An In-Depth Study on the Metabolite Profile and Biological Properties of Primula auriculata Extracts: A Fascinating Sparkle on the Way from Nature to Functional Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Preparation of Extracts
2.2. Profile of Bioactive Compounds
2.3. Chemicals
2.4. Ultra-High-Performance Liquid Chromatography Coupled with Hybrid Quadrupole-Orbitrap High-Resolution Mass Spectrometry (UHPLC–HRMS)
2.5. Determination of Antioxidant and Enzyme Inhibitory Effects
2.6. Molecular Docking
2.7. Cell Culture
2.8. Cell Viability and Cytotoxicity Determination with WST-1
2.9. Advanced Glycation End Products Inhibition Assay
2.10. Western Blot
2.11. Gelatin Zymography Assay
2.12. Real-Time Polymerase Chain Reaction (RT-PCR)
2.13. Statistical Analysis
3. Results and Discussion
3.1. Chemical Composition
3.1.1. Phenolic Acids and Glycosides
3.1.2. Flavones and Flavonols
3.1.3. Chalcones and Dihydrochalcones
3.1.4. Saponins
3.2. Antioxidant Properties
3.3. Enzyme Inhibitory Properties
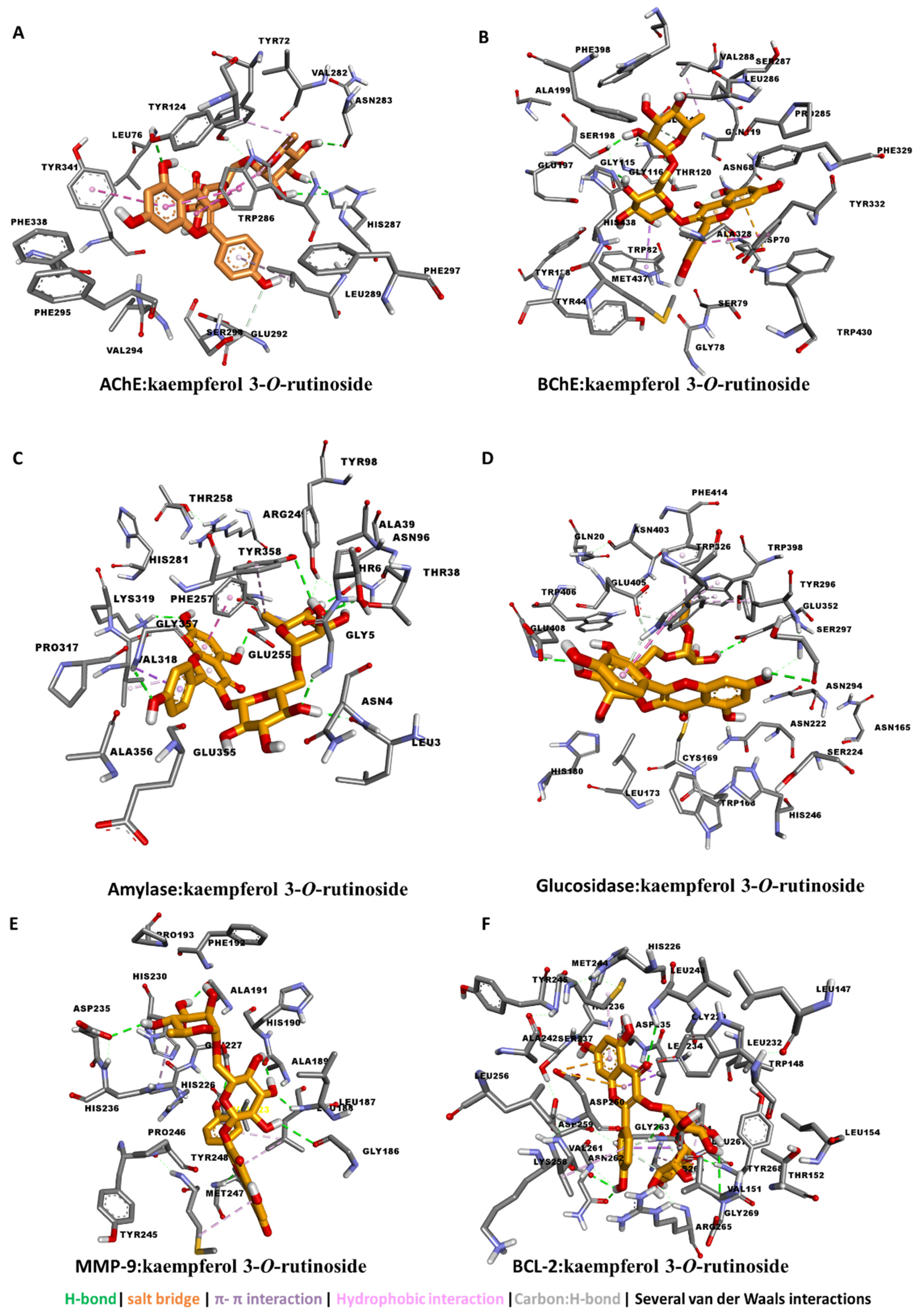
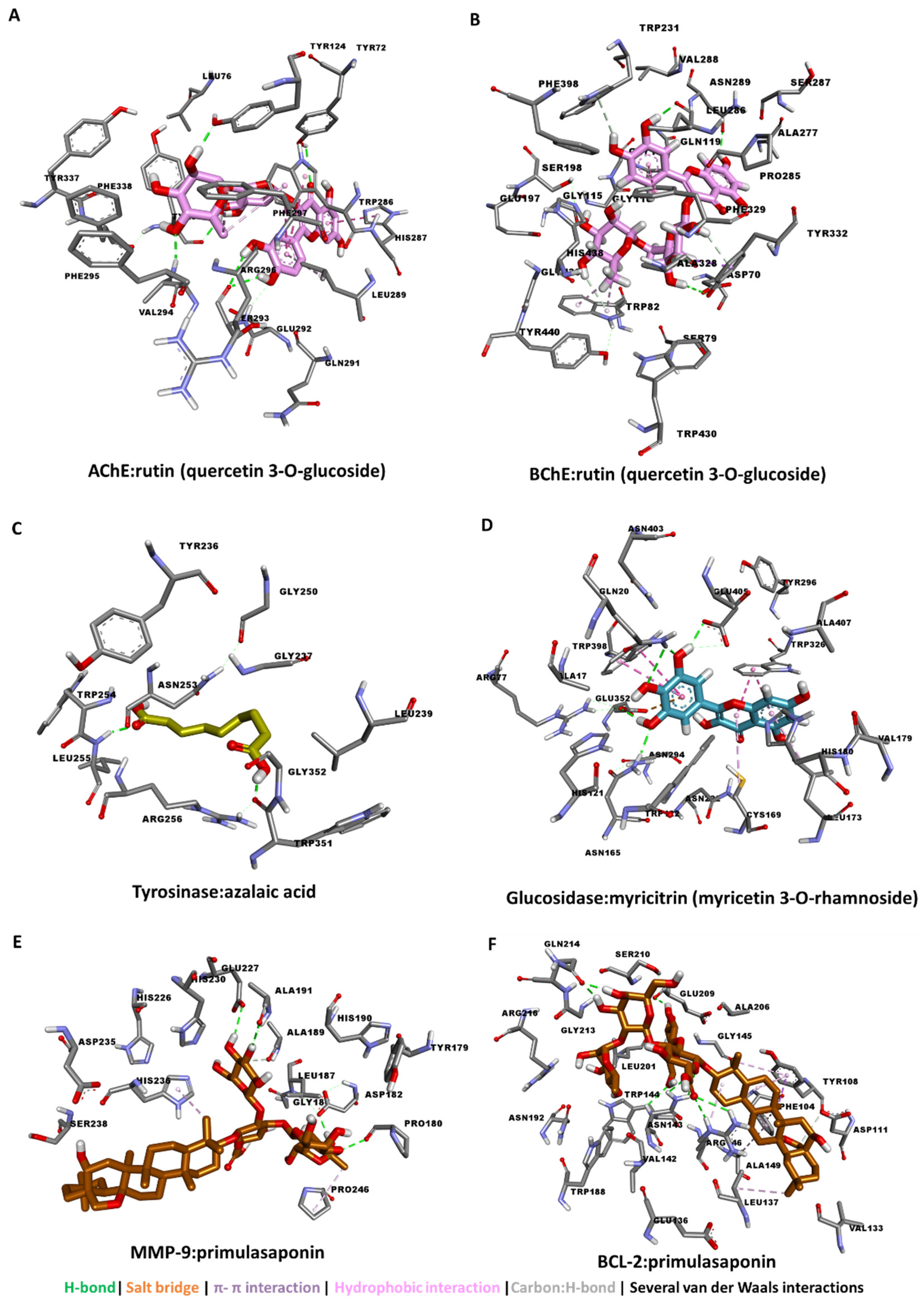
3.4. Molecular Docking
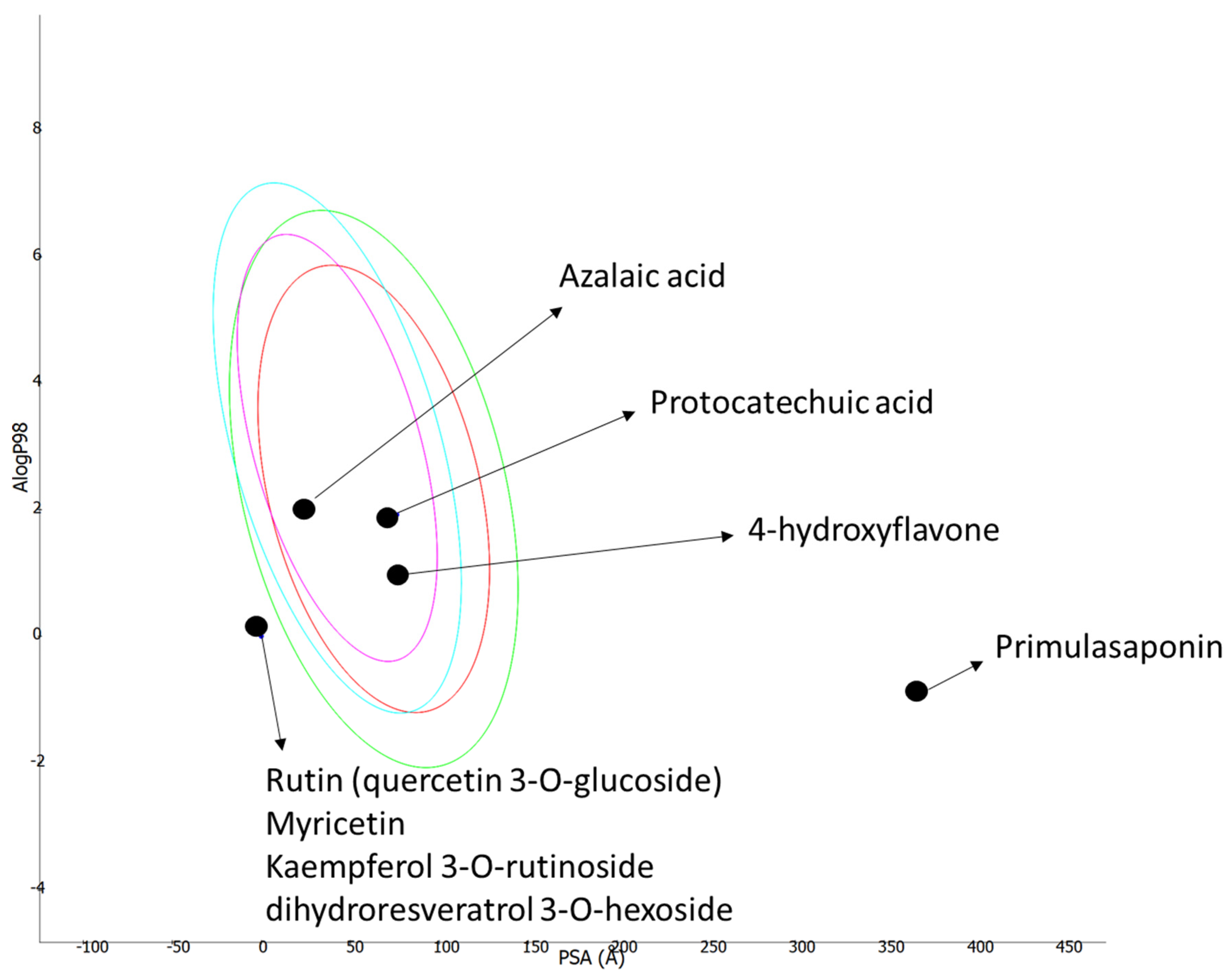
3.5. ADMET Prediction
3.6. Molecular Approach in Apoptotic Pathway
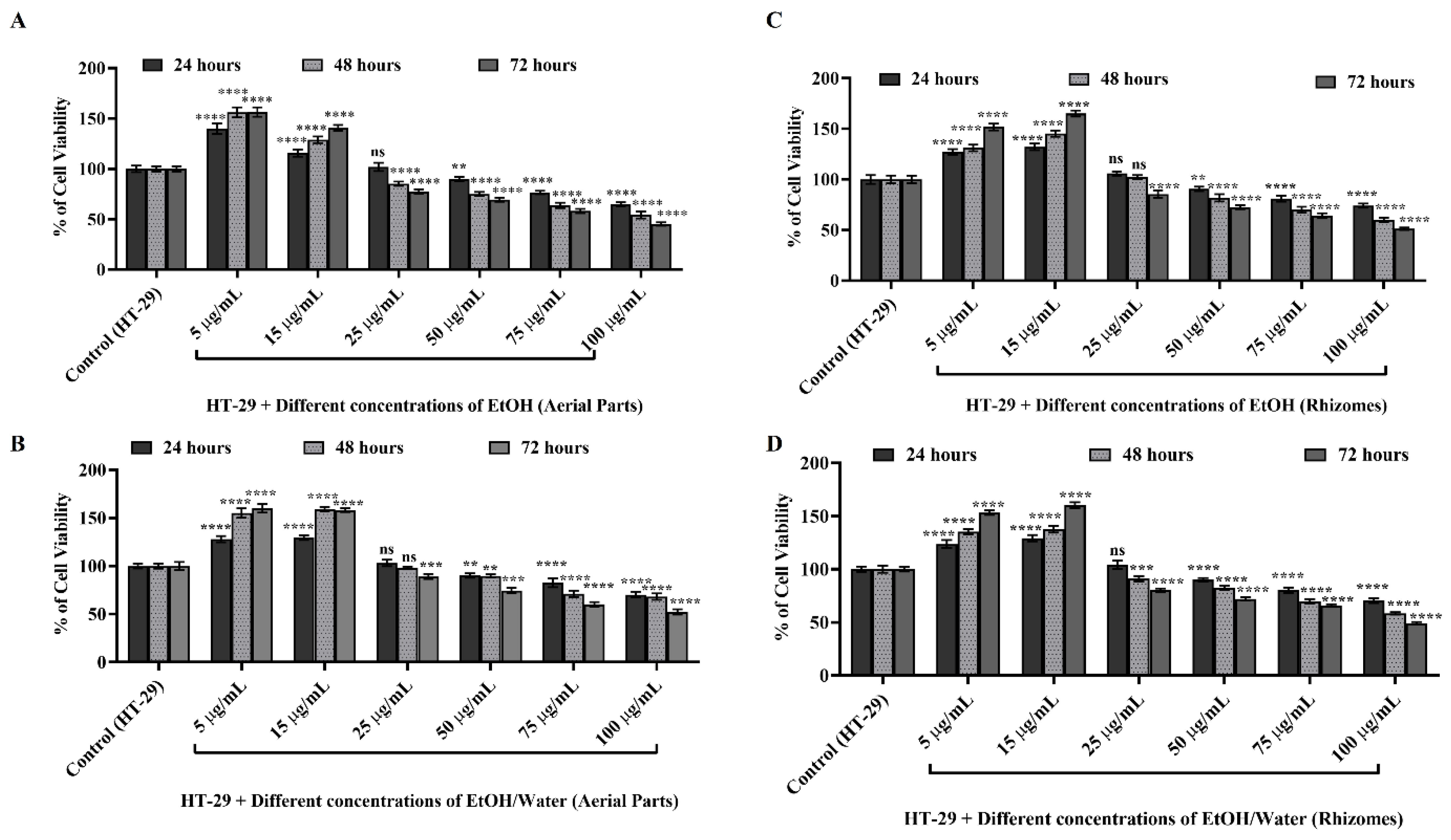
3.6.1. Cell Viability and Cytotoxicity Determination with WST-1
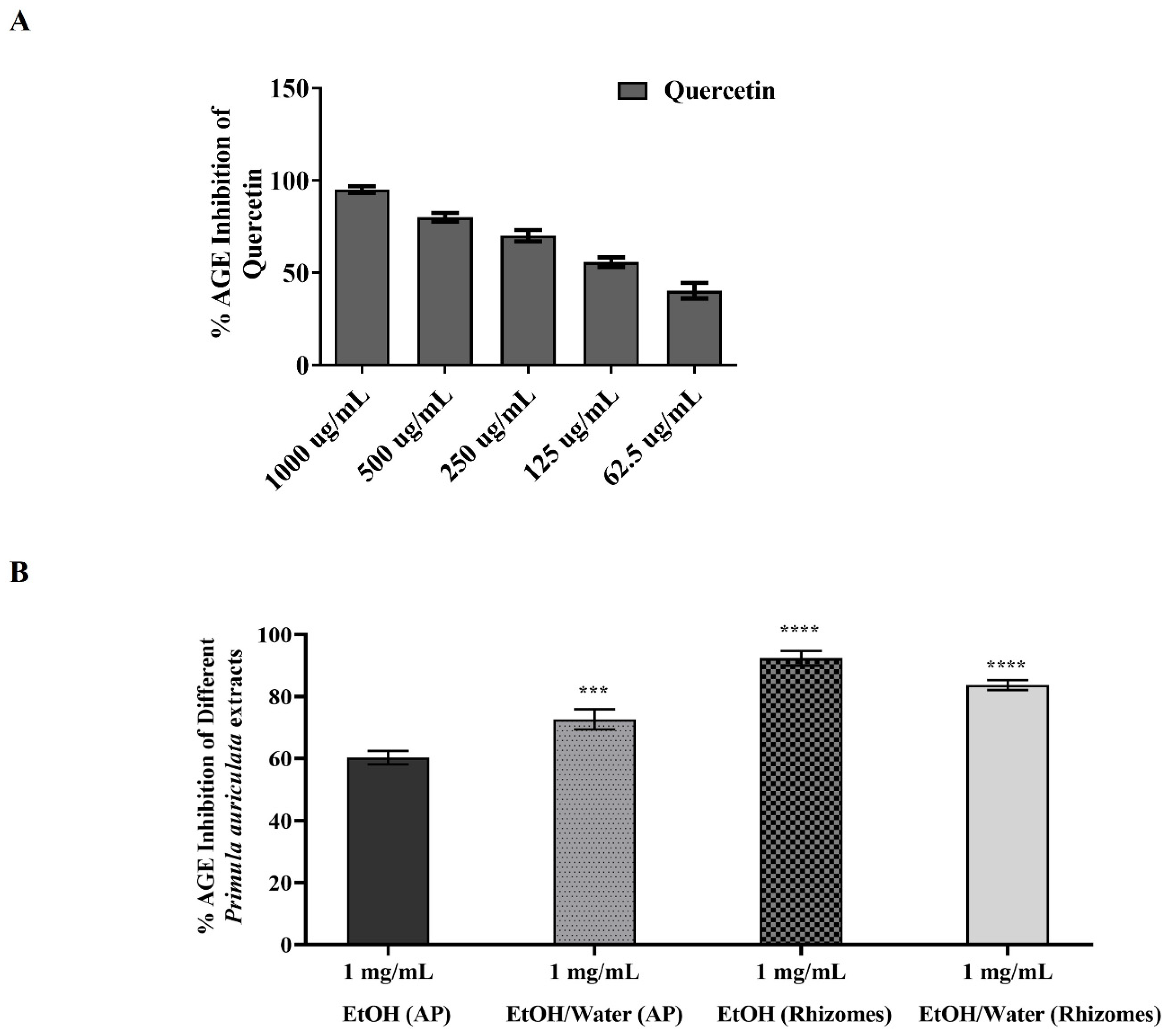
3.6.2. AGE Inhibition Activity of Primula auriculata
3.6.3. Western Blot and Protein Analysis
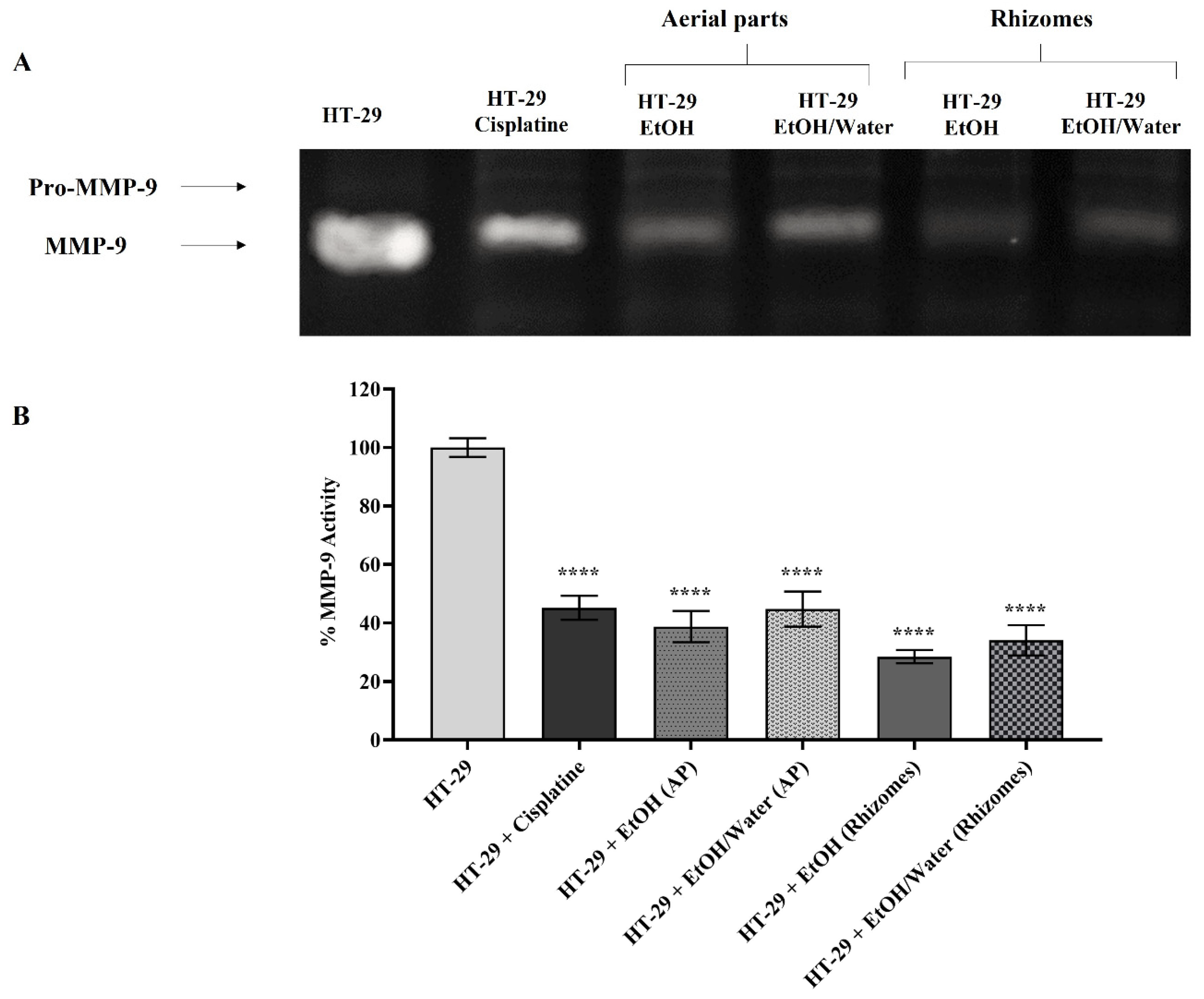
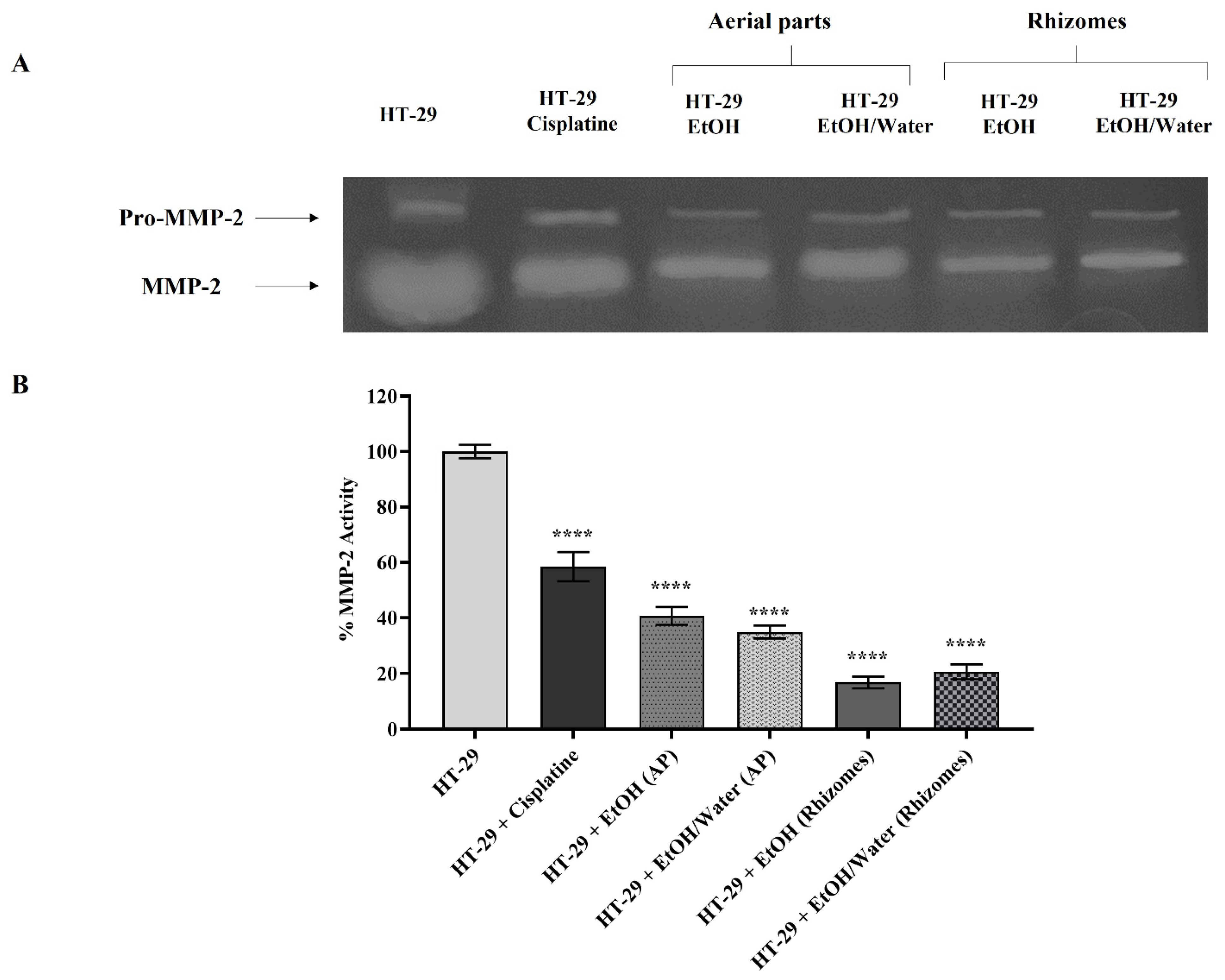
3.6.4. Matrix Metalloproteinase Activation and Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.N.; Tsitsilin, A.N.; Pozharitskaya, O.N.; Makarov, V.G.; Heinrich, M. Traditional and current food use of wild plants listed in the Russian Pharmacopoeia. Front. Pharmacol. 2017, 8, 841. [Google Scholar] [CrossRef] [PubMed]
- Nirmala, C.; Shahar, B.; Dolma, N.; Santosh, O. Promising underutilized wild plants of cold desert Ladakh, India for nutritional security and health benefits. Appl. Food Res. 2022, 2, 100145. [Google Scholar] [CrossRef]
