Evaluation of Proanthocyanidins from Kiwi Leaves (Actinidia chinensis) against Caco-2 Cells Oxidative Stress through Nrf2-ARE Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Extraction and Purificantion of PAs from Kiwifruit Leaves
2.3. Reversed-Phase HPLC-QTOF-MS/MS Analysis of PKLPs
2.4. Cell Culture and Treatment
2.5. Injured Cell Model Induced by H2O2
2.6. Intracellular Antioxidant Activity Assay
2.7. Determination of Reactive Oxygen Species (ROS)
2.8. Determination of Oxidative Stress Indices
2.9. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
2.10. Immunofluorescence
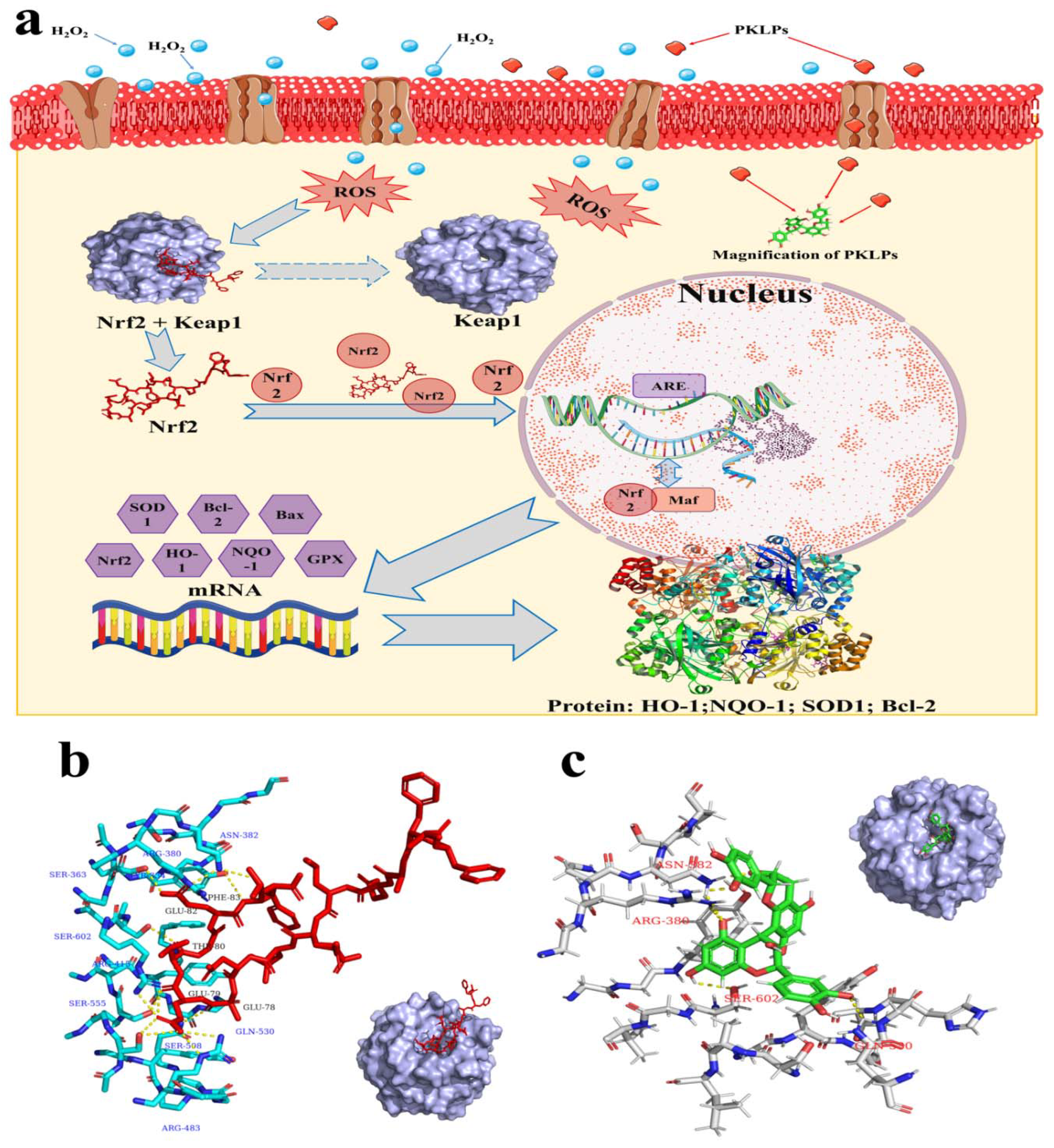
2.11. Molecular Docking
2.12. Western Blot
2.13. Statistical Analysis
3. Results and Discussion
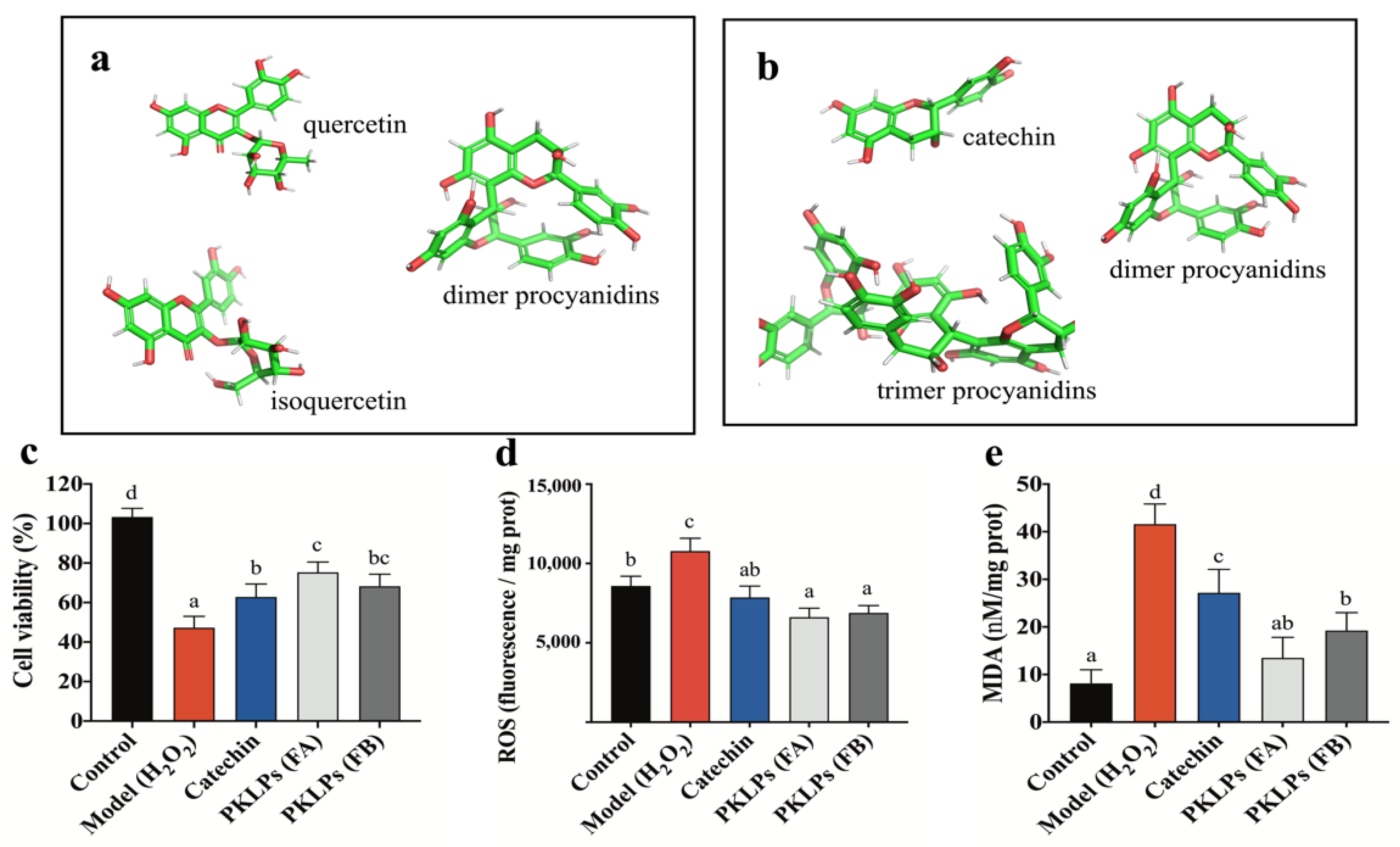
3.1. Comparation of the Chemical Composition of PKLPs (FA and FB) by Reversed-Phase HPLC-QTOF-MS/MS
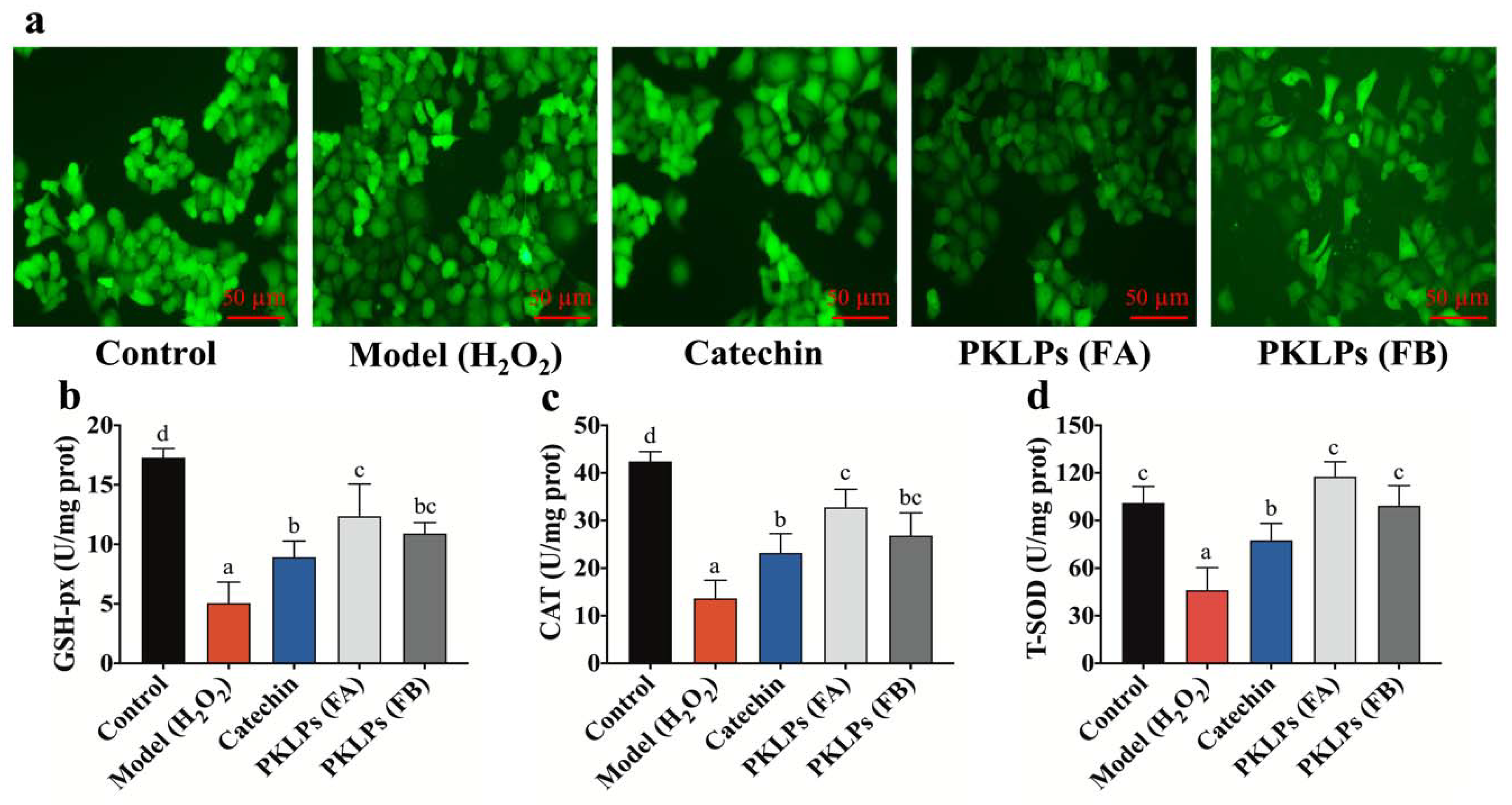
3.2. PKLPs (FA and FB) Suppressed H2O2-Induced Oxidative Stress in Caco-2 Cells
3.3. Protective Effect of PKLPs (FA and FB) in H2O2-Induced Caco-2 Cells
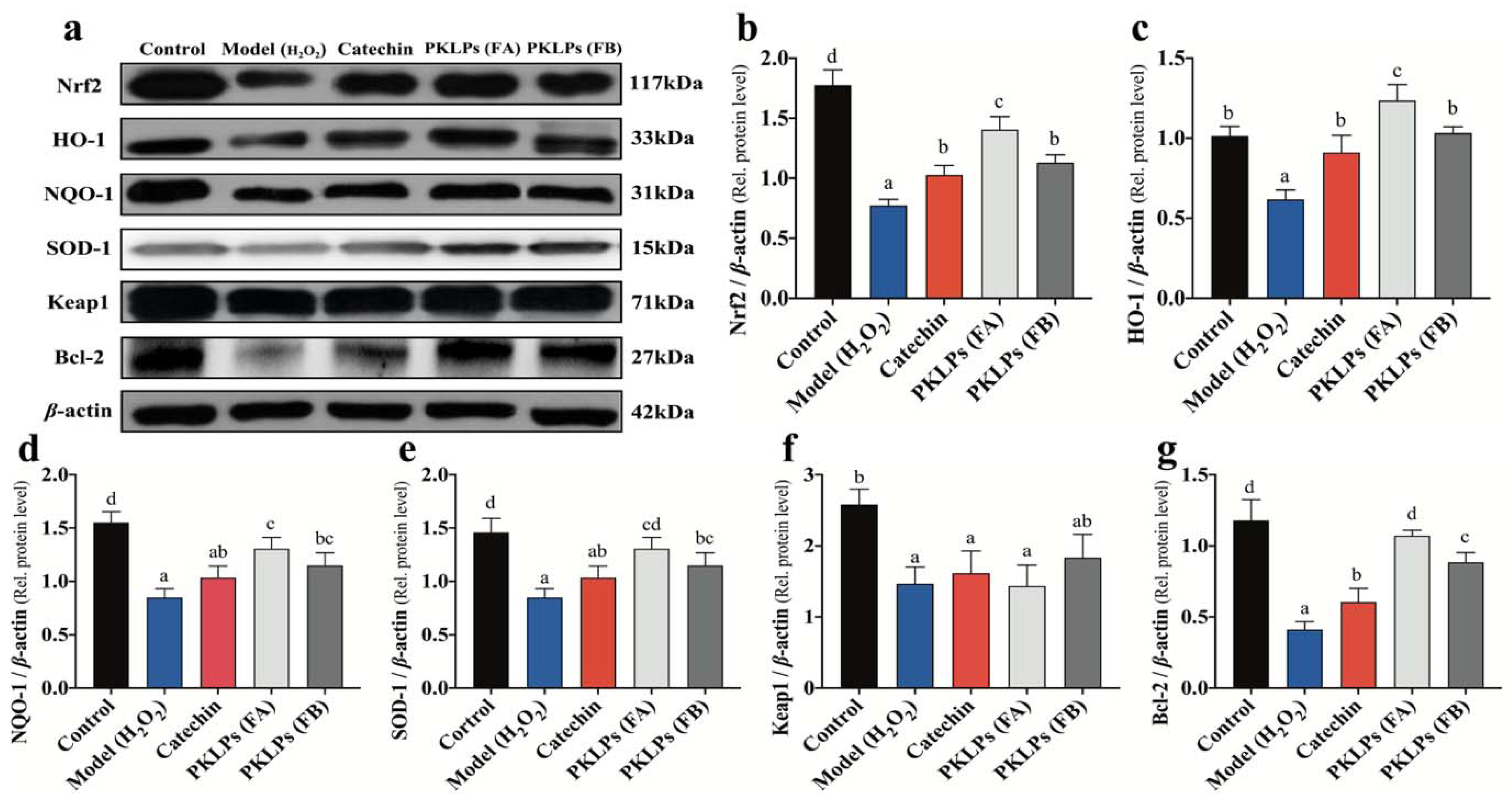
3.4. Effect ofPKLPs (FA and FB) on the Nrf2 and Its Downstream Target Genes Transcription
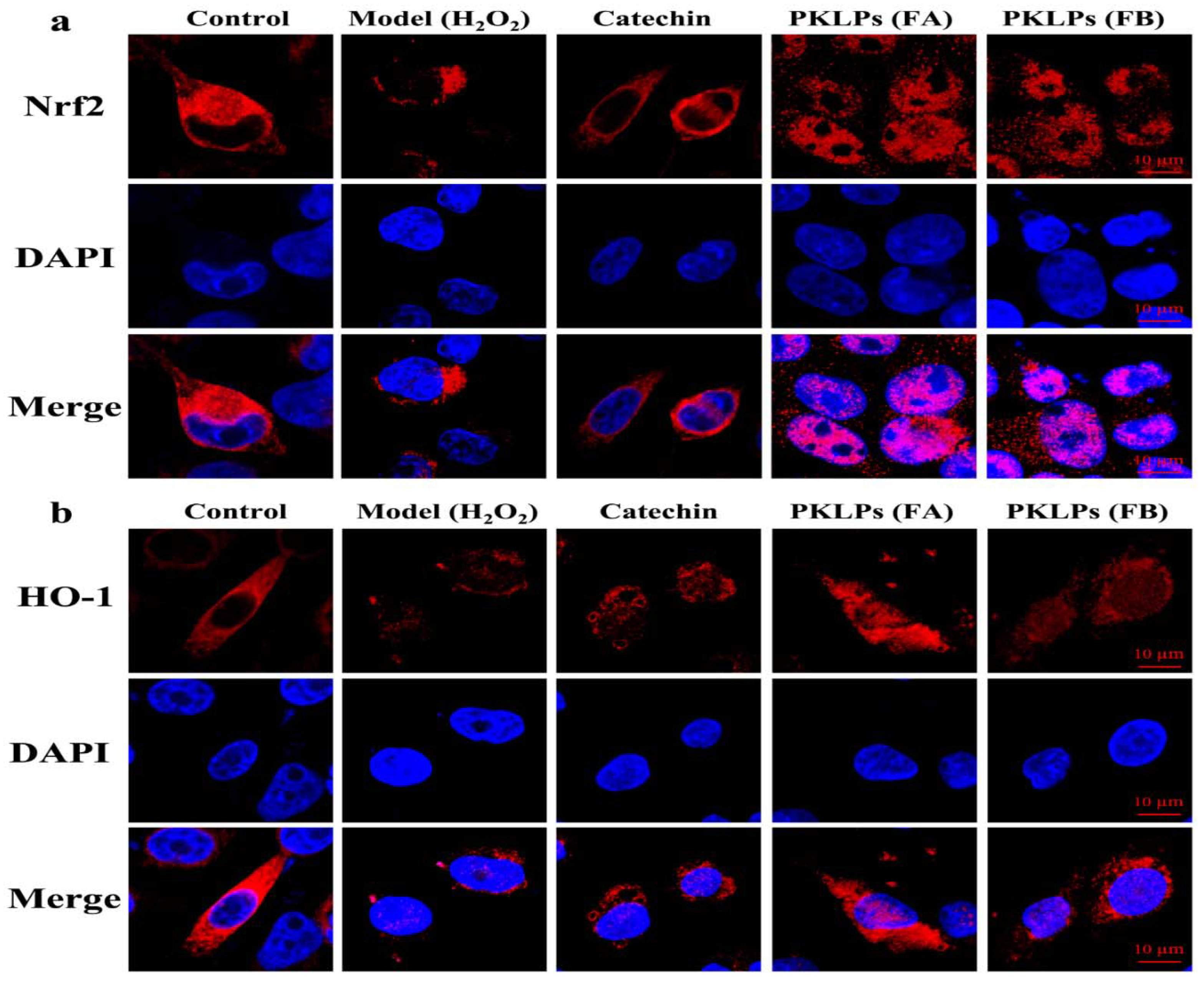
3.5. Effect of PKLPs (FA and FB) on the Nrf2 and Its Downstream Protein Expression
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hou, K.; Bao, M.; Wang, L.; Zhang, H.; Yang, L.; Zhao, H.; Wang, Z. Aqueous enzymatic pretreatment ionic liquid–lithium salt based microwave–assisted extraction of essential oil and procyanidins from pinecones of pinus koraiensis. J. Clean. Prod. 2019, 236, 117581. [Google Scholar]
- Li, Q.; Zhang, N.; Ni, L.; Wei, Z.; Quan, H.; Zhou, Y. One-pot high efficiency low temperature ultrasonic-assisted strategy for fully bio-based coloristic, anti-pilling, antistatic, bioactive and reinforced cashmere using grape seed proanthocyanidins. J. Clean. Prod. 2021, 315, 128148. [Google Scholar] [CrossRef]
- Zhou, L.; Chang, J.; Gao, Y.; Wang, C. Procyanidin b2 protects neurons from cypermethrin-induced oxidative stress through the p13k/akt/nrf2 signaling pathway. Nan Fang Yi Ke Da Xue Xue Bao J. South. Med. Univ. 2021, 41, 1158–1164. [Google Scholar]
