Novel Probiotic Lactic Acid Bacteria Were Identified from Healthy Infant Feces and Exhibited Anti-Inflammatory Capacities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Strain Isolation
2.2. Morphological and Biochemical Characterization
2.3. Preliminary Selection of LAB Using Specific Primers
2.4. Antimicrobial Activity Assessment
2.5. Tolerance to Simulated Digestive Tract Conditions
2.6. Adherence to Intestinal Epithelial Cells
2.7. Antibiotic Resistance Assay
2.8. Strain Identification Using 16S rRNA Sequence Analyses
2.9. Cytokine Measurement
2.10. Determination of Antioxidant Enzymes and H2O2 Production
2.11. Western Blotting
2.12. Statistical Analysis
2.13. Data Availability
3. Results
3.1. Morphological and Biochemical Test Results
3.2. Specific Primers Amplification for LAB
3.3. Antimicrobial Test Results
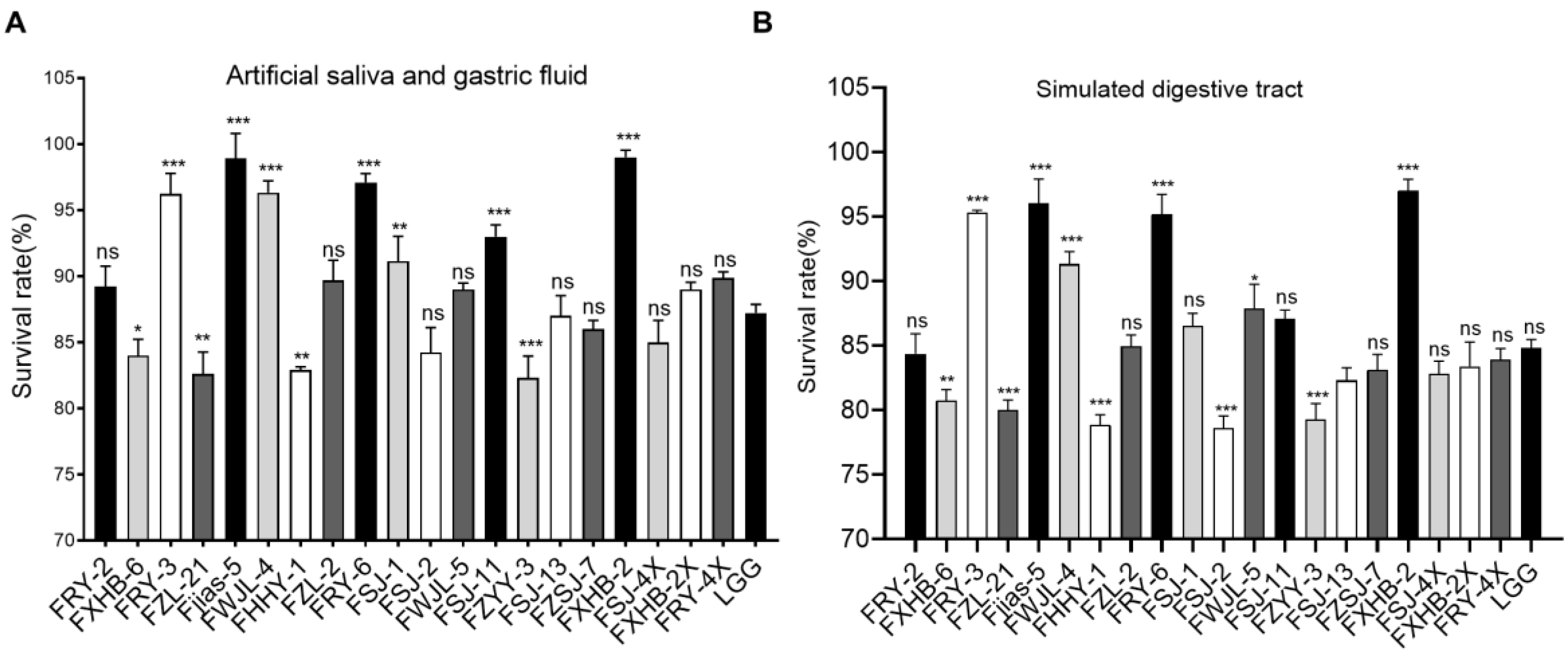
3.4. Tolerance to Simulated Digestive Tract Condition
3.5. Adherence to Caco-2 Cells
3.6. Antibiotic Resistance
3.7. 16S rRNA Sequencing and Phylogenetic Tree Results
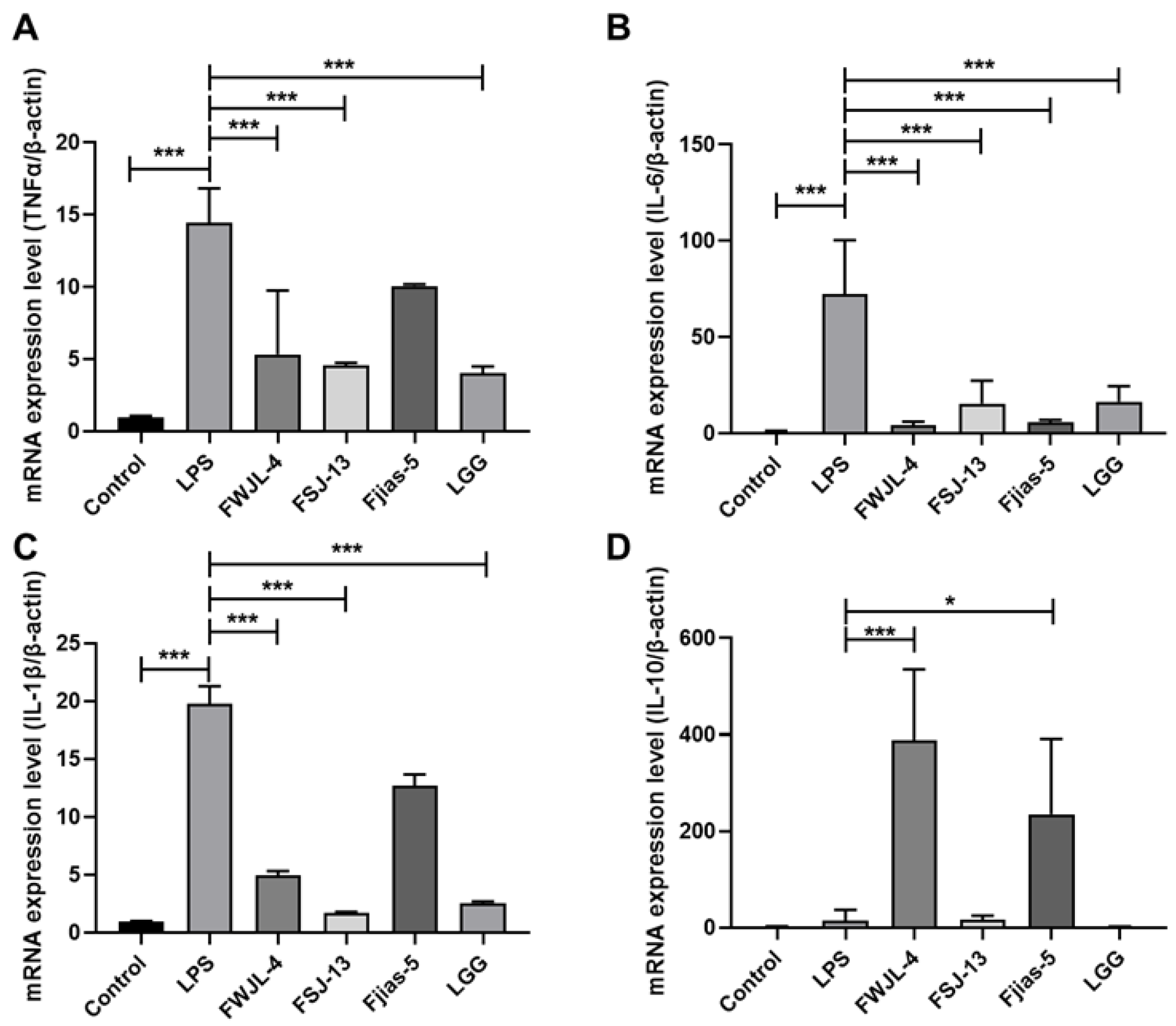
3.8. LAB Treatments Inhibit Inflammation in LPS-Treated RAW 264.7 Murine Macrophages
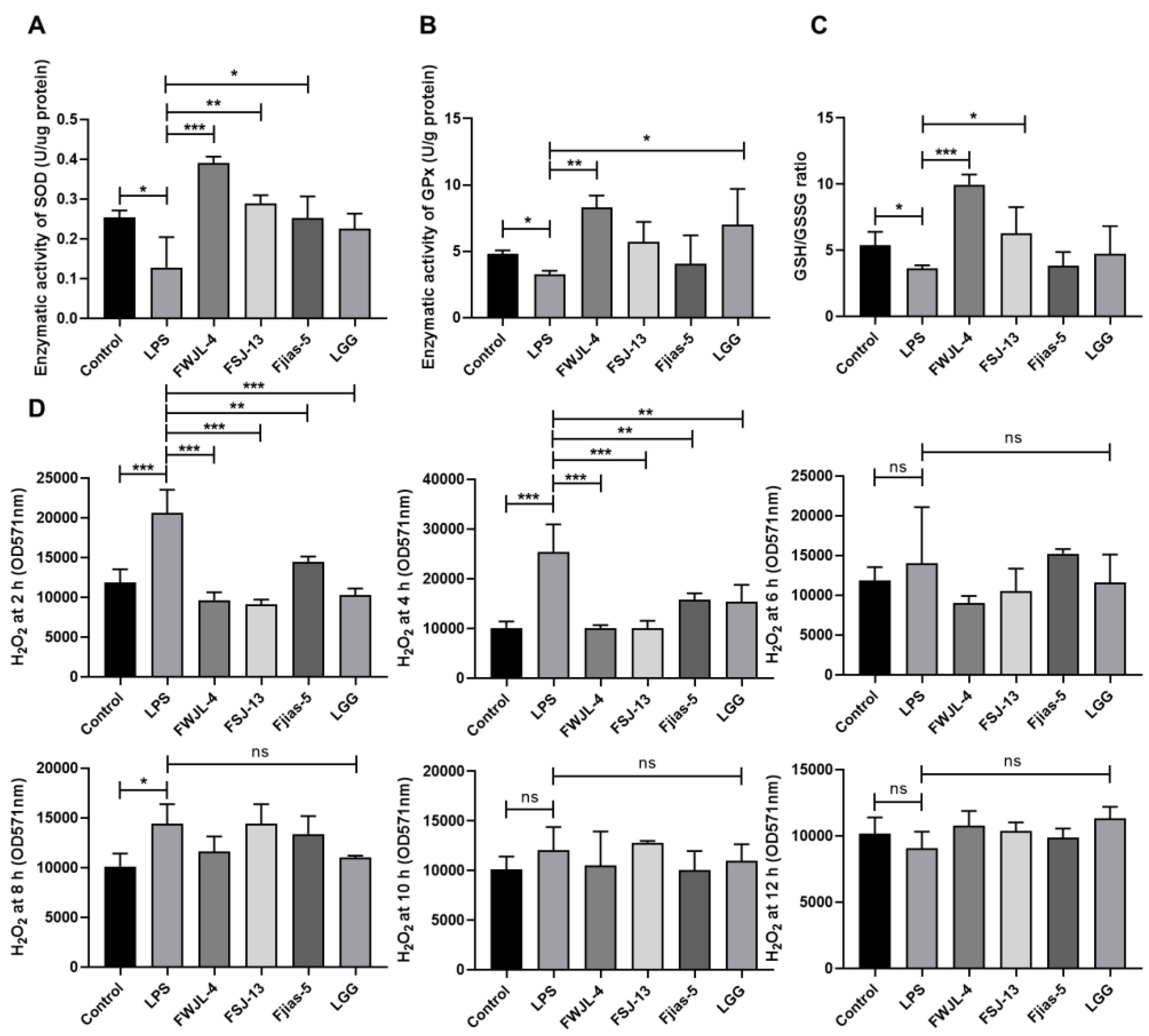
3.9. LAB Treatments Reduce ROS Production in LPS-Treated RAW 264.7 Murine Macrophages
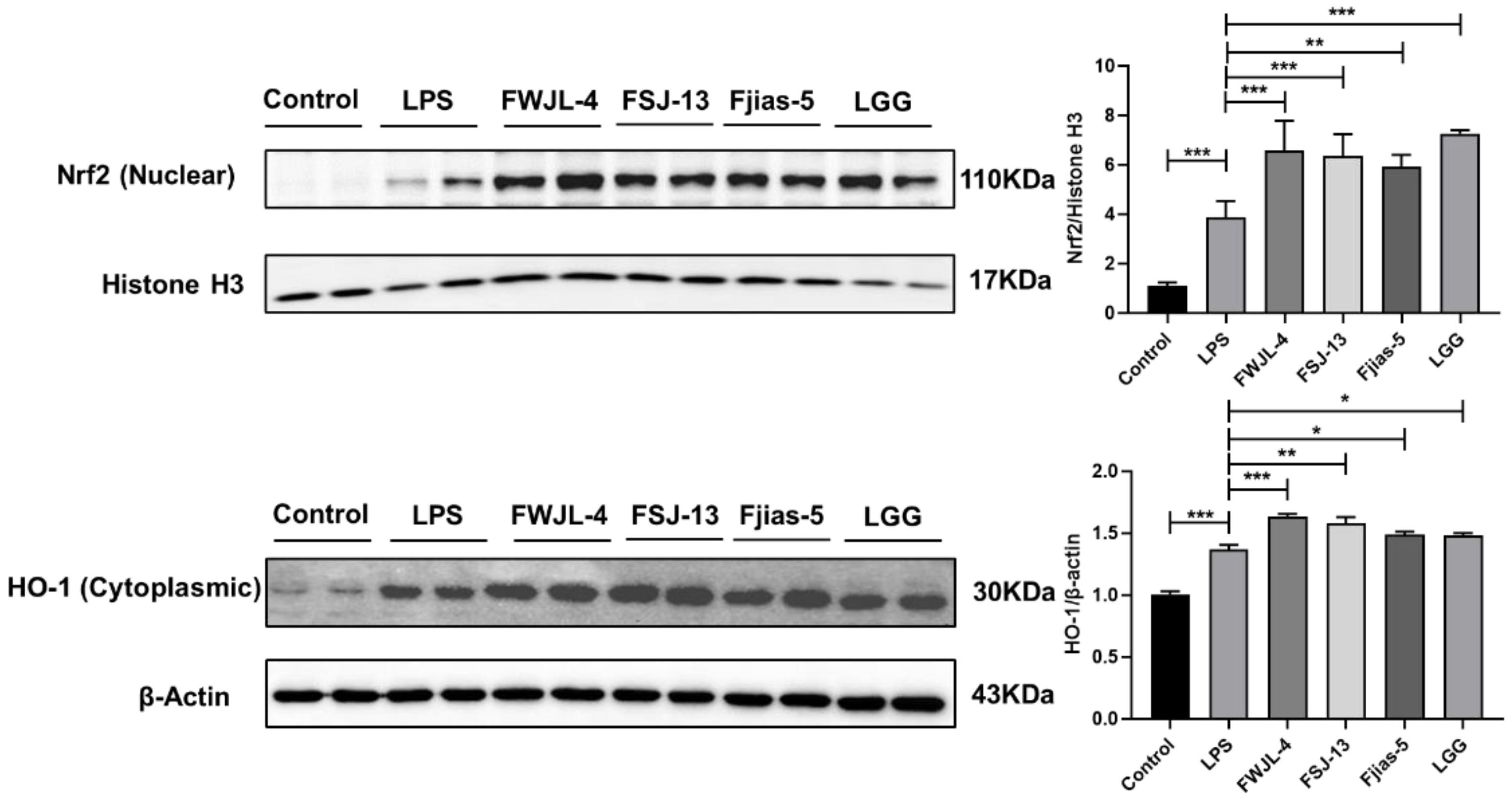
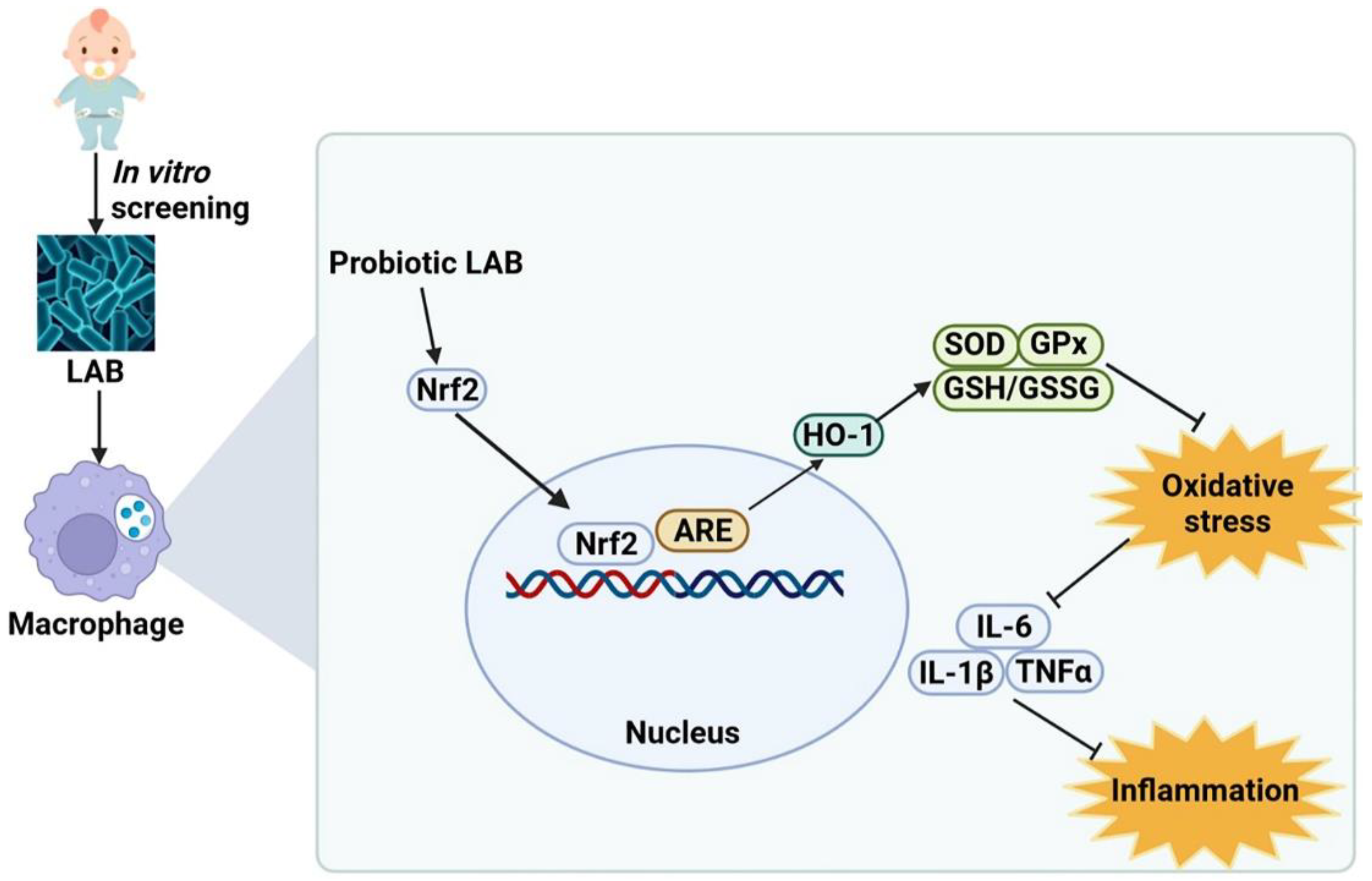
3.10. Selected Probiotic LAB Induce Nuclear Factor-Erythroid 2 Related Factor 2 (Nrf2)-Mediated Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Doron, S.; Snydman, D.R. Risk and safety of probiotics. Clin. Infect. Dis. 2015, 60 (Suppl. S2), S129–S134. [Google Scholar] [CrossRef] [Green Version]
- Prado, F.C.; Lindner, J.D.D.; Inaba, J.; Thomaz-Soccol, V.; Brar, S.K.; Soccol, C.R. Development and evaluation of a fermented coconut water beverage with potential health benefits. J. Funct. Foods 2015, 12, 489–497. [Google Scholar] [CrossRef]
- Maria Rosa Machado, C.R.S. Current Developments in Probiotics. J. Microb. Biochem. Technol. 2015, 7, 11–020. [Google Scholar] [CrossRef]
- KKumar, R.; Sood, U.; Gupta, V.; Singh, M.; Scaria, J.; Lal, R. Recent Advancements in the Development of Modern Probiotics for Restoring Human Gut Microbiome Dysbiosis. Indian J. Microbiol. 2019, 60, 12–25. [Google Scholar] [CrossRef]
- Kerry, R.G.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.-S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Dixit, Y.; Wagle, A.; Vakil, B. Patents in the Field of Probiotics, Prebiotics, Synbiotics: A Review. J. Food Microbiol. Saf. Hyg. 2016, 1, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, N.; Wang, Q.; Liu, Z.; Lee, Y.-K.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Microbial diversity and volatile profile of traditional fermented yak milk. J. Dairy Sci. 2020, 103, 87–97. [Google Scholar] [CrossRef]