- Singh, P.A.; Bajwa, N.; Chinnam, S.; Chandan, A.; Baldi, A. An overview of some important deliberations to promote medicinal plants cultivation. J. Appl. Res. Med. Aromat. Plants 2022, 31, 100400. [Google Scholar] [CrossRef]
- Mohammed, A.; Tajuddeen, N. Antidiabetic compounds from medicinal plants traditionally used for the treatment of diabetes in Africa: A review update (2015–2020). S. Afr. J. Bot. 2022, 146, 585–602. [Google Scholar] [CrossRef]
- Prasathkumar, M.; Anisha, S.; Dhrisya, C.; Becky, R.; Sadhasivam, S. Therapeutic and pharmacological efficacy of selective Indian medicinal plants—A review. Phytomedicine Plus 2021, 1, 100029. [Google Scholar] [CrossRef]
- Chukwuma, C.I.; Matsabisa, M.G.; Ibrahim, M.A.; Erukainure, O.L.; Chabalala, M.H.; Islam, M.S. Medicinal plants with concomitant anti-diabetic and anti-hypertensive effects as potential sources of dual acting therapies against diabetes and hypertension: A review. J. Ethnopharmacol. 2019, 235, 329–360. [Google Scholar] [CrossRef]
- Hao, D.-C.; Xiao, P.-G. Pharmaceutical resource discovery from traditional medicinal plants: Pharmacophylogeny and pharmacophylogenomics. Chin. Herb. Med. 2020, 12, 104–117. [Google Scholar] [CrossRef]
- Tojibaev, K.; Karimov, F.; Khassanov, F.; Sennikov, A.; Usmonov, M. An updated checklist of Primula species (Primulaceae) in Uzbekistan. J. Asia-Pac. Biodivers. 2020, 13, 667–678. [Google Scholar] [CrossRef]
- Richards, J. Primula: Illustrated by B. Edwards; Timber Press: Portland, OR, USA, 1993. [Google Scholar]
- Khan, S.; Shaheen, H.; Mehmood, A.; Nasar, S.; Khan, T. Ethnobotanical and antibacterial study of Primula plants traditionally used in the indigenous communities of Western Himalaya, Pakistan. Saudi J. Biol. Sci. 2022, 29, 3244–3254. [Google Scholar] [CrossRef]
- Thakur, K.S.; Kumar, M.; Bawa, R.; Bussmann, R.W. Ethnobotanical study of herbaceous flora along an altitudinal gradient in Bharmour Forest Division, District Chamba of Himachal Pradesh, India. J. Evid. Based Complement. Altern. Med. 2014, 2014, 946870. [Google Scholar] [CrossRef]