- Gouda, M.; El-Din Bekhit, A.; Tang, Y.; Huang, Y.; Huang, L.; He, Y.; Li, X. Recent innovations of ultrasound green technology in herbal phytochemistry: A review. Ultrason. Sonochem. 2021, 73, 105538. [Google Scholar] [PubMed]
- Gouda, M.; Huang, Z.; Liu, Y.; He, Y.; Li, X. Physicochemical impact of bioactive terpenes on the microalgae biomass structural characteristics. Bioresour. Technol. 2021, 334, 125232. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.M.; Gouda, M.; Zhu, Y.Y.; Ye, X.Q.; Chen, J.C. Ultrasound-assisted extraction optimization of proanthocyanidins from kiwi (Actinidia chinensis) leaves and evaluation of its antioxidant activity. Antioxidants 2021, 10, 1317. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.-M.; Gouda, M.; El-Din Bekhit, A.; He, Y.-K.; Ye, X.-Q.; Chen, J.-C. Identification of novel bioactive proanthocyanidins with potent antioxidant and anti-proliferative activities from kiwifruit leaves. Food Biosci. 2022, 46, 101554. [Google Scholar] [CrossRef]
- Li, Q.; Liu, C.; Li, T.; McClements, D.J.; Fu, Y.; Liu, J. Comparison of phytochemical profiles and antiproliferative activities of different proanthocyanidins fractions from choerospondias axillaris fruit peels. Food Res. Int. 2018, 113, 298–308. [Google Scholar] [CrossRef]
- Marangi, F.; Pinto, D.; de Francisco, L.; Alves, R.C.; Puga, H.; Sut, S.; Dall’Acqua, S.; Rodrigues, F.; Oliveira, M. Hardy kiwi leaves extracted by multi-frequency multimode modulated technology: A sustainable and promising by-product for industry. Food Res. Int. 2018, 112, 184–191. [Google Scholar] [CrossRef]
- Ou, K.; Gu, L. Absorption and metabolism of proanthocyanidins. J. Funct. Foods 2014, 7, 43–53. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Li, T.; Liu, C.; Liu, W.; Liu, J. Comparison of bioactivities and phenolic composition of choerospondias axillaris peels and fleshes. J. Sci. Food Agric. 2016, 96, 2462–2471. [Google Scholar] [PubMed]
- Hernansanz-Agustín, P.; Enríquez, J.A. Generation of reactive oxygen species by mitochondria. Antioxidants 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Lian, P.; Braber, S.; Garssen, J.; Wichers, H.J.; Folkerts, G.; Fink-Gremmels, J.; Varasteh, S. Beyond heat stress: Intestinal integrity disruption and mechanism-based intervention strategies. Nutrients 2020, 12, 734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herb, M.; Schramm, M. Functions of ros in macrophages and antimicrobial immunity. Antioxidants 2021, 10, 313. [Google Scholar] [CrossRef]
- Koudoufio, M.; Feldman, F.; Ahmarani, L.; Delvin, E.; Spahis, S.; Desjardins, Y.; Levy, E. Intestinal protection by proanthocyanidins involves anti-oxidative and anti-inflammatory actions in association with an improvement of insulin sensitivity, lipid and glucose homeostasis. Sci. Rep. 2021, 11, 3878. [Google Scholar] [CrossRef]
- Kellett, M.E.; Greenspan, P.; Pegg, R.B. Modification of the cellular antioxidant activity (caa) assay to study phenolic antioxidants in a caco-2 cell line. Food Chem. 2018, 244, 359–363. [Google Scholar] [CrossRef]
- Zhao, H.; Eguchi, S.; Alam, A.; Ma, D. The role of nuclear factor-erythroid 2 related factor 2 (nrf-2) in the protection against lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L155–L162. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhao, Y.; Pan, F.; Zhu, M.; Yao, L.; Liu, Y.; Feng, J.; Xiong, J.; Chen, X.; Ren, F.; et al. Inhibition of the nrf2-trxr axis sensitizes the drug-resistant chronic myelogenous leukemia cell line k562/g01 to imatinib treatments. BioMed Res. Int. 2019, 2019, 6502793. [Google Scholar] [CrossRef]
- Arroyave-Ospina, J.C.; Wu, Z.; Geng, Y.; Moshage, H. Role of oxidative stress in the pathogenesis of non-alcoholic fatty liver disease: Implications for prevention and therapy. Antioxidants 2021, 10, 174. [Google Scholar] [CrossRef]
- Gulati, S.; Yadav, A.; Kumar, N.; Priya, K.; Aggarwal, N.K.; Gupta, R. Phenotypic and genotypic characterization of antioxidant enzyme system in human population exposed to radiation from mobile towers. Mol. Cell. Biochem. 2017, 440, 1–9. [Google Scholar] [CrossRef]
