kim, W.Y.; Yun, D.J.; et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 2084–2089. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.T.; Lu, S.J.; Wang, M.J.; Bi, D.L.; Sun, L.; Zhou, S.F.; Song, Z.T.; Liu, J.X. A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 2014, 79, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Bi, D.L.; Sun, L.; Song, Z.T.; Zhang, S.S.; Zhou, S.F.; Liu, J.X. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014, 10, e1004243. [Google Scholar] [CrossRef] [PubMed]
- Howell, S.H. Evolution of the unfolded protein response in plants. Plant Cell Environ. 2021, 44, 2625–2635. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Pu, Y.; Yu, X.; Gregory, B.D.; Srivastava, R.; Howell, S.H.; Bassham, D.C. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 2018, 14, 1562–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishiba, K.; Nagashima, Y.; Suzuki, E.; Hayashi, N.; Ogata, Y.; Shimada, Y.; Koizumi, N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. USA 2013, 110, 5713–5718. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, Y.; Choi, B.Y.; Kim, H.; Shin, S.; Kim, Y.; Jang, S.; Song, W.Y.; Cho, C.H.; Yoon, H.S.; Kohno, K.; et al. Identification and functional study of the endoplasmic reticulum stress sensor IRE1 in Chlamydomonas reinhardtii. Plant J. 2018, 94, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Zhang, S.J.; Zhao, Q.; Long, Y.; Guo, H.; Jia, H.F.; Yang, Y.X.; Zhang, H.Y.; Ye, X.F.; Zhang, S.T. Overexpression of tobacco GCN2 stimulates multiple physiological changes associated with stress tolerance. Front. Plant Sci. 2018, 9, 725. [Google Scholar] [CrossRef]
- Zhao, L.; Deng, L.; Zhang, Q.; Jing, X.; Ma, M.; Yi, B.; Wen, J.; Ma, C.; Tu, J.; Fu, T.; et al. Autophagy contributes to sulfonylurea herbicide tolerance via GCN2-independent regulation of amino acid homeostasis. Autophagy 2018, 14, 702–714. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Afrin, T.; Pajerowska-Mukhtar, K.M. Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun. Biol. 2019, 2, 302. [Google Scholar] [CrossRef] [Green Version]
- Pajerowska-Mukhtar, K.M.; Wang, W.; Tada, Y.; Oka, N.; Tucker, C.L.; Fonseca, J.P.; Dong, X. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 2012, 22, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Selles, B.; Jacquot, J.P.; Rouhier, N. Comparative genomic study of protein disulfide isomerases from photosynthetic organisms. Genomics 2011, 97, 37–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, C.Y.; Matsumoto, K.O.; Christopher, D.A. Variation in the subcellular localization and protein folding activity among Arabidopsis thaliana homologs of protein disulfide isomerase. Biomolecules 2013, 3, 848–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamauchi, S.; Wadahama, H.; Iwasaki, K.; Nakamoto, Y.; Nishizawa, K.; Ishimoto, M.; Kawada, T.; Urade, R. Molecular cloning and characterization of two soybean protein disulfide isomerases as molecular chaperones for seed storage proteins. FEBS J. 2008, 275, 2644–2658. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Higashino, Y.; Kitao, Y.; Masuda, T.; Urade, R. Expression and characterization of protein disulfide isomerase family proteins in bread wheat. BMC Plant Biol. 2015, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birk, J.; Meyer, M.; Aller, I.; Hansen, H.G.; Odermatt, A.; Dick, T.P.; Meyer, A.J.; Appenzeller-Herzog, C. Endoplasmic reticulum: Reduced and oxidized glutathione revisited. J. Cell Sci. 2013, 126, 1604–1617. [Google Scholar] [CrossRef] [Green Version]
- Matsusaki, M.; Okuda, A.; Matsuo, K.; Gekko, K.; Masuda, T.; Naruo, Y.; Hirose, A.; Kono, K.; Tsuchi, Y.; Urade, R. Regulation of plant ER oxidoreductin 1 (ERO1) activity for efficient oxidative protein folding. J. Biol. Chem. 2019, 294, 18820–18835. [Google Scholar] [CrossRef]
- Gross, E.; Sevier, C.S.; Heldman, N.; Vitu, E.; Bentzur, M.; Kaiser, C.A.; Thorpe, C.; Fass, D. Generating disulfides enzymatically: Reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA 2006, 103, 299–304. [Google Scholar] [CrossRef] [Green Version]
- Birk, J.; Ramming, T.; Odermatt, A.; Appenzeller-Herzog, C. Green fluorescent protein-based monitoring of endoplasmic reticulum redox poise. Front. Genet. 2013, 4, 108. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, K.; Jakob, U. The role of thiols in antioxidant systems. Free Radic. Biol. Med. 2019, 140, 14–27. [Google Scholar] [CrossRef]
- Uzilday, B.; Ozgur, R.; Sekmen, A.H.; Turkan, I. Endoplasmic reticulum stress regulates glutathione metabolism and activities of glutathione related enzymes in Arabidopsis. Funct. Plant Biol. FPB 2018, 45, 284–296. [Google Scholar] [CrossRef]
- Dorion, S.; Ouellet, J.C.; Rivoal, J. Glutathione metabolism in plants under stress: Beyond reactive oxygen species detoxification. Metabolites 2021, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Tolin, S.; Arrigoni, G.; Trentin, A.R.; Veljovic-Jovanovic, S.; Pivato, M.; Zechman, B.; Masi, A. Biochemical and quantitative proteomics investigations in Arabidopsis ggt1 mutant leaves reveal a role for the gamma-glutamyl cycle in plant’s adaptation to environment. Proteomics 2013, 13, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Ponsero, A.J.; Igbaria, A.; Darch, M.A.; Miled, S.; Outten, C.E.; Winther, J.R.; Palais, G.; D’autreaux, B.; Delaunay-Moisan, A.; Toledano, M.B. Endoplasmic reticulum transport of glutathione by Sec61 is regulated by Ero1 and Bip. Mol. Cell 2017, 67, 962–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, S.S.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 2014, 21, 396–413. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Janku, M.; Luhova, L.; Petrivalsky, M. On the origin and Fate of reactive oxygen species in plant cell compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Angelos, E.; Brandizzi, F. NADPH oxidase activity is required for ER stress survival in plants. Plant J. 2018, 96, 1106–1120. [Google Scholar] [CrossRef] [Green Version]
- Noirot, E.; Der, C.; Lherminier, J.; Robert, F.; Moricova, P.; Kieu, K.; Leborgne-Castel, N.; Simon-Plas, F.; Bouhidel, K. Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 2014, 65, 5011–5022. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2021, 105, 942–956. [Google Scholar] [CrossRef]
- Tu, B.P.; Weissman, J.S. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; van Aken, O.; de Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; El-Shetehy, M.; Shine, M.B.; Yu, K.; Navarre, D.; Wendehenne, D.; Kachroo, A.; Kachroo, P. Free radicals mediate systemic acquired resistance. Cell Rep. 2014, 7, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Wendehenne, D.; Gao, Q.M.; Kachroo, A.; Kachroo, P. Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 2014, 20, 127–134. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, C. S-Nitrosylation control of ROS and RNS homeostasis in plants: The switching function of catalase. Mol. Plant 2020, 13, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Wang, Z.; Wang, X.; Takano, T.; Liu, S. A peroxisomal APX from Puccinellia tenuiflora improves the abiotic stress tolerance of transgenic Arabidopsis thaliana through decreasing of H2O2 accumulation. J. Plant Physiol. 2015, 175, 183–191. [Google Scholar] [CrossRef]
- Lu, D.P.; Christopher, D.A. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genom. 2008, 280, 199–210. [Google Scholar] [CrossRef]
- Puigpinos, J.; Casas, C.; Herrero, E. Altered intracellular calcium homeostasis and endoplasmic reticulum redox state in Saccharomyces cerevisiae cells lacking Grx6 glutaredoxin. Mol. Biol. Cell 2015, 26, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Ramming, T.; Hansen, H.G.; Nagata, K.; Ellgaard, L.; Appenzeller-Herzog, C. GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 2014, 70, 106–116. [Google Scholar] [CrossRef]
- Murphy, M.P. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab. 2013, 18, 145–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozgur, R.; Uzilday, B.; Sekmen, A.H.; Turkan, I. The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in Arabidopsis. Ann. Bot. 2015, 116, 541–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrshahi, P.; Stefano, G.; Andaloro, J.M.; Brandizzi, F.; Froehlich, J.E.; Dellapenna, D. Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12126–12131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, A.M.; Trobacher, C.P.; Mathur, N.; Greenwood, J.S.; Mathur, J. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 2009, 59, 231–242. [Google Scholar] [CrossRef]
- Mullen, R.T.; Trelease, R.N. The ER-peroxisome connection in plants: Development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim. Biophys. Acta 2006, 1763, 1655–1668. [Google Scholar] [CrossRef] [Green Version]
- Bhattarai, K.R.; Riaz, T.A.; Kim, H.R.; Chae, H.J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 2021, 53, 151–167. [Google Scholar] [CrossRef]
- Medina, E.; Kim, S.H.; Yun, M.; Choi, W.G. Recapitulation of the function and role of ROS generated in response to heat stress in plants. Plants 2021, 10, 371. [Google Scholar] [CrossRef]
- Che, P.; Bussell, J.D.; Zhou, W.; Estavillo, G.M.; Pogson, B.J.; Smith, S.M. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 2010, 3, ra69. [Google Scholar] [CrossRef]
- Gao, H.; Brandizzi, F.; Benning, C.; Larkin, R.M. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 16398–16403. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, R.; Takahashi, M.; Suzuki, N. Coordination between bZIP28 and HSFA2 in the regulation of heat response signals in Arabidopsis. Plant Signal. Behav. 2017, 12, e1376159. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levings, D.C.; Lacher, S.E.; Palacios-Moreno, J.; Slattery, M. Transcriptional reprogramming by oxidative stress occurs within a predefined chromatin accessibility landscape. Free Radic. Biol. Med. 2021, 171, 319–331. [Google Scholar] [CrossRef]
- Poidevin, L.; Forment, J.; Unal, D.; Ferrando, A. Transcriptome and translatome changes in germinated pollen under heat stress uncover roles of transporter genes involved in pollen tube growth. Plant Cell Environ. 2021, 44, 2167–2184. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.B.; Lohani, N.; Bhalla, P.L. The role of endoplasmic reticulum stress response in pollen development and heat stress tolerance. Front. Plant Sci. 2021, 12, 661062. [Google Scholar] [CrossRef]
- Park, C.J.; Park, J.M. Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant Sci. 2019, 10, 399. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhang, T.; Liu, Y.; Li, Z.; Wang, Y.; Fu, S.; Xu, Y.; Zhou, T.; Wu, J.; Zhou, X. Rice stripe virus activates the bZIP17/28 branch of the unfolded protein response signalling pathway to promote viral infection. Mol. Plant Pathol. 2022, 23, 447–458. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, H.; Brandizzi, F.; Verchot, J.; Wang, A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015, 11, e1005164. [Google Scholar] [CrossRef] [Green Version]
- Poor, P.; Czekus, Z.; Tari, I.; Ordog, A. The multifaceted roles of plant hormone salicylic acid in endoplasmic reticulum stress and unfolded protein response. Int. J. Mol. Sci. 2019, 20, 5842. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Li, S.; Wang, L.; Li, Y.; Li, Y.; Zhang, C.; Hou, X. Turnip mosaic virus induces expression of the LRR II subfamily genes and regulates the salicylic acid signaling pathway in non-heading Chinese cabbage. Physiol. Mol. Plant Pathol. 2013, 82, 64–72. [Google Scholar] [CrossRef]
- Saleem, M.; Fariduddin, Q.; Castroverde, C.D.M. Salicylic acid: A key regulator of redox signalling and plant immunity. Plant Physiol. Biochem. 2021, 168, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Ruberti, C.; Gong, Z.; Brandizzi, F. CPR5 modulates salicylic acid and the unfolded protein response to manage tradeoffs between plant growth and stress responses. Plant J. 2017, 89, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasch, C.M.; Sonnewald, U. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Wang, C.; Hao, N.; Fujiwara, T.; Wu, T. Endoplasmic Reticulum Stress and Reactive Oxygen Species in Plants. Antioxidants 2022, 11, 1240. https://doi.org/10.3390/antiox11071240
Cao J, Wang C, Hao N, Fujiwara T, Wu T. Endoplasmic Reticulum Stress and Reactive Oxygen Species in Plants. Antioxidants. 2022; 11(7):1240. https://doi.org/10.3390/antiox11071240
Chicago/Turabian StyleCao, Jiajian, Chunhua Wang, Ning Hao, Toru Fujiwara, and Tao Wu. 2022. "Endoplasmic Reticulum Stress and Reactive Oxygen Species in Plants" Antioxidants 11, no. 7: 1240. https://doi.org/10.3390/antiox11071240
APA StyleCao, J., Wang, C., Hao, N., Fujiwara, T., & Wu, T. (2022). Endoplasmic Reticulum Stress and Reactive Oxygen Species in Plants. Antioxidants, 11(7), 1240. https://doi.org/10.3390/antiox11071240