- Başbülbül, G.; Özmen, A.; Biyik, H.H.; Şen, Ö. Antimitotic and antibacterial effects of the Primula veris L. flower extracts. Caryologia 2008, 61, 88–91. [Google Scholar] [CrossRef]
- Jaberian, H.; Piri, K.; Nazari, J. Phytochemical composition and in vitro antimicrobial and antioxidant activities of some medicinal plants. Food Chem. 2013, 136, 237–244. [Google Scholar] [CrossRef]
- Behzad, S.; Ebrahim, K.; Mosaddegh, M.; Haeri, A. Primula auriculata Extracts Exert Cytotoxic and Apoptotic Effects against HT-29 Human Colon Adenocarcinoma Cells. Iran. J. Pharm Res. 2016, 15, 311–322. [Google Scholar]
- Sarikurkcu, C.; Zengin, G. Polyphenol Profile and Biological Activity Comparisons of Different Parts of Astragalus macrocephalus subsp. finitimus from Turkey. Biology 2020, 9, 231. [Google Scholar] [CrossRef]
- Ak, G.; Zengin, G.; Mahomoodally, M.F.; Llorent-Martínez, E.; Orlando, G.; Chiavaroli, A.; Brunetti, L.; Recinella, L.; Leone, S.; Di Simone, S.C. Shedding Light into the Connection between Chemical Components and Biological Effects of Extracts from Epilobium hirsutum: Is It a Potent Source of Bioactive Agents from Natural Treasure? Antioxidants 2021, 10, 1389. [Google Scholar] [CrossRef]
- Uba, A.I.; Zengin, G.; Montesano, D.; Cakilcioglu, U.; Selvi, S.; Ulusan, M.D.; Caprioli, G.; Sagratini, G.; Angeloni, S.; Jugreet, S.; et al. Antioxidant and Enzyme Inhibitory Properties, and HPLC–MS/MS Profiles of Different Extracts of Arabis carduchorum Boiss.: An Endemic Plant to Turkey. Appl. Sci. 2022, 12, 6561. [Google Scholar] [CrossRef]
- Zengin, G.; Aktumsek, A. Investigation of antioxidant potentials of solvent extracts from different anatomical parts of Asphodeline anatolica E. Tuzlaci: An endemic plant to Turkey. Afr. J. Tradit. Complement. Altern. Med. 2014, 11, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Gevrenova, R.; Zheleva-Dimitrova, D.; Balabanova, V.; Voynikov, Y.; Sinan, K.I.; Mahomoodally, M.F.; Zengin, G. Integrated