Transcriptome Analysis of Glutathione Response: RNA-Seq Provides Insights into Balance between Antioxidant Response and Glucosinolate Metabolism
Abstract
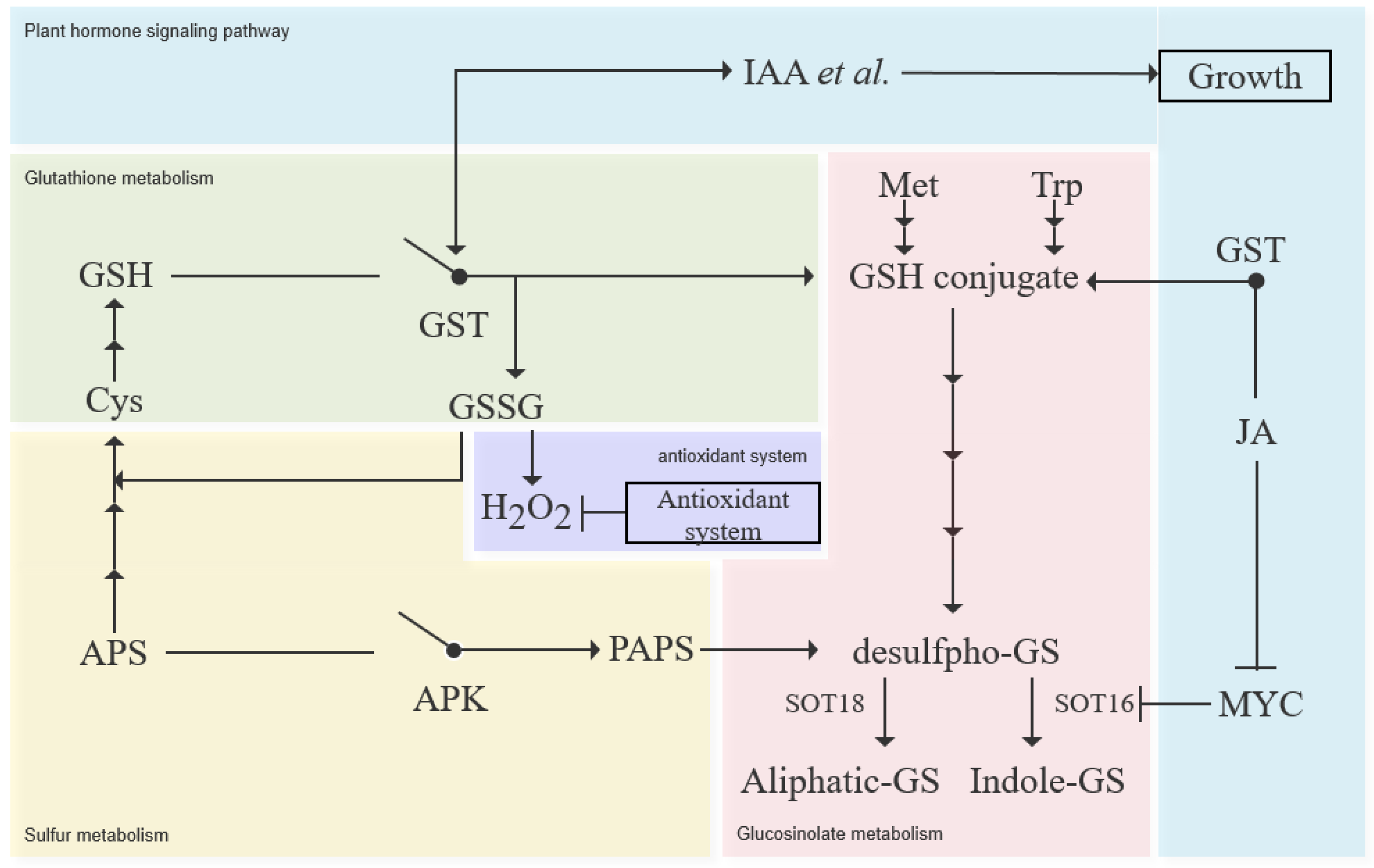
:1. Introduction
2. Materials and Methods
2.1. Growth Condition and Treatments
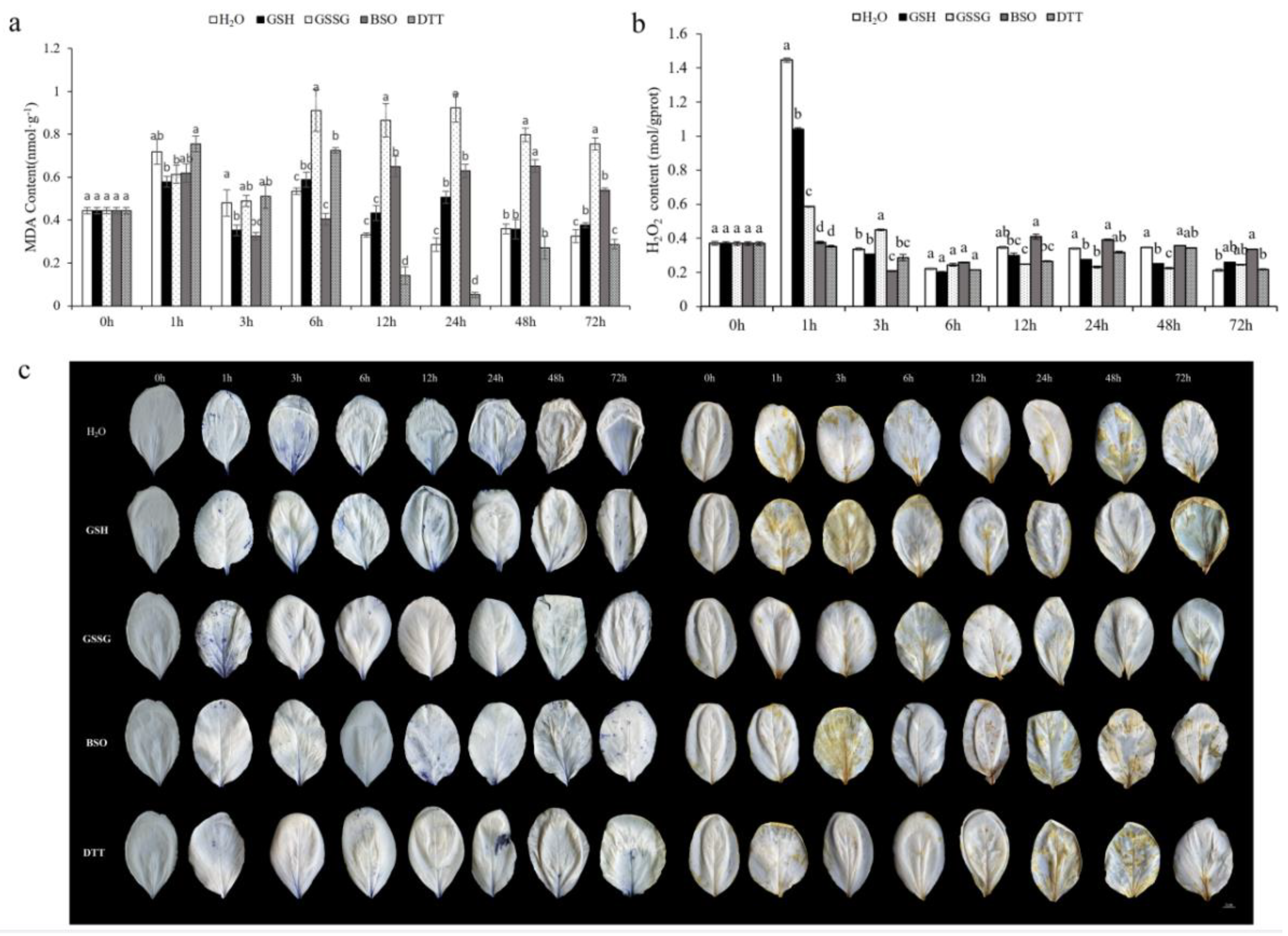
2.2. Measurement of Lipid Peroxidation
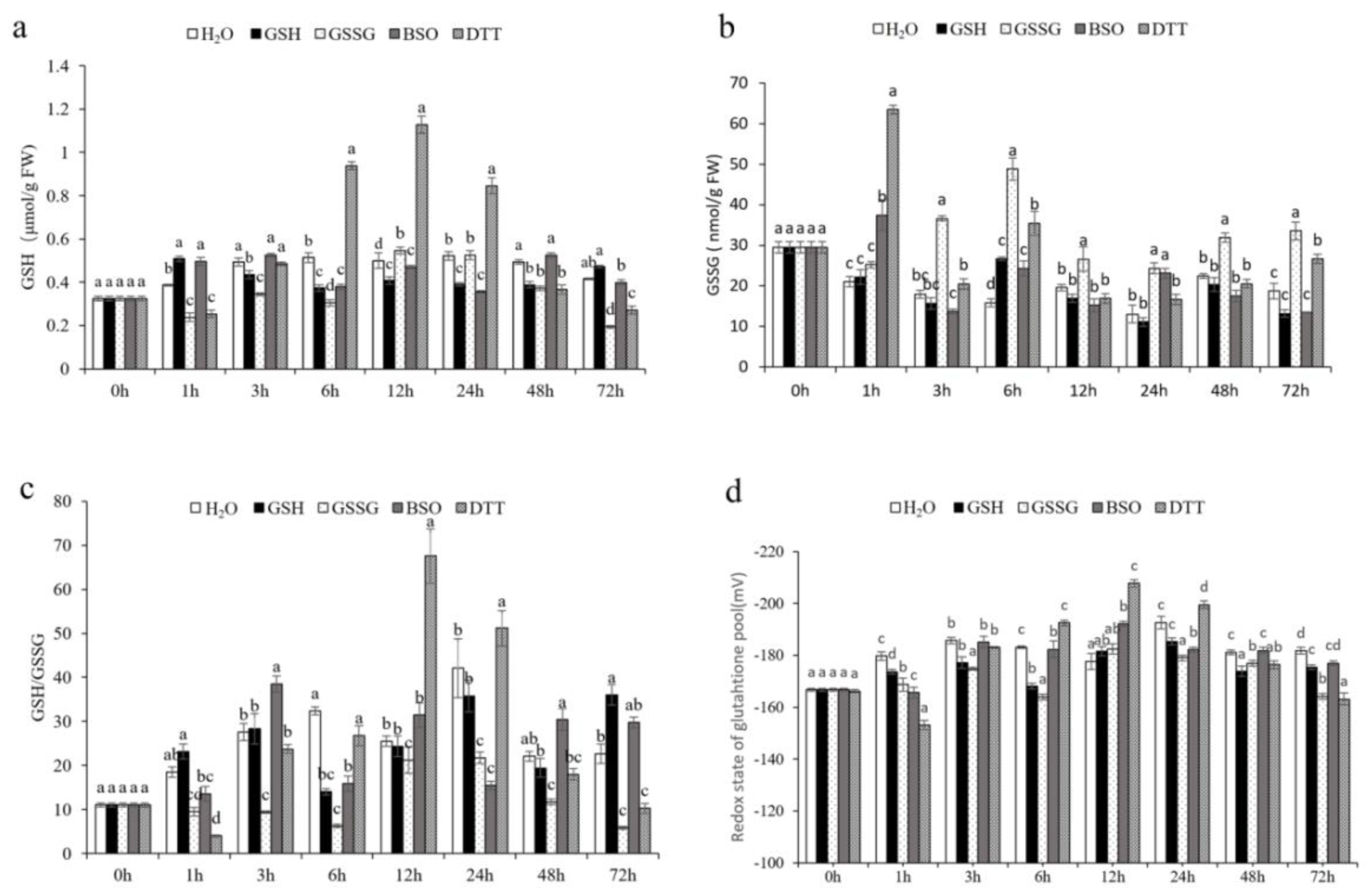
2.3. Glutathione Content Assay
2.4. Enzymatic Antioxidant Systems
2.5. Estimation of Reactive Oxygen Species (ROS)
2.6. RNA Isolation, cDNA Library Preparation, Transcriptome Sequencing, RNA Sequencing Data Analysis, and Quantitative Real-Time PCR Analysis
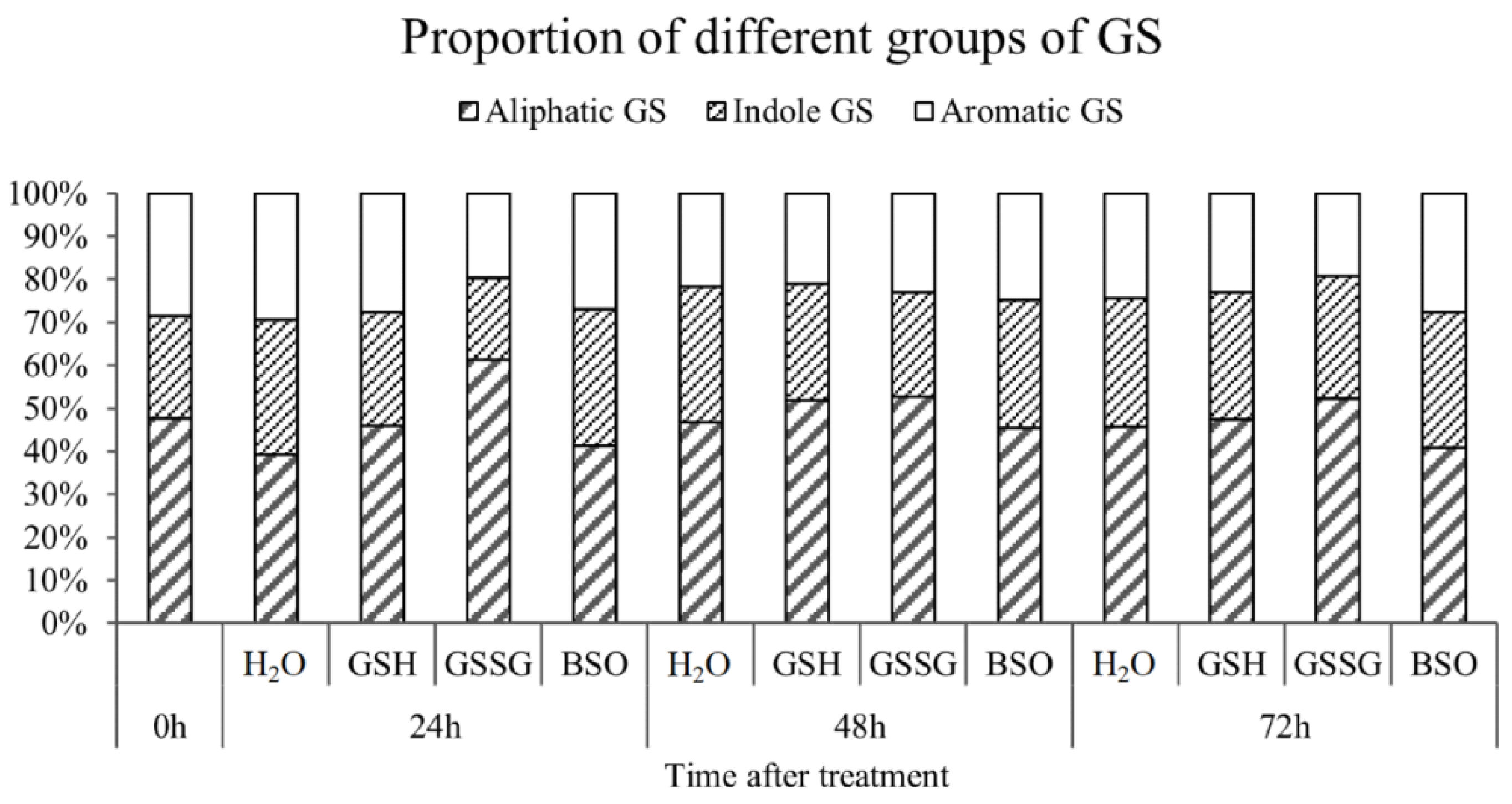
2.7. Measurement of Glucosinolates
2.8. Statistical Analysis
3. Results
3.1. Identification of Pakchoi Transcripts in Response to GSH
3.2. GSH Increased the Oxidative Resistance of Pakchoi
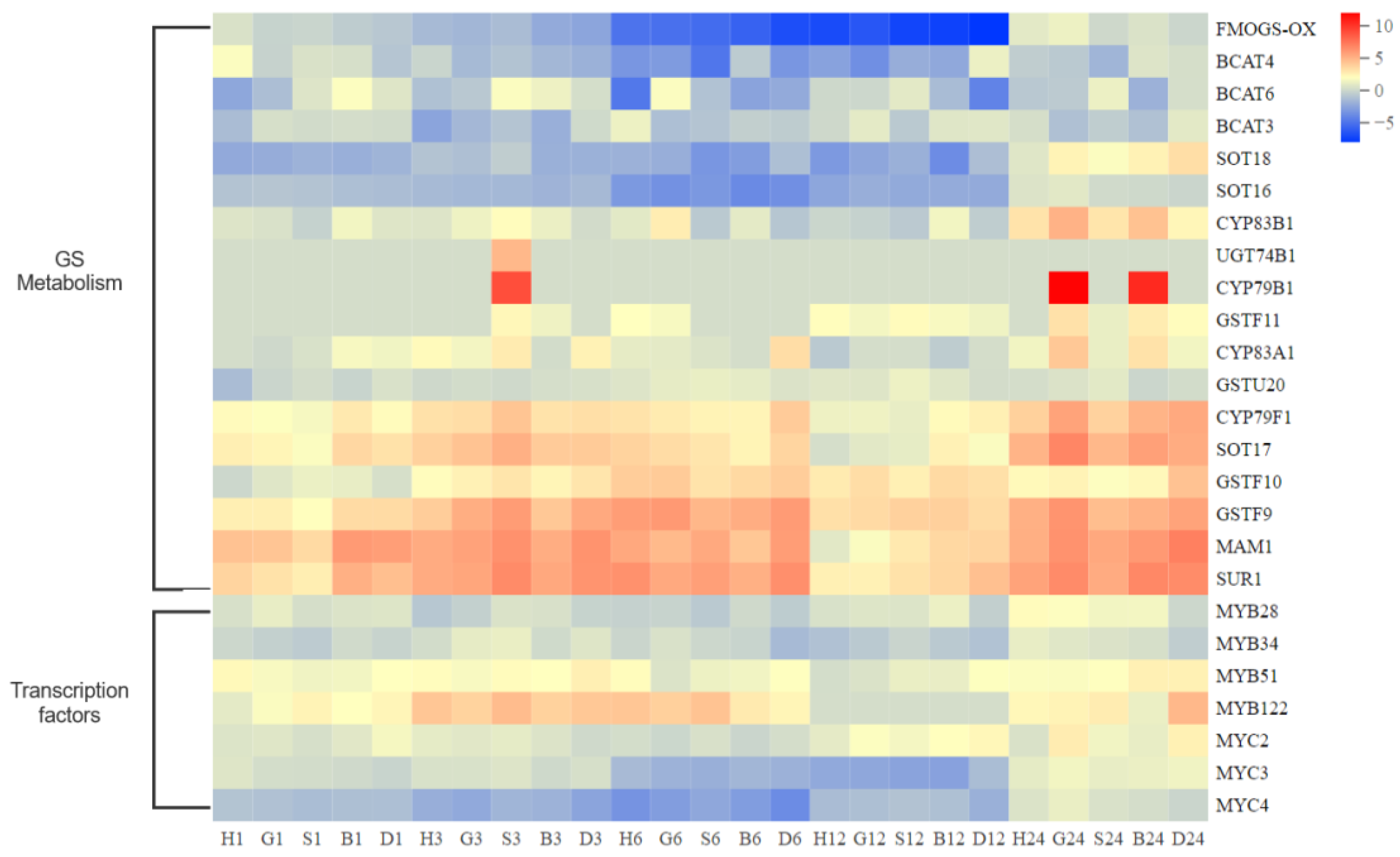
3.3. Coexpression Analysis Reveals GSH Involved in GS Metabolism and Antioxidant System in Pakchoi
3.4. GSH Increased the Resistance of Oxidative Stress in Pakchoi
3.4.1. Effect of GSH on Oxidative Signaling in Pakchoi Leaves
3.4.2. Effect of GSH on Oxidative Stress Markers of Pakchoi Leaves
3.4.3. Effect of GSH on Antioxidant Enzymes Activities of Pakchoi Leaves
3.4.4. Effect of GSH on Antioxidant System Genes of Pakchoi Leaves
3.5. GSH Inhibit the Secondary Metabolism during Oxidative Stress Response in Pakchoi
3.6. Effect of High Antioxidant Expression on GS Metabolic Pathway in Pakchoi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GSH | Glutathione |
| ROS | Reactive oxygen species |
| GS | Glucosinolate |
| AsA | Ascorbate |
| SOD | Superoxide dismutase |
| CAT | Catalase |
| APX | Ascorbate peroxidase |
| DHAR | Dehydroascorbate reductase |
| GR | Glutathione reductase |
| GPX | Glutathione peroxidase |
| GST | Glutathione S-transferase |
| POD | Peroxidases |
| GSSG | Glutathione disulfide |
| JA | Jasmonic acid |
| BSO | L-Buthionine-sulfoximine |
| DTT | Dithiothreitol |
| MDA | Malondialdehyde |
| H2O2 | Hydrogen peroxide |
| O2•− | Superoxide |
| PCA | Principal component analysis |
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| SA | Salicylic acid |
| ET | Ethylene |
| JA | Jasmonate |
| ABA | Abscisic acid |
| IAA | Indole-3-acetic acid |
References
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, Oxidative Damage and Oxygen Deprivation Stress: A Review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnocka, W.; Karpiński, S. Friend or Foe? Reactive Oxygen Species Production, Scavenging and Signaling in Plant Response to Environmental Stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, D.; Singh, S.; Kumar, V.; Romero, R.; Prasad, R.; Singh, J. Antioxidant Enzymes Regulation In Plants In Reference To Reactive Oxygen Species (ROS) And Reactive Nitrogen Species (RNS). Plant Gene 2019, 19, 100182. [Google Scholar] [CrossRef]
- Somayanda, I.; Nadaradjan, S.; Jagadish, K. Abiotic Stress Responses in Plants; Metabolism, Productivity and Sustainability; Springer: New York, NY, USA, 2012. [Google Scholar]
- Ma, Q.; Yue, L.-J.; Zhang, J.-L.; Wu, G.-Q.; Bao, A.-K.; Wang, S.-M. Sodium Chloride Improves Photosynthesis And Water Status in the Succulent Xerophyte Zygophyllum Xanthoxylum. Tree Physiol. 2011, 32, 4–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.-J. The Role of the Plant Antioxidant System in Drought Tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species And Antioxidant Defense in Plants Under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, C.; Chen, J.; Qiu, J.; Huang, Z.; Wang, Q.; Ye, Y. Cold Acclimation Improves Photosynthesis by Regulating the Ascorbate–Glutathione Cycle in Chloroplasts Of Kandelia obovata. J. For. Res. 2019, 30, 755–765. [Google Scholar] [CrossRef]
- Wu, X.; Chen, J.; Yue, X.; Wei, X.; Zou, J.; Chen, Y.; Su, N.; Cui, J. The Zinc-Regulated Protein (ZIP) Family Genes and Glutathione S-Transferase (GST) Family Genes Play Roles in Cd Resistance And Accumulation Of Pak Choi (Brassica Campestris Ssp. Chinensis). Ecotoxicol. Environ. Saf. 2019, 183, 109571. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant Glutathione Peroxidases: Emerging Role of the Antioxidant Enzymes in Plant Development And Stress Responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and Glutathione Reductase: A Boon in Disguise for Plant Abiotic Stress Defense Operations. Plant Physiol. Biochem. PPB 2013, 70, 204–212. [Google Scholar] [CrossRef]
- Rao, A.S.V.C.; Reddy, A.R. Glutathione Reductase: A Putative Redox Regulatory System in Plant Cells. In Sulfur Assimilation and Abiotic Stress in Plants; Khan, N.A., Singh, S., Umar, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 111–147. [Google Scholar]
- Ding, H.; Wang, B.; Han, Y.; Li, S. The Pivotal Function of Dehydroascorbate Reductase in Glutathione Homeostasis in Plants. J. Exp. Bot. 2020, 71, 3405–3416. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Cummins, I.; Cole, D.J.; Edwards, R. Glutathione-Mediated Detoxification Systems in Plants. Curr. Opin. Plant Biol. 1998, 1, 258–266. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants Under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in Plants: Biosynthesis and Physiological Role in Environmental Stress Tolerance. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Aslam, S.; Gul, N.; Mir, M.A.; Asgher, M.; Al-Sulami, N.; Abulfaraj, A.A.; Qari, S. Role Of Jasmonates, Calcium, And Glutathione In Plants To Combat Abiotic Stresses Through Precise Signaling Cascade. Front. Plant Sci. 2021, 12, 1172. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role In Combating Abiotic Stresses Including Arsenic Detoxification In Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Li, J.W.; Wang, J.H.; Yang, W.T.; Yang, Y.X. Identification And Characterization of the Glutathione Peroxidase (GPX) Gene Family in Watermelon and Its Expression under Various Abiotic Stresses. Agronomy 2018, 8, 206. [Google Scholar] [CrossRef] [Green Version]
- Dutta, R. Glutathione as a Crucial Modulator of Phytohormone Signalling During Pathogen Defence in Plants. Proc. Indian Natl. Sci. Acad. 2018, 84, 581–597. [Google Scholar] [CrossRef]
- Schnaubelt, D. The Roles of Glutathione in the Control of Plant Growth, Development and Signalling in Arabidopsis thaliana. Ph.D. Thesis, University of Leeds, Leeds, UK, 2013. [Google Scholar]
- Ghanta, S.; Bhattacharyya, D.; Chattopadhyay, S. Glutathione Signaling Acts Through NPR1-Dependent SA-Mediated Pathway To Mitigate Biotic Stress. Plant Signal. Behav. 2011, 6, 607–609. [Google Scholar] [CrossRef] [Green Version]
- Gomez, L.D.; Noctor, G.; Knight, M.R.; Foyer, C.H. Regulation Of Calcium Signalling and Gene Expression By Glutathione. J. Exp. Bot. 2004, 55, 1851–1859. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.-Q.; Lin, H.-X. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, X.; Zhang, X.; Zhang, L.; Zhang, Z.; Zhang, Y.; Yang, Q. Linking Environmental Signals to Plant Metabolism: The Combination of Field Trials And Environment Simulators. Mol. Plant 2021, 15, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Camara-Martos, F.; Obregon-Cano, S.; Mesa-Plata, O.; Cartea-Gonzalez, M.E.; De Haro-Bailon, A. Quantification and In Vitro Bioaccessibility of Glucosinolates And Trace Elements in Brassicaceae Leafy Vegetables. Food Chem. 2021, 339, 127860. [Google Scholar] [CrossRef] [PubMed]
- Bohinc, T.; Ban, G.; Ban, D.; Trdan, S. Glucosinolates In Plant Protection Strategies: A Review. Arch. Biol. Sci. 2012, 64, 821–828. [Google Scholar] [CrossRef]
- Burow, M.; Halkier, B.A. How Does A Plant Orchestrate Defense in Time and Space? Using Glucosinolates in Arabidopsis as Case Study. Curr. Opin. Plant Biol. 2017, 38, 142–147. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foryer, C.H. Glutathione In Plants: An Integrated Overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Mitreiter, S.; Gigolashvili, T. Regulation Of Glucosinolate Biosynthesis. J. Exp. Bot. 2020, 72, 70–91. [Google Scholar] [CrossRef]
- Ji, Y.; Ren, Y.; Han, C.; Zhu, W.; Gu, J.; He, J. Application Of Exogenous Glycinebetaine Alleviates Lead Toxicity In Pakchoi (Brassica chinensis L.) By Promoting Antioxidant Enzymes And Suppressing Pb Accumulation. Environ. Sci. Pollut. Res. 2022, 29, 25568–25580. [Google Scholar] [CrossRef]
- Zhang, X.T.; Zhang, M.; Devahastin, S.; Guo, Z.M. Effect Of Combined Ultrasonication And Modified Atmosphere Packaging On Storage Quality Of Pakchoi (Brassica chinensis L.). Food Bioprocess Technol. 2019, 12, 1573–1583. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf Senescence: Correlated With Increased Levels Of Membrane Permeability And Lipid Peroxidation, And Decreased Levels Of Superoxide Dismutase And Catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence In Higher Plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Hemeda, H.; Klein, B. Effects Of Naturally Occurring Antioxidants On Peroxidase Activity Of Vegetable Extracts. J. Food Sci. 2006, 55, 184–185. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Alam, M.M.; Nahar, K.; Mohsin, S.M.; Bhuyan, M.H.M.B.; Parvin, K.; Hawrylak-Nowak, B.; Fujita, M. Silicon-Induced Antioxidant Defense And Methylglyoxal Detoxification Works Coordinately in Alleviating Nickel Toxicity in Oryza sativa L. Ecotoxicology 2019, 28, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen Peroxide Is Scavenged By Ascorbate-Specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Foyer, C.H.; Halliwell, B. The Presence Of Glutathione And Glutathione Reductase in Chloroplasts: A Proposed Role in Ascorbic Acid Metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef]
- Rao, S.-Q.; Chen, X.-Q.; Wang, K.-H.; Zhu, Z.-J.; Yang, J.; Zhu, B. Effect Of Short-Term High Temperature On The Accumulation Of Glucosinolates In Brassica Rapa. Plant Physiol. Biochem. PPB 2021, 161, 222–233. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses In Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [Green Version]
- Nahar, K.; Hasanuzzaman, M.; Alam, M.M.; Fujita, M. Roles Of Exogenous Glutathione In Antioxidant Defense System And Methylglyoxal Detoxification During Salt Stress In Mung Bean. Biol. Plant. 2015, 59, 745–756. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, And Signaling Mechanisms Of Malondialdehyde And 4-Hydroxy-2-Nonenal. Oxid. Med. Cell Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species In Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [Green Version]
- Locato, V.; Cimini, S.; De Gara, L. Glutathione As A Key Player In Plant Abiotic Stress Responses And Tolerance. In Glutathione in Plant Growth, Development, and Stress Tolerance; Hossain, M.A., Mostofa, M.G., Diaz Vivancos, P., Burritt, D.J., Fujita, M., Tran, L.S.P., Eds.; Springer: Cham, Switzerland, 2017; pp. 127–145. [Google Scholar]
- Halkier, B.A. Chapter One—General Introduction To Glucosinolates. In Advances in Botanical Research; Kopriva, S., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 80, pp. 1–14. [Google Scholar]
- Kwon, E.; Feechan, A.; Yun, B.-W.; Hwang, B.-H.; Pallas, J.A.; Kang, J.-G.; Loake, G.J. Atgsnor1 Function Is Required For Multiple Developmental Programs In Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Blum, R.; Beck, A.; Korte, A.; Stengel, A.; Letzel, T.; Lendzian, K.; Grill, E. Function Of Phytochelatin Synthase In Catabolism Of Glutathione-Conjugates. Plant J. 2007, 49, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.C.; Sun, S.H.; Ke, Y.T.; Xie, C.P.; Chen, F.X. Effects of Glutathione on Chloroplast Membrane Fluidity and the Glutathione Circulation System in Young Loquat Fruits Under Low Temperature Stress. In Proceedings of the III International Symposium on Loquat, Antakya-Hatay, Turkey, 3–6 May 2010; International Society For Horticultural Science (ISHS): Leuven, Belgium, 2011; pp. 221–225. [Google Scholar]
- Chung, W.H. Unraveling New Functions Of Superoxide Dismutase Using Yeast Model System: Beyond Its Conventional Role In Superoxide Radical Scavenging. J. Microbiol. 2017, 55, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Lukesh, J.; Palte, M.; Raines, R. A Potent, Versatile Disulfide-Reducing Agent From Aspartic Acid. J. Am. Chem. Soc. 2012, 134, 4057–4059. [Google Scholar] [CrossRef]
- Blazevic, I.; Montaut, S.; Burcul, F.; Olsen, C.E.; Burow, M.; Rollin, P.; Agerbirk, N. Glucosinolate Structural Diversity, Identification, Chemical Synthesis And Metabolism In Plants. Phytochemistry 2020, 169, 112100. [Google Scholar] [CrossRef]
- Harun, S.; Abdullah-Zawawi, M.R.; Goh, H.H.; Mohamed-Hussein, Z.A. A Comprehensive Gene Inventory For Glucosinolate Biosynthetic Pathway In Arabidopsis Thaliana. J. Agric. Food Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef]
- Sanchez-Pujante, P.J.; Borja-Martinez, M.; Pedreno, M.A.; Almagro, L. Biosynthesis And Bioactivity Of Glucosinolates And Their Production In Plant In Vitro Cultures. Planta 2017, 246, 19–32. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.H. Jasmonic Acid Signaling Pathway In Response To Abiotic Stresses In Plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [Green Version]
- Zang, Y.X.; Ge, J.L.; Huang, L.H.; Gao, F.; Lv, X.S.; Zheng, W.W.; Hong, S.B.; Zhu, Z.J. Leaf And Root Glucosinolate Profiles Of Chinese Cabbage (Brassica Rapa Ssp. Pekinensis) As A Systemic Response To Methyl Jasmonate And Salicylic Acid Elicitation. J. Zhejiang Univ. Sci. B 2015, 16, 696–708. [Google Scholar] [CrossRef] [Green Version]
- Bakhtiari, M.; Glauser, G.; Rasmann, S. Root JA Induction Modifies Glucosinolate Profiles And Increases Subsequent Aboveground Resistance To Herbivore Attack In Cardamine Hirsuta. Front. Plant. Sci. 2018, 9, 1230. [Google Scholar] [CrossRef]
- Guo, J.; Pang, Q.; Wang, L.; Yu, P.; Li, N.; Yan, X. Proteomic Identification Of MYC2-Dependent Jasmonate-Regulated Proteins In Arabidopsis Thaliana. Proteome Sci. 2012, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, R.; Huang, Z.; Deng, Y.; Chen, X.; Xuhan, X.; Lai, Z. Comparative Transcriptome Analyses Reveal A Special Glucosinolate Metabolism Mechanism In Brassica Alboglabra Sprouts. Front. Plant. Sci 2016, 7, 1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-Transferase: A Versatile Protein Family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Callaway, E.C. High Cellular Accumulation Of Sulphoraphane, A Dietary Anticarcinogen, Is Followed By Rapid Transporter-Mediated Export As A Glutathione Conjugate. Biochem. J. 2002, 364 Pt 1, 301–307. [Google Scholar] [CrossRef] [Green Version]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General Detoxification And Stress Responses Are Mediated By Oxidized Lipids Through TGA Transcription Factors In Arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef] [Green Version]
- Skopelitou, K.; Muleta, A.W.; Papageorgiou, A.C.; Chronopoulou, E.; Labrou, N.E. Catalytic Features And Crystal Structure Of A Tau Class Glutathione Transferase From Glycine Max Specifically Upregulated In Response To Soybean Mosaic Virus Infections. Biochim. Biophys. Acta 2015, 1854, 166–177. [Google Scholar] [CrossRef] [Green Version]
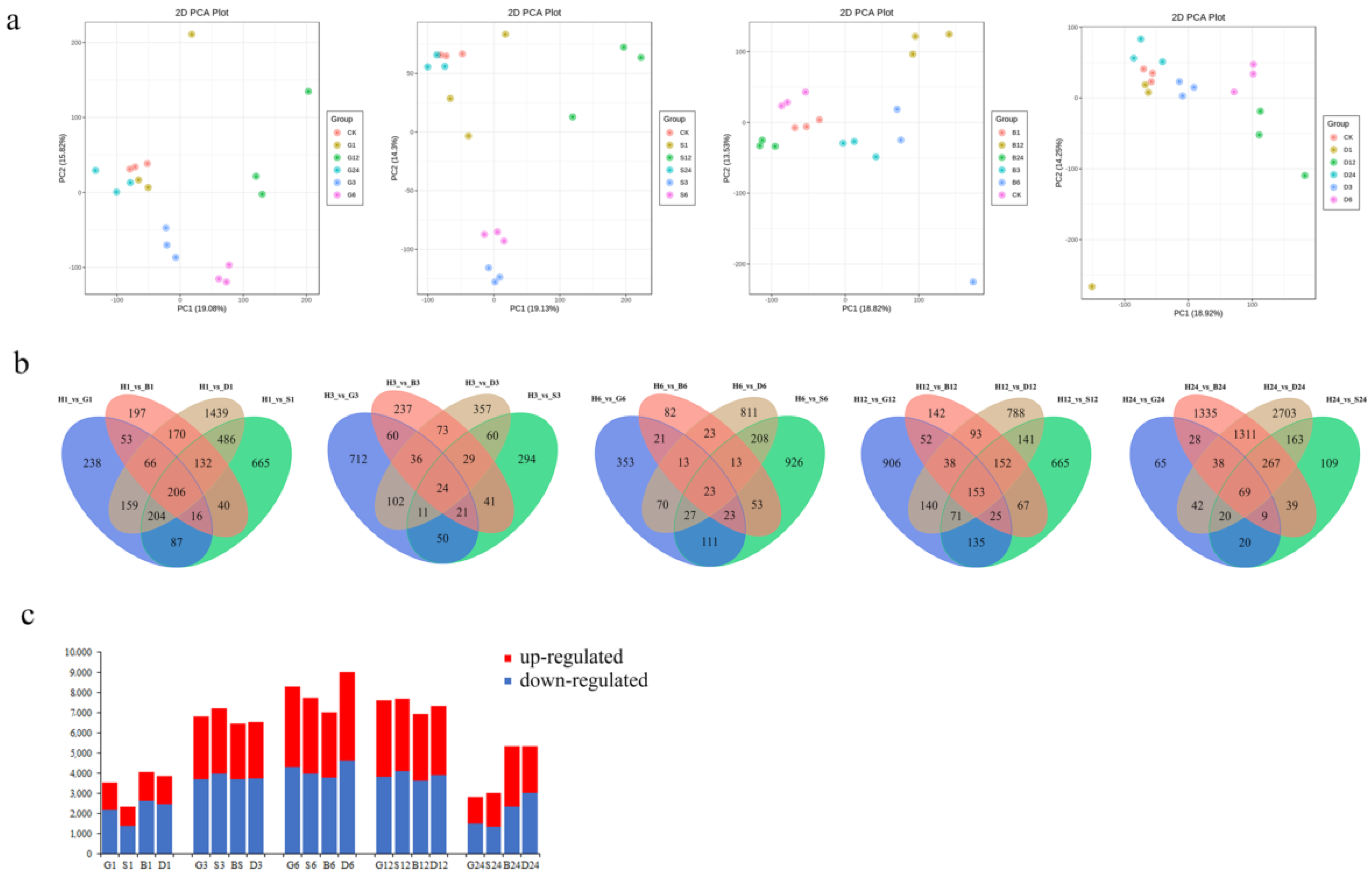
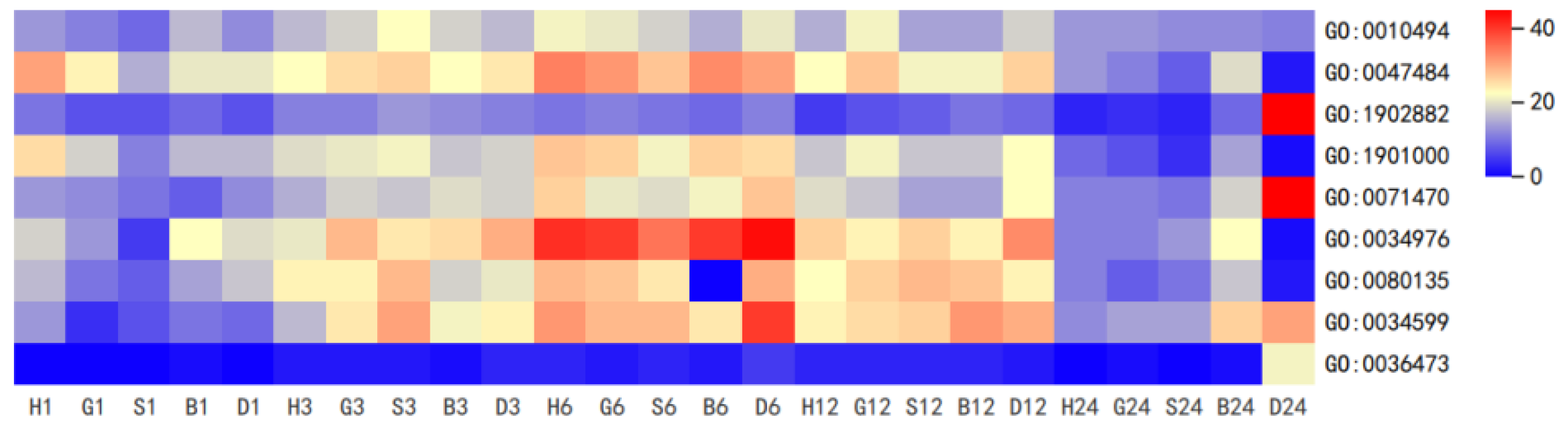
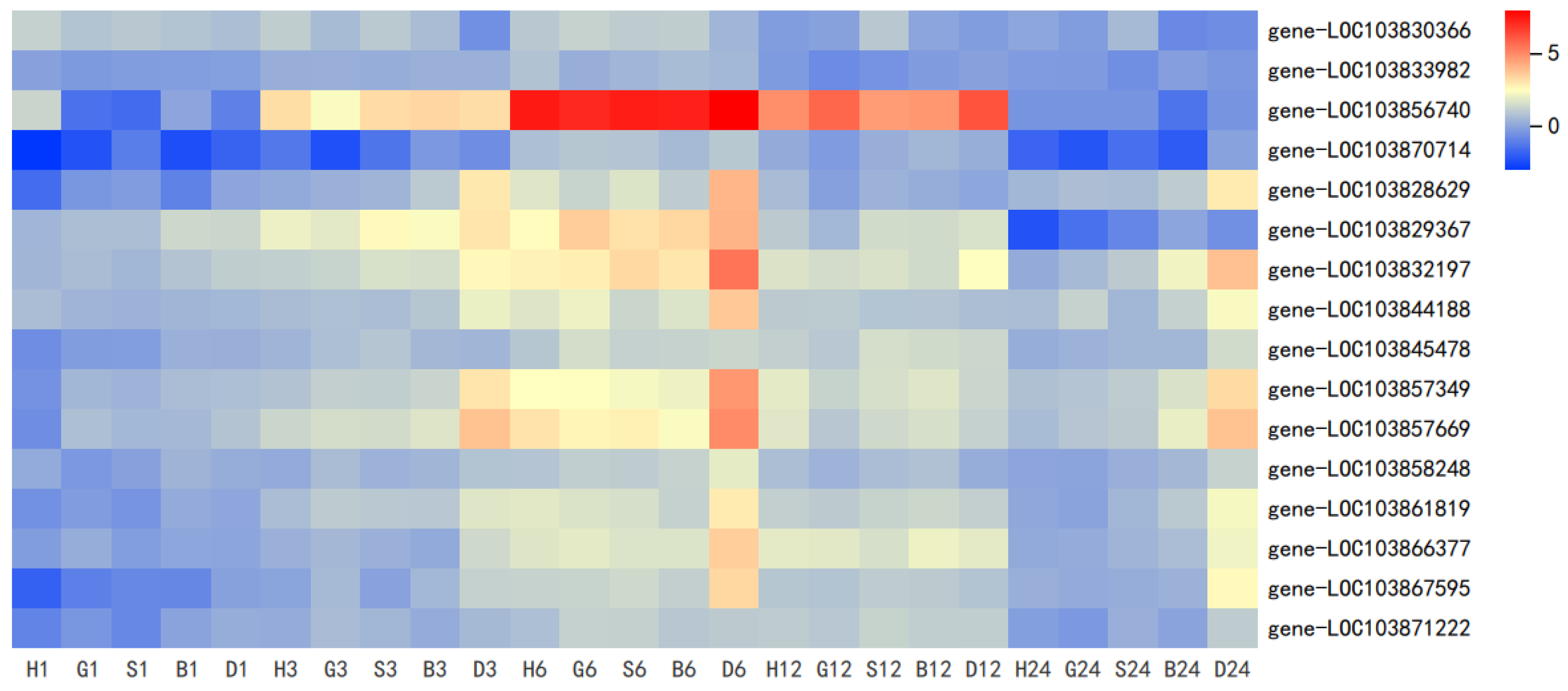
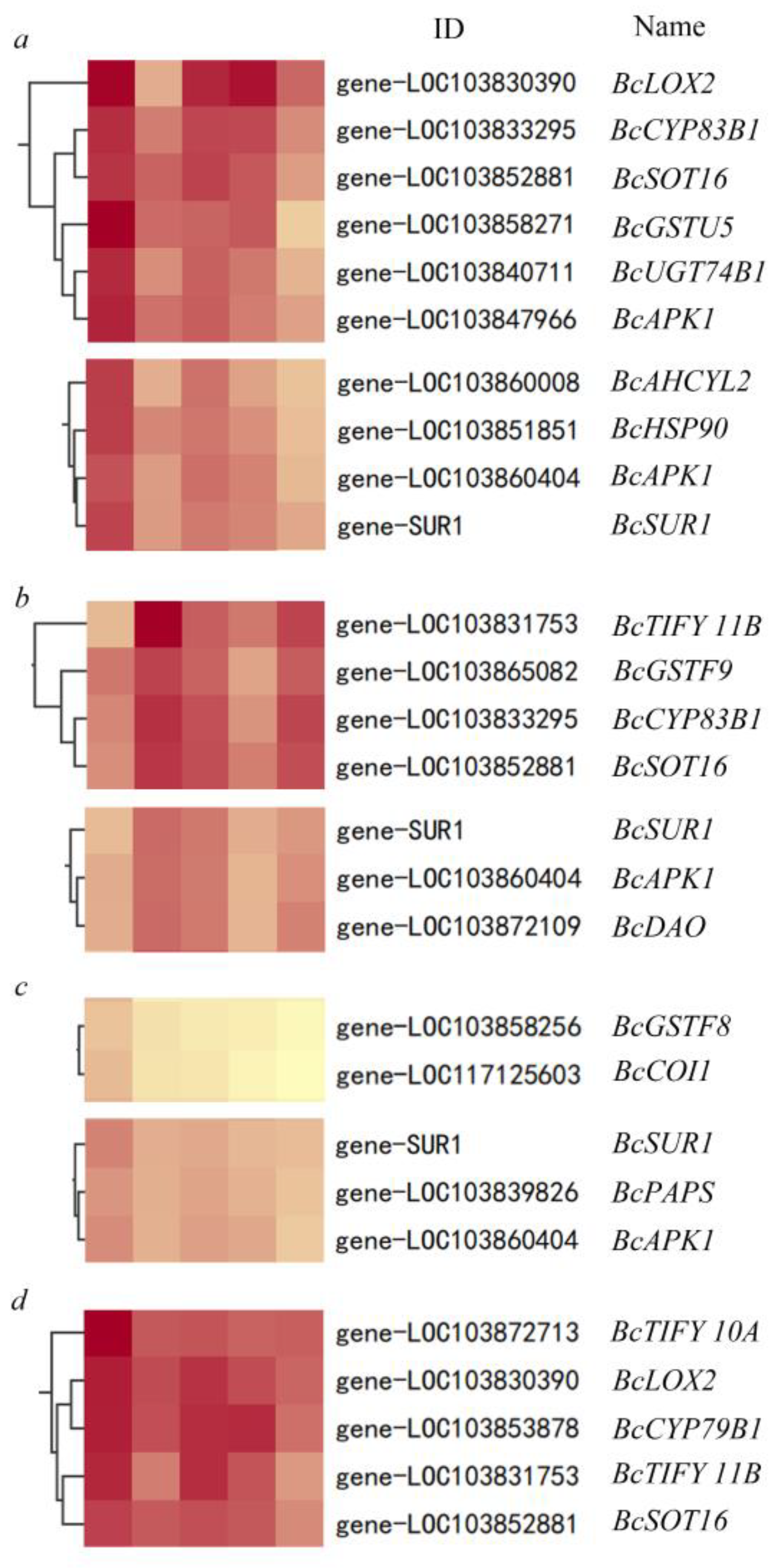




| Gene Name | Treatment | log2FoldChange (Relative to 0 h) | ||||
|---|---|---|---|---|---|---|
| 1 h | 3 h | 6 h | 12 h | 24 h | ||
| CXXS1 | H2O | 1.245 | −0.41 | 2.155 | 0.728 | 0.227 |
| GSH | 0.467 | 2.380 | 2.180 | 1.362 | 0.158 | |
| GSSG | 1.191 | 1.995 | 2.249 | 1.156 | 0.463 | |
| BSO | 0.963 | 1.127 | 2.733 | 1.031 | 0.620 | |
| DTT | 1.576 | 2.301 | 2.503 | 1.086 | 0.605 | |
| CAT1 | H2O | 0.702 | 0.599 | 0.933 | −1.635 | −0.912 |
| GSH | −0.785 | 0.975 | 1.027 | −0.259 | 1.228 | |
| GSSG | 0.237 | 0.654 | 0.052 | −0.191 | 1.645 | |
| BSO | 0.096 | 0.338 | 1.448 | −0.016 | 1.845 | |
| DTT | 0.584 | 0.492 | 1.027 | 0.828 | 1.759 | |
| RbohD | H2O | 3.632 | 2.442 | 2.230 | −0.81 | 1.640 |
| GSH | 3.018 | 1.736 | 2.226 | −1.32 | 1.983 | |
| GSSG | 2.873 | 2.918 | 0.231 | −0.35 | 1.580 | |
| BSO | 3.705 | 3.304 | 2.198 | 0.095 | 2.692 | |
| DTT | 2.908 | 2.530 | 2.508 | 0.852 | 2.665 | |
| CSD1 | H2O | −0.94 | −0.85 | −1.99 | −0.61 | 0.324 |
| GSH | −0.71 | 1.162 | 0.659 | −1.31 | 1.667 | |
| GSSG | 0.548 | −1.68 | 1.047 | 0.494 | 0.631 | |
| BSO | −0.81 | −1.26 | −1.54 | 0.791 | −0.29 | |
| DTT | 0.897 | −0.35 | −1.38 | −2.27 | 0.990 | |
| FSD3 | H2O | −1.60 | −0.05 | 1.351 | 1.671 | −0.36 |
| GSH | −0.63 | −1.68 | 1.191 | 1.200 | 0.156 | |
| GSSG | 0.112 | −0.98 | 2.273 | 2.016 | 1.559 | |
| BSO | −0.12 | 0.378 | 0.689 | 1.849 | −0.58 | |
| DTT | 0.513 | 0.061 | 0.903 | 0.040 | 1.526 | |
| GPX6 | H2O | 1.051 | 1.291 | 1.559 | −0.28 | 0.562 |
| GSH | 0.895 | 1.430 | 1.589 | −0.97 | 1.112 | |
| GSSG | 0.753 | 1.916 | 0.802 | −0.51 | 0.963 | |
| BSO | 0.774 | 1.500 | 1.654 | −0.62 | 1.873 | |
| DTT | 0.890 | 1.430 | 1.269 | 0.336 | 1.779 | |
| DHAR1 | H2O | −0.24 | −0.11 | 0.297 | 0.321 | 2.152 |
| GSH | −0.06 | 0.583 | 0.603 | −0.12 | 1.874 | |
| GSSG | −0.39 | 0.149 | 0.050 | −0.40 | 1.711 | |
| BSO | 0.095 | −0.01 | 0.276 | −0.79 | 2.278 | |
| DTT | 0.890 | 1.430 | 1.269 | 0.336 | 1.779 | |
| GR | H2O | −0.30 | 0.344 | 0.832 | 0.437 | 0.032 |
| GSH | 0.467 | 1.721 | 1.202 | 0.515 | 0.669 | |
| GSSG | 0.343 | 1.263 | 1.620 | 0.245 | −0.10 | |
| BSO | 1.018 | 0.183 | 1.031 | 0.093 | 0.693 | |
| DTT | 0.416 | 0.059 | 1.214 | 0.673 | 0.465 | |
| APX1 | H2O | 0.167 | −0.16 | 0.995 | 0.277 | 1.729 |
| GSH | 0.486 | 0.460 | 1.011 | 0.297 | 2.586 | |
| GSSG | 0.484 | 0.630 | 1.441 | 0.762 | 2.398 | |
| BSO | 0.127 | 0.636 | 1.189 | 0.803 | 3.094 | |
| DTT | 0.661 | 1.054 | 2.085 | 0.689 | 3.178 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Wang, K.; Liang, Z.; Zhu, Z.; Yang, J. Transcriptome Analysis of Glutathione Response: RNA-Seq Provides Insights into Balance between Antioxidant Response and Glucosinolate Metabolism. Antioxidants 2022, 11, 1322. https://doi.org/10.3390/antiox11071322
Zhu B, Wang K, Liang Z, Zhu Z, Yang J. Transcriptome Analysis of Glutathione Response: RNA-Seq Provides Insights into Balance between Antioxidant Response and Glucosinolate Metabolism. Antioxidants. 2022; 11(7):1322. https://doi.org/10.3390/antiox11071322
Chicago/Turabian StyleZhu, Biao, Kuanhong Wang, Zhile Liang, Zhujun Zhu, and Jing Yang. 2022. "Transcriptome Analysis of Glutathione Response: RNA-Seq Provides Insights into Balance between Antioxidant Response and Glucosinolate Metabolism" Antioxidants 11, no. 7: 1322. https://doi.org/10.3390/antiox11071322