- Pathak, R.K.; Kim, D.Y.; Lim, B.; Kim, J.M. Investigating multi-target antiviral compounds by screening of phytochemicals from neem (azadirachta indica) against prrsv: A vetinformatics approach. Front. Vet. Sci. 2022, 9, 854528. [Google Scholar] [CrossRef] [PubMed]
- Gabr, S.K.; Bakr, R.O.; Mostafa, E.S.; El-Fishawy, A.M.; El-Alfy, T.S. Antioxidant activity and molecular docking study of erythrina × neillii polyphenolics. S. Afr. J. Bot. 2019, 121, 470–477. [Google Scholar] [CrossRef]
- Chai, W.M.; Shi, Y.; Feng, H.L.; Xu, L.; Xiang, Z.H.; Gao, Y.S.; Chen, Q.X. Structure characterization and anti-tyrosinase mechanism of polymeric proanthocyanidins fractionated from kiwifruit pericarp. J. Agric. Food Chem. 2014, 62, 6382–6389. [Google Scholar] [CrossRef]
- Tsolmon, B.; Fang, Y.; Yang, T.; Guo, L.; He, K.; Li, G.Y.; Zhao, H. Structural identification and uplc-esi-qtof-ms(2) analysis of flavonoids in the aquatic plant landoltia punctata and their in vitro and in vivo antioxidant activities. Food Chem. 2021, 343, 128392. [Google Scholar] [CrossRef]
- Tang, W.; Shen, M.; Xie, J.; Liu, D.; Du, M.; Lin, L.; Gao, H.; Hamaker, B.R.; Xie, M. Physicochemical characterization, antioxidant activity of polysaccharides from mesona chinensis benth and their protective effect on injured nctc-1469 cells induced by h2o2. Carbohydr. Polym. 2017, 175, 538–546. [Google Scholar] [PubMed]
- Rajput, S.A.; Shaukat, A.; Wu, K.; Rajput, I.R.; Baloch, D.M.; Akhtar, R.W.; Raza, M.A.; Najda, A.; Rafal, P.; Albrakati, A.; et al. Luteolin alleviates aflatoxinb1-induced apoptosis and oxidative stress in the liver of mice through activation of nrf2 signaling pathway. Antioxidants 2021, 10, 1268. [Google Scholar] [PubMed]
- Gao, X.; Xiao, Z.; Li, C.; Zhang, J.; Zhu, L.; Sun, L.; Zhang, N.; Khalil, M.M.; Rajput, S.A.; Qi, D. Prenatal exposure to zearalenone disrupts reproductive potential and development via hormone-related genes in male rats. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2018, 116, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides exhibit antioxidant activity through the keap1-nrf2 signaling pathway. J. Funct. Foods 2020, 64, 103696. [Google Scholar] [CrossRef]
- Jiang, C.S.; Zhuang, C.L.; Zhu, K.; Zhang, J.; Muehlmann, L.A.; Figueiro Longo, J.P.; Azevedo, R.B.; Zhang, W.; Meng, N.; Zhang, H. Identification of a novel small-molecule keap1-nrf2 ppi inhibitor with cytoprotective effects on lps-induced cardiomyopathy. J. Enzym. Inhib. Med. Chem. 2018, 33, 833–841. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Li, Y.; Hu, D.; Xie, L.; Ke, H.; Zheng, X.; Chen, W. Procyanidin b2 ameliorates free fatty acids-induced hepatic steatosis through regulating tfeb-mediated lysosomal pathway and redox state. Free Radic. Biol. Med. 2018, 126, 269–286. [Google Scholar] [CrossRef]
- Miranda-Hernandez, A.M.; Muniz-Marquez, D.B.; Wong-Paz, J.E.; Aguilar-Zarate, P.; de la Rosa-Hernandez, M.; Larios-Cruz, R.; Aguilar, C.N. Characterization by hplc-esi-ms(2) of native and oxidized procyanidins from litchi (litchi chinensis) pericarp. Food Chem. 2019, 291, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Zenin, V.; Ivanova, J.; Pugovkina, N.; Shatrova, A.; Aksenov, N.; Tyuryaeva, I.; Kirpichnikova, K.; Kuneev, I.; Zhuravlev, A.; Osyaeva, E.; et al. Resistance to h2o2-induced oxidative stress in human cells of different phenotypes. Redox Biol. 2022, 50, 102245. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Iahtisham Ul, H.; Patel, S.; Pan, X.; Naz, S.; Sanches Silva, A.; Saeed, F.; Rasul Suleria, H.A. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Wu, P.; Ma, G.; Li, N.; Deng, Q.; Yin, Y.; Huang, R. Investigation of in vitro and in vivo antioxidant activities of flavonoids rich extract from the berries of rhodomyrtus tomentosa(ait.) hassk. Food Chem. 2015, 173, 194–202. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, T.; Li, P.; Chen, J.; Nepovimova, E.; Long, M.; Wu, W.; Kuca, K. Bacillus amyloliquefaciens b10 can alleviate aflatoxin b1-induced kidney oxidative stress and apoptosis in mice. Ecotoxicol. Environ. Saf. 2021, 218, 112286. [Google Scholar] [CrossRef]
- Zhou, Q.; Han, X.; Li, R.; Zhao, W.; Bai, B.; Yan, C.; Dong, X. Anti-atherosclerosis of oligomeric proanthocyanidins from rhodiola rosea on rat model via hypolipemic, antioxidant, anti-inflammatory activities together with regulation of endothelial function. Phytomedicine Int. J. Phytother. Phytopharm. 2018, 51, 171–180. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Nabi, X. Protective effect of ziziphora clinopodioides flavonoids against h2o2-induced oxidative stress in huvec cells. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 117, 109156. [Google Scholar] [CrossRef]
- Fujimaki, T.; Mori, S.; Horikawa, M.; Fukui, Y. Isolation of proanthocyanidins from red wine, and their inhibitory effects on melanin synthesis in vitro. Food Chem. 2018, 248, 61–69. [Google Scholar] [CrossRef]
- Ahmed, F.E.; Gouda, M.M.; Hussein, L.A.; Ahmed, N.C.; Vos, P.W.; Mohammad, M.A. Role of melt curve analysis in interpretation of nutrigenomics’ microrna expression data. Cancer Genom. Proteom. 2017, 14, 469–481. [Google Scholar]
- Shaw, P.; Chattopadhyay, A. Nrf2-are signaling in cellular protection: Mechanism of action and the regulatory mechanisms. J. Cell. Physiol. 2020, 235, 3119–3130. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Nie, R.; Maimaiti, T.; Yao, F.; Li, C. Comparison of the inhibition on cellular 22-nbd-cholesterol accumulation and transportation of monomeric catechins and their corresponding a-type dimers in caco-2 cell monolayers. J. Funct. Foods 2016, 27, 343–351. [Google Scholar] [CrossRef]
- Siddiqui, W.A.; Ahad, A.; Ahsan, H. The mystery of bcl2 family: Bcl-2 proteins and apoptosis: An update. Arch. Toxicol. 2015, 89, 289–317. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Xiao, G.; Xu, Y.; Wu, J.; Yu, Y.; Fu, M. Persimmon vinegar polyphenols protect against hydrogen peroxide-induced cellular oxidative stress via nrf2 signalling pathway. Food Chem. 2018, 255, 23–30. [Google Scholar] [CrossRef]
- Hilary, S.; Tomas-Barberan, F.A.; Martinez-Blazquez, J.A.; Kizhakkayil, J.; Souka, U.; Al-Hammadi, S.; Habib, H.; Ibrahim, W.; Platat, C. Polyphenol characterisation of phoenix dactylifera l. (date) seeds using hplc-mass spectrometry and its bioaccessibility using simulated in-vitro digestion/caco-2 culture model. Food Chem. 2020, 311, 125969. [Google Scholar] [CrossRef]
- Song, M.Y.; Lee, D.Y.; Chun, K.S.; Kim, E.H. The role of nrf2/keap1 signaling pathway in cancer metabolism. Int. J. Mol. Sci. 2021, 22, 4376. [Google Scholar] [CrossRef]
- Wang, M.; Chen, J.; Ye, X.; Liu, D. In vitro inhibitory effects of chinese bayberry (myrica rubra sieb. Et zucc.) leaves proanthocyanidins on pancreatic alpha-amylase and their interaction. Bioorganic Chem. 2020, 101, 104029. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, H.; Chen, F.; Fu, J.; Xu, Y.; Hou, Y.; Kou, H.H.; Zhai, C.; Nelson, M.B.; Zhang, Q.; et al. An overview of chemical inhibitors of the nrf2-are signaling pathway and their potential applications in cancer therapy. Free Radic. Biol. Med. 2016, 99, 544–556. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.Z.; Shen, Y.; Lin, J.; Wang, H.R.; Talukder, M.; Li, J.L. Lycopene prevents dehp-induced leydig cell damage with the nrf2 antioxidant signaling pathway in mice. J. Agric. Food Chem. 2020, 68, 2031–2040. [Google Scholar] [CrossRef]
- Uddin, M.J.; Faraone, I.; Haque, M.A.; Rahman, M.M.; Halim, M.A.; Sonnichsen, F.D.; Cicek, S.S.; Milella, L.; Zidorn, C. Insights into the leaves of ceriscoides campanulata: Natural proanthocyanidins alleviate diabetes, inflammation, and esophageal squamous cell cancer via in vitro and in silico models. Fitoterapia 2022, 158, 105164. [Google Scholar] [CrossRef]
- Ma, G.; Tang, C.; Sun, X.; Zhang, J. The interaction mechanism of β-casein with oligomeric proanthocyanidins and its effect on proanthocyanidin bioaccessibility. Food Hydrocoll. 2021, 113, 106485. [Google Scholar] [CrossRef]

| Gene | Forward Sequence | Reverse Sequence | NCBI No. |
|---|---|---|---|
| Nrf2 | TCACACGAGATGAGCTTAGGGCAA | TACAGTTCTGGGCGGCGACTTTAT | NM_010902.4 |
| Keap1 | CAGCAACTCTGTGACGTGACC | TCAATAAGCCTTTCCATGACCT | NM_016679.4 |
| SOD-1 | TGGTTTGCGTCGTAGTCTCC | CTTCGTCGCCATAACTCGCT | NM_000454.4 |
| NQO-1 | GGTGAGCTGAAGGACTCGAA | ACCACTGCAATGGGAACTGAA | NM_008706.5 |
| HO-1 | ATGGCCTCCCTGTACCACATC | TGTTGCGCTCAATCTCCTCCT | NM_002133.2 |
| CAT | CCATTATAAGACTGACCAGGGC | AGTCCAGGAGGGGTACTTTCC | NM_001752.3 |
| Bcl-2 | ATGTGTGTGGAGCGTCAACC | CAGAGACAGCCAGGAGAAATC | NM_000633.3 |
| Bax | GAGCTGCAGAGGATGATTGCT | TGATCAGCTCGGGCACTTTA | NM_007527.3 |
| β-actin | CAAGAGAGGTATCCTGACCT | TGATCTGGGTCATCTTTTCAC | NM_007393.5 |
| Fractions | [M − H]− (m/z) | Typical MS2 Ions (m/z) | Molecular Formula | Compound Name | Rt (min) |
|---|---|---|---|---|---|
| Fraction A (FA) | |||||
| FA | 289 | 137; 163; 245 | C15H14O6 | Catechin | 8.7 |
| FA | 289 | 137; 163; 179; 289 | C15H14O6 | Epicatechin | 11.2 |
| FA | 447 | 109; 301; 447 | C21H20O11 | Quercetin | 13.3 |
| FA | 463 | 109; 301; 463 | C21H20O12 | Isoquercetin | 14.7 |
| FA | 577 | 287; 289; 425; 451; 577 | C30H26O12 | Dimer procyanidins | 18.2 |
| FA | 865 | 289; 577; 739; 711; 865 | C45H38O18 | Trimer procyanidins | 27.1 |
| Fraction B (FB) | |||||
| FB | 289 | 137; 163; 245 | C15H14O6 | Catechin | 7.7 |
| FB | 289 | 137; 163; 179; 289 | C15H14O6 | Epicatechin | 9.7 |
| FB | 561 | 271; 289; 409; 561 | C30H26O11 | Dimer propelargonidins | 14.1 |
| FB | 577 | 287; 289; 451; 425; 577 | C30H26O12 | Dimer procyanidins | 16.7 |
| FB | 593 | 287; 305; 467; 593 | C30H26O13 | Dimer prodelphindins | 21.9 |
| FB | 865 | 289; 577; 739; 711; 865 | C45H38O18 | Trimer procyanidins | 27.5 |
| FB | 1153 | 289; 577; 865; 1153 | C60H50O24 | Tetramer procyanidins | 37.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, J.-M.; Gouda, M.; Ye, X.-Q.; Shao, Z.-P.; Chen, J.-C. Evaluation of Proanthocyanidins from Kiwi Leaves (Actinidia chinensis) against Caco-2 Cells Oxidative Stress through Nrf2-ARE Signaling Pathway. Antioxidants 2022, 11, 1367. https://doi.org/10.3390/antiox11071367
Lv J-M, Gouda M, Ye X-Q, Shao Z-P, Chen J-C. Evaluation of Proanthocyanidins from Kiwi Leaves (Actinidia chinensis) against Caco-2 Cells Oxidative Stress through Nrf2-ARE Signaling Pathway. Antioxidants. 2022; 11(7):1367. https://doi.org/10.3390/antiox11071367
Chicago/Turabian StyleLv, Ji-Min, Mostafa Gouda, Xing-Qian Ye, Zhi-Peng Shao, and Jian-Chu Chen. 2022. "Evaluation of Proanthocyanidins from Kiwi Leaves (Actinidia chinensis) against Caco-2 Cells Oxidative Stress through Nrf2-ARE Signaling Pathway" Antioxidants 11, no. 7: 1367. https://doi.org/10.3390/antiox11071367