phytochemistry, bio-functional potential and multivariate analysis of Tanacetum macrophyllum (Waldst. & Kit.) Sch. Bip. and Telekia speciosa (Schreb.) Baumg. (Asteraceae). Ind. Crop. Prod. 2020, 155, 112817. [Google Scholar]
- Grochowski, D.M.; Uysal, S.; Aktumsek, A.; Granica, S.; Zengin, G.; Ceylan, R.; Locatelli, M.; Tomczyk, M. In vitro enzyme inhibitory properties, antioxidant activities, and phytochemical profile of Potentilla thuringiaca. Phytochem. Lett. 2017, 20, 365–372. [Google Scholar] [CrossRef]
- Uysal, S.; Zengin, G.; Locatelli, M.; Bahadori, M.B.; Mocan, A.; Bellagamba, G.; De Luca, E.; Mollica, A.; Aktumsek, A. Cytotoxic and enzyme inhibitory potential of two Potentilla species (P. speciosa L. and P. reptans Willd.) and their chemical composition. Front. Pharmacol. 2017, 8, 290. [Google Scholar] [CrossRef]
- Gerlits, O.; Ho, K.-Y.; Cheng, X.; Blumenthal, D.; Taylor, P.; Kovalevsky, A.; Radić, Z. A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies. Chem. Biol. Interact. 2019, 309, 108698. [Google Scholar] [CrossRef]