- Zhai, Q.; Shen, X.; Cen, S.; Zhang, C.; Tian, F.; Zhao, J.; Zhang, H.; Xue, Y.; Chen, W. Screening of Lactobacillus salivarius strains from the feces of Chinese populations and the evaluation of their effects against intestinal inflammation in mice. Food Funct. 2020, 11, 221–235. [Google Scholar] [CrossRef]
- Ooi, M.F.; Mazlan, N.; Foo, H.L.; Loh, T.C.; Rosfarizan, M.; Rahim, R.A.; Ariff, A. Effects of carbon and nitrogen sources on bacteriocin-inhibitory activity of postbiotic metabolites produced by Lactobacillus plantarum I-UL4. Malays. J. Microbiol. 2015, 176–184. [Google Scholar]
- Wang, G.; Yang, S.; Sun, S.; Si, Q.; Wang, L.; Zhang, Q.; Wu, G.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus rhamnosus Strains Relieve Loperamide-Induced Constipation via Different Pathways Independent of Short-Chain Fatty Acids. Front. Cell. Infect. Microbiol. 2020, 10, 423. [Google Scholar] [CrossRef]
- Tsilingiri, K.; Rescigno, M. Postbiotics: What else? Benef. Microbes 2013, 4, 101–107. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, K.; Yin, S.; Liu, S.; Zhu, Y.; Yang, Y.; Wang, C. Purification and characterization of an exopolysaccharide produced by Lactobacillus plantarum HY isolated from home-made Sichuan Pickle. Int. J. Biol. Macromol. 2019, 134, 516–526. [Google Scholar] [CrossRef]
- Valladares, R.S.D.; Li, N.; Williams, E.; Lai, K.K.; Abdelgeliel, A.S.; Gonzalez, C.F. Lactobacillus johnsonii N6.2 Mitigates the Developmentof Type 1 Diabetes in BB-DP Rats. PLoS ONE 2010, 5, e10507. [Google Scholar]
- Alberda, C.G.L.; Meddings, J.; Field, C.; McCargar, L.; Kutsogiannis, D.; Fedorak, R. Effects of probiotic therapy in critically ill patients a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 2007, 85, 816–823. [Google Scholar] [CrossRef] [Green Version]
- García-Díez, E.; López-Oliva, M.E.; Caro-Vadillo, A.; Pérez-Vizcaíno, F.; Pérez-Jiménez, J.; Ramos, S.; Martín, M. Supplementation with a Cocoa–Carob Blend, Alone or in Combination with Metformin, Attenuates Diabetic Cardiomyopathy, Cardiac Oxidative Stress and Inflammation in Zucker Diabetic Rats. Antioxidants 2022, 11, 432. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Zhao, S.; Cheng, C.K.; Zhang, C.-L.; Huang, Y. Interplay Between Oxidative Stress, Cyclooxygenases, and Prostanoids in Cardiovascular Diseases. Antioxid. Redox Signal. 2021, 34, 784–799. [Google Scholar] [CrossRef]
- Carroll, I.M.; Andrus, J.M.; Bruno-Barcena, J.M.; Klaenhammer, T.R.; Hassan, H.M.; Threadgill, D.S. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am. J. Physiol. Gastrointest Liver Physiol. 2007, 293, G729–G738. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yang, J.; Ye, Z.; Zhang, G.; Nie, W.; Cheng, H.; Peng, M.; Zhang, K.; Liu, J.; Zhang, Z.; et al. Physiologically Inspired Mucin Coated Escherichia coli Nissle 1917 Enhances Biotherapy by Regulating the Pathological Microenvironment to Improve Intestinal Colonization. ACS Nano 2022, 16, 4041–4058. [Google Scholar] [CrossRef]
- Finamore, A.; Ambra, R.; Nobili, F.; Garaguso, I.; Raguzzini, A.; Serafini, M. Redox Role of Lactobacillus casei Shirota Against the Cellular Damage Induced by 2,2’-Azobis (2-Amidinopropane) Dihydrochloride-Induced Oxidative and Inflammatory Stress in Enterocytes-Like Epithelial Cells. Front. Immunol. 2018, 9, 1131. [Google Scholar] [CrossRef] [Green Version]
- Jomehzadeh, N.; Javaherizadeh, H.; Amin, M.; Saki, M.; Al-Ouqaili, M.T.; Hamidi, H.; Seyedmahmoudi, M.; Gorjian, Z. Isolation and identification of potential probiotic Lactobacillus species from feces of infants in southwest Iran. Int. J. Infect. Dis. 2020, 96, 524–530. [Google Scholar] [CrossRef]
- BBazireh, H.; Shariati, P.; Jamalkandi, S.A.; Ahmadi, A.; Boroumand, M.A. Isolation of Novel Probiotic Lactobacillus and Enterococcus Strains From Human Salivary and Fecal Sources. Front. Microbiol. 2020, 11, 597946. [Google Scholar] [CrossRef]
- Lu, W.; Chen, W.; Zhai, Q.; Pan, M.; Xie, M.; Cui, S.; Zhao, J.; Zhang, H. A Method for Screening and/or Identifying Lactobacillus and Application. CN109971871A, 5 July 2019. [Google Scholar]
- Shang, J.; Wan, F.; Zhao, L.; Meng, X.; Li, B. Potential Immunomodulatory Activity of a Selected Strain Bifidobacterium bifidum H3-R2 as Evidenced in vitro and in Immunosuppressed Mice. Front. Microbiol. 2020, 11, 2089. [Google Scholar] [CrossRef]
- Balthazar, C.F.; Silva, H.L.; Esmerino, E.A.; Rocha, R.S.; Moraes, J.; Carmo, M.A.; Azevedo, L.; Camps, I.; Abud, Y.K.; Sant’Anna, C.; et al. The addition of inulin and Lactobacillus casei 01 in sheep milk ice cream. Food Chem. 2018, 246, 464–472. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Young, S.; Rohr, J.R.; Harwood, V.J. Vancomycin resistance plasmids affect persistence of Enterococcus faecium in water. Water Res. 2019, 166, 115069. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Fabrícioz, L.T.; Winkelströter, L.K.; De Martini, E.C.P. Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum ft259, a bacteriocinogenic strain isolated from brazilian semi-hard artisanal cheese. Anaerobe 2013, 22, 57–63. [Google Scholar]
- Yan, S.; Zhang, X.; Zheng, H.; Hu, D.; Zhang, Y.; Guan, Q.; Liu, L.; Ding, Q.; Li, Y. Clematichinenoside inhibits VCAM-1 and ICAM-1 expression in TNF-alpha-treated endothelial cells via NADPH oxidase-dependent IkappaB kinase/NF-kappaB pathway. Free Radic. Biol. Med. 2015, 78, 190–201. [Google Scholar] [CrossRef]
- Dong, X.; Wu, D.; Zhang, Y.; Jia, L.; Pan, X.; Sun, J.; Pan, L.-L. Cathelicidin Modulates Vascular Smooth Muscle Cell Phenotypic Switching through ROS/IL-6 Pathway. Antioxidants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, Y.; Li, L.; Liu, S.; Wang, C.; Yuan, Y.; Yang, G.; Chen, Y.; Cheng, J.; Lu, Y.; et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics 2021, 11, 1845–1863. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Li, R.; Dai, P.; Li, Z.; Li, Y.; Li, C. Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ. Pollut. 2019, 251, 689–698. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, J.; Duan, H.; Li, R.; Peng, W.; Wu, C. Activation of Nrf2/HO-1 signaling: An important molecular mechanism of herbal medicine in the treatment of atherosclerosis via the protection of vascular endothelial cells from oxidative stress. J. Adv. Res. 2021, 34, 43–63. [Google Scholar] [CrossRef]
- Xing, J.; Wang, F.; Xu, Q.; Yin, B.; Fang, D.; Zhao, J.; Zhang, H.; Chen, Y.Q.; Wang, G.; Chen, W. Screening of potential probiotic lactic acid bacteria based on gastrointestinal properties and perfluorooctanoate toxicity. Appl. Microbiol. Biotechnol. 2016, 100, 6755–6766. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Shaukat, A.; Levitt, M.D.; Taylor, B.C.; MacDonald, R.; Shamliyan, T.A.; Kane, R.L.; Wilt, T.J. Systematic review: Effective management strategies for lactose intolerance. Ann. Intern. Med. 2010, 152, 797–803. [Google Scholar] [CrossRef]
- Munoz-Quezada, S.; Chenoll, E.; Vieites, J.M.; Genoves, S.; Maldonado, J.; Bermudez-Brito, M.; Gomez-Llorente, C.; Matencio, E.; José Bernal, M.; Romero, F.; et al. Isolation, identification and characterisation of three novel probiotic strains (Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036) from the faeces of exclusively breast-fed infants. Br. J. Nutr. 2013, 109 (Suppl. S2), S51–S62. [Google Scholar] [CrossRef] [Green Version]
- Gheziel, C.; Russo, P.; Arena, M.P.; Spano, G.; Ouzari, H.-I.; Kheroua, O.; Saidi, D.; Fiocco, D.; Kaddouri, H.; Capozzi, V. Evaluating the Probiotic Potential of Lactobacillus plantarum Strains from Algerian Infant Feces: Towards the Design of Probiotic Starter Cultures Tailored for Developing Countries. Probiotics Antimicrob. Proteins 2018, 11, 113–123. [Google Scholar] [CrossRef]
- El-Deeb, W.M.; Fayez, M.; Elsohaby, I.; Ghoneim, I.; Al-Marri, T.; Kandeel, M.; Gioushy, M. Isolation and characterization of vaginal Lactobacillus spp. in dromedary camels (Camelus dromedarius): In vitro evaluation of probiotic potential of selected isolates. PeerJ 2020, 8, e8500. [Google Scholar] [CrossRef] [Green Version]
- Colomer-Winter, C.; Flores-Mireles, A.L.; Kundra, S.; Hultgren, S.J.J.A.L. (p)ppGpp and CodY Promote Enterococcus faecalis Virulence in a Murine Model of Catheter-Associated Urinary Tract Infection. mSphere 2019, 24, e00392-19. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Sun, Z.; Wang, F.; Liu, Y.; Zhu, Y.; Du, L.; Wang, D.; Xu, W. Inhibition of biofilm formation and exopolysaccharide synthesis of Enterococcus faecalis by phenyllactic acid. Food Microbiol. 2019, 86, 103344. [Google Scholar] [CrossRef]
- Kaur, M.; Singh, H.; Jangra, M.; Kaur, L.; Jaswal, P.; Dureja, C.; Nandanwar, H.; Chaudhuri, S.R.; Raje, M.; Mishra, S.; et al. Lactic acid bacteria isolated from yak milk show probiotic potential. Appl. Microbiol. Biotechnol. 2017, 101, 7635–7652. [Google Scholar] [CrossRef]
- Liu, W.; Chen, M.; Duo, L.; Wang, J.; Guo, S.; Sun, H.; Menghe, B.; Zhang, H. Characterization of potentially probiotic lactic acid bacteria and bifidobacteria isolated from human colostrum. J. Dairy Sci. 2020, 103, 4013–4025. [Google Scholar] [CrossRef]
- Talib, N.; Mohamad, N.E.; Yeap, S.K.; Hussin, Y.; Aziz, M.N.M.; Masarudin, M.J.; Sharifuddin, S.A.; Hui, Y.W.; Ho, C.L.; Alitheen, N.B. Isolation and Characterization of Lactobacillus spp. from Kefir Samples in Malaysia. Molecules 2019, 24, 2606. [Google Scholar] [CrossRef] [Green Version]
- Ruas-Madiedo, P.; Gueimonde, M.; Arigoni, F.; Reyes-Gavilán, C.G.D.L.; Margolles, A. Bile Affects the Synthesis of Exopolysaccharides by Bifidobacterium animalis. Appl. Environ. Microbiol. 2009, 75, 1204–1207. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, L.; Sanchez, B.; Ruas-Madiedo, P.; de Los Reyes-Gavilan, C.G.; Margolles, A. Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol. Lett. 2007, 274, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Gueimonde, M.; Margolles, A.; de los Reyes-Gavilán, C.G.; Salminen, S. Competitive exclusion of enteropathogens from human intestinal mucus by Bifidobacterium strains with acquired resistance to bile—A preliminary study. Int. J. Food Microbiol. 2007, 113, 228–232. [Google Scholar] [CrossRef]
- Temmerman, R.; Pot, B.; Huys, G.; Swings, J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 2003, 81, 1–10. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; de Los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M.Á. In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- Izuddin, W.I.; Humam, A.M.; Loh, T.C.; Foo, H.L.; Samsudin, A.A. Dietary Postbiotic Lactobacillus plantarum Improves Serum and Ruminal Antioxidant Activity and Upregulates Hepatic Antioxidant Enzymes and Ruminal Barrier Function in Post-Weaning Lambs. Antioxidants 2020, 9, 250. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Joishy, T.; Das, S.; Kalita, M.C.; Mukherjee, A.K.; Khan, M.R. A Potential Probiotic Lactobacillus plantarum JBC5 Improves Longevity and Healthy Aging by Modulating Antioxidative, Innate Immunity and Serotonin-Signaling Pathways in Caenorhabditis elegans. Antioxidants 2022, 11, 268. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, B.; Tang, L.; Zhou, Y.; Wang, Q.; Gong, L.; Ni, J.; Li, W. Probiotic Bacillus Alleviates Oxidative Stress-Induced Liver Injury by Modulating Gut-Liver Axis in a Rat Model. Antioxidants 2022, 11, 291. [Google Scholar] [CrossRef]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.-F.; Shen, X.-Y.; Lio, C.-K.; Dai, Y.; Cheng, C.-S.; Liu, J.-X.; Yao, Y.-D.; Yu, Y.; Xie, Y.; Luo, P.; et al. Activation of Nrf2/HO-1 Pathway by Nardochinoid C Inhibits Inflammation and Oxidative Stress in Lipopolysaccharide-Stimulated Macrophages. Front. Pharmacol. 2018, 9, 911. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Yuan, T.; Yang, T.; Chen, H.; Fu, D.; Hu, Y.; Wang, J.; Yuan, Q.; Yu, H.; Xu, W.; Xie, X. New insights into oxidative stress and inflammation during diabetes mellitus-accelerated atherosclerosis. Redox Biol. 2019, 20, 247–260. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Zhao, L.; Zhao, Y.; Yang, G.; Wang, C.; Gao, L.; Niu, C.; Li, S. Lactobacillus plantarum DP189 Reduces alpha-SYN Aggravation in MPTP-Induced Parkinson’s Disease Mice via Regulating Oxidative Damage, Inflammation, and Gut Microbiota Disorder. J. Agric. Food Chem. 2022, 70, 1163–1173. [Google Scholar] [CrossRef]
- Gao, D.; Gao, Z.; Zhu, G. Antioxidant effects of Lactobacillus plantarum via activation of transcription factor Nrf2. Food Funct. 2013, 4, 982–989. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Gao, N.; Wang, Z.; Li, F.; Li, J.; Shan, A. Exopolysaccharides produced by Lactobacillus rhamnosus GG alleviate hydrogen peroxide-induced intestinal oxidative damage and apoptosis through the Keap1/Nrf2 and Bax/Bcl-2 pathways in vitro. Food Funct. 2021, 12, 9632–9641. [Google Scholar] [CrossRef]
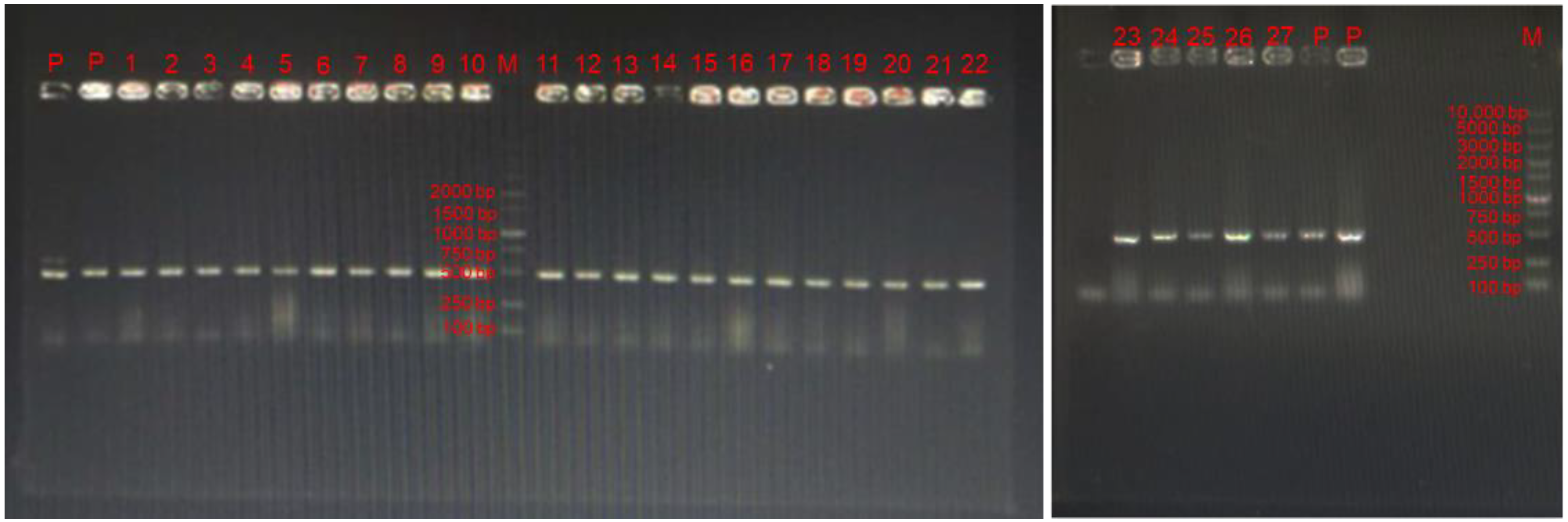

| Gene | Sequence (5′-3′) | Product Size (bp) | Accession Number | |
|---|---|---|---|---|
| β-Actin | Forward | ATGACCCAAGCCGAGAAGG | 185 | NM_027493 |
| Reverse | CGGCCAAGTCTTAGAGTTGTTG | |||
| Tnfα | Forward | CCACGCTCTTCTGTCTACTG | 169 | NM_010851.2 |
| Reverse | ACTTGGTGGTTTGCTACGAC | |||
| Il-10 | Forward | GGACCAGCTGGACAACATACTGCTA | 80 | NM_010548.2 |
| Reverse | CCGATAAGGCTTGGCAACCCAAGT | |||
| Il-6 | Forward | GAGTCACAGAAGGAGTGGCTAAGG | 106 | NM_031168.1 |
| Reverse | CGCACTAGGTTTGCCGAGTAGATCT | |||
| Il-1β | Forward | TTGAAAGTCCACCTCCTTACAGA | 129 | NM_008756 |
| Reverse | CCGGATAAAAAGAGTACGCTGG |
| Strains | Staphylococcus aureus ATCC 25923 | Salmonella enterica ATCC 14028 | Escherichia coli ATCC 25922 | Listeria monocytogenes ATCC 13932 | Enterococcus faecalis E27 |
|---|---|---|---|---|---|
| FRY-2 | 10.67 ± 0.5 | 14.3 ± 1 | 11.67 ± 0.5 | 12.7 ± 0.5 | 14.7 ± 1.53 |
| FXHB-6 | 11.33 ± 0 | 12.67 ± 1.1 | 10.3 ± 0.4 | 13.7 ± 0.5 | 15.7 ± 0.5 |
| FRY-3 | 8.7 ± 1.0 | 8.3 ± 0.5 | 10.7 ± 0.5 | 11.3 ± 0.5 | 15.3 ± 1.1 |
| FMM-1 | - | - | - | - | 11.3 ± 0.5 |
| FZL-21 | 11.7 ± 1.1 | 10.7 ± 0.5 | 12.3 ± 1.1 | 11 ± 1.0 | 17 ± 1.0 |
| Fjias-5 | 19.3 ± 1.0 | 14.33 ± 0.5 | 20.3 ± 0.5 | 19.67 ± 0.4 | 19.3 ± 1.1 |
| FWJL-4 | 14.67 ± 0.5 | 19.7 ± 1.1 | 19.33 ± 0.5 | 14.7 ± 0.5 | 21.7 ± 0.5 |
| FLY-17 | - | - | - | - | 10.7 ± 1.1 |
| FZL-2 | 16.67 ± 1.7 | 12.7 ± 1.1 | 15 ± 0 | 12.67 ± 0.5 | 15 ± 1.0 |
| FRY-6 | 15.3 ± 0.5 | 12.3 ± 1.1 | 11.67 ± 0.5 | 14.33 ± 0.5 | 18.7 ± 0.5 |
| FMM-7 | - | - | - | - | - |
| FSJ-1 | 12.3 ± 0.5 | 11.3 ± 0.5 | 10.6 ± 0.5 | 9.33 ± 0.5 | 17.3 ± 1.1 |
| FWJL-5 | 13.7 ± 0.58 | 17 ± 1.0 | 14.7 ± 0.5 | 13.6 ± 0.5 | 20.7 ± 1.1 |
| FHHY-7 | - | - | - | - | 15.3 ± 1.1 |
| FSJ-11 | 14.33 ± 1.0 | 15.7 ± 1.1 | 12.67 ± 0.5 | 18.3 ± 0.5 | 18.7 ± 1.5 |
| FSJ-13 | 18.7 ± 1.5 | 19.7 ± 0.5 | 18.3 ± 0.5 | 17.3 ± 1.1 | 20.7 ± 1.1 |
| FZSJ-7 | 11.3 ± 0.4 | 12.7 ± 0.5 | 14.33 ± 0.5 | 12.7 ± 0.5 | 18 ± 1.0 |
| FSJ-6 | - | - | - | 8.7 ± 1.1 | - |
| FXHB-2 | 12.67 ± 0.5 | 17 ± 1.0 | 19 ± 1.0 | 17.3 ± 1.15 | 18.67 ± 0.5 |
| FSJ-4X | 14.33 ± 0.5 | 17 ± 0.5 | 13.3 ± 0.5 | 17 ± 1.0 | 19 ± 1.0 |
| FLT6-11 | 8.67 ± 0.4 | - | - | 8.3 ± 0.5 | - |
| FXHB-2X | 9.3 ± 1.1 | 8.3 ± 0.5 | 16 ± 1.0 | 17.3 ± 1.1 | 15.3 ± 0.5 |
| FRY-4X | 9.7 ± 0.5 | 14.3 ± 1.15 | 13.7 ± 0.5 | 11 ± 1.0 | 20.7 ± 1.1 |
| FSJ-2 | 10.3 ± 0.4 | 9.7 ± 0.5 | 9 ± 1.0 | 8.7 ± 1.53 | 15.3 ± 0.5 |
| FHHY-1 | 16.3 ± 0.5 | 15 ± 1.0 | 12.7 ± 0.5 | 15.7 ± 0.5 | 20.67 ± 1.15 |
| FZYY-3 | 10.7 ± 0.5 | 9.67 ± 0.5 | 8.33 ± 0.5 | 10.7 ± 0.5 | 15.67 ± 0.5 |
| FQM-3X | - | - | - | - | - |
| Penicillin | 32.7 ± 0.5 | 30.3 ± 1.15 | 35.7 ± 0.5 | 33.3 ± 0.4 | 38.67 ± 0 |
| Strains | Name | a MIC (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| K | P | V | E | A | S | C | PO | ||
| FRY-2 | Lactobacillus gasseri | 16 | 4 | 4 | <2 | <2 | 2 | 2 | ≥1024 |
| FXHB-6 | Lactobacillus gasseri | 16 | 32 | 2 | <2 | <2 | 2 | 2 | ≥1024 |
| FRY-3 | Lactobacillus gasseri | 16 | 4 | 2 | <2 | 2 | 2 | 2 | 1024 |
| FZL-21 | Lactobacillus gasseri | 64 | 4 | 2 | <2 | 2 | 2 | 2 | ≥1024 |
| Fjias-5 | Lactiplantibacillus plantarum | 512 | <2 | 2 | <2 | <2 | 16 | 8 | ≥1024 |
| FRY-4X | Lactobacillus gasseri | <2 | 8 | 2 | <2 | <2 | 2 | <2 | ≥1024 |
| FWJL-4 | Lactobacillus gasseri | 64 | 8 | 2 | <2 | 8 | 16 | 2 | 1024 |
| FZL-2 | Lactobacillus gasseri | 64 | 16 | 4 | <2 | 64 | 2 | 2 | ≥1024 |
| FRY-6 | Lactobacillus gasseri | 64 | 16 | 8 | 8 | 4 | 8 | 2 | ≥1024 |
| FSJ-1 | Lacticaseibacillus rhamnosus | 512 | 512 | ≥1024 | 8 | 16 | 32 | 2 | ≥1024 |
| FWJL-5 | Lactobacillus gasseri | 64 | 32 | 2 | <2 | 8 | 2 | 2 | 1024 |
| FSJ-11 | Lacticaseibacillus rhamnosus | 512 | 2 | ≥1024 | 2 | <2 | 32 | <2 | ≥1024 |
| FSJ-13 | Lacticaseibacillus rhamnosus | ≥1024 | <2 | ≥1024 | 2 | 2 | 128 | <2 | ≥1024 |
| FZSJ-7 | Lactobacillus gasseri | 16 | 4 | ≥1024 | <2 | <2 | <2 | <2 | ≥1024 |
| FXHB-2 | Lactobacillus gasseri | 16 | 4 | ≥1024 | <2 | <2 | <2 | <2 | ≥1024 |
| FSJ-4X | Lacticaseibacillus rhamnosus | 32 | <2 | ≥1024 | 2 | <2 | 32 | <2 | ≥1024 |
| FXHB-2X | Lactobacillus gasseri | 128 | 4 | ≥1024 | <2 | 8 | 4 | <2 | ≥1024 |
| LGG | Lacticaseibacillus rhamnosus | 256 | 8 | ≥1024 | <2 | 32 | 64 | 8 | ≥1024 |
| Names | Sequencing Results | Identity | Accession Number |
|---|---|---|---|
| FRY-2 | Lactobacillus gasseri | 97.90% | MZ220367 |
| FXHB-6 | Lactobacillus gasseri | 99.79% | MZ220368 |
| Fjias-5 | Lactiplantibacillus plantarum | 99.93% | MZ220369 |
| FRY-4X | Lactobacillus gasseri | 99.79% | MZ220370 |
| FWJL-4 | Lactobacillus gasseri | 99.79% | MZ220371 |
| FZL-2 | Lactobacillus gasseri | 99.79% | MZ220372 |
| FRY-6 | Lactobacillus gasseri | 93.73% | MZ220373 |
| FSJ-13 | Lacticaseibacillus rhamnosus | 99.80% | MZ220374 |
| FZSJ-7 | Lactobacillus gasseri | 99.79% | MZ220375 |
| FXHB-2 | Lactobacillus gasseri | 99.86% | MZ220376 |
| FWJL-5 | Lactobacillus gasseri | 99.79% | MZ220377 |
| FSJ-4X | Lacticaseibacillus rhamnosus | 99.86% | MZ220378 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Pan, L.-L.; Sun, J. Novel Probiotic Lactic Acid Bacteria Were Identified from Healthy Infant Feces and Exhibited Anti-Inflammatory Capacities. Antioxidants 2022, 11, 1246. https://doi.org/10.3390/antiox11071246
Li B, Pan L-L, Sun J. Novel Probiotic Lactic Acid Bacteria Were Identified from Healthy Infant Feces and Exhibited Anti-Inflammatory Capacities. Antioxidants. 2022; 11(7):1246. https://doi.org/10.3390/antiox11071246
Chicago/Turabian StyleLi, Binbin, Li-Long Pan, and Jia Sun. 2022. "Novel Probiotic Lactic Acid Bacteria Were Identified from Healthy Infant Feces and Exhibited Anti-Inflammatory Capacities" Antioxidants 11, no. 7: 1246. https://doi.org/10.3390/antiox11071246
APA StyleLi, B., Pan, L.-L., & Sun, J. (2022). Novel Probiotic Lactic Acid Bacteria Were Identified from Healthy Infant Feces and Exhibited Anti-Inflammatory Capacities. Antioxidants, 11(7), 1246. https://doi.org/10.3390/antiox11071246