- Rosenberry, T.; Brazzolotto, X.; Macdonald, I.; Wandhammer, M.; Trovaslet-Leroy, M.; Darvesh, S.; Nachon, F. Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study. Molecules 2017, 22, 2098. [Google Scholar] [CrossRef] [Green Version]
- Božić, N.; Rozeboom, H.J.; Lončar, N.; Slavić, M.Š.; Janssen, D.B.; Vujčić, Z. Characterization of the starch surface binding site on Bacillus paralicheniformis α-amylase. Int. J. Biol. Macromol. 2020, 165, 1529–1539. [Google Scholar] [CrossRef]
- Fujieda, N.; Umakoshi, K.; Ochi, Y.; Nishikawa, Y.; Yanagisawa, S.; Kubo, M.; Kurisu, G.; Itoh, S. Copper–Oxygen Dynamics in the Tyrosinase Mechanism. Angew. Chem. Int. Ed. 2020, 59, 13385–13390. [Google Scholar] [CrossRef]
- Karade, S.S.; Hill, M.L.; Kiappes, J.L.; Manne, R.; Aakula, B.; Zitzmann, N.; Warfield, K.L.; Treston, A.M.; Mariuzza, R.A. N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum α-Glucosidases I and II with Antiviral Activity. J. Med. Chem. 2021, 64, 18010–18024. [Google Scholar] [CrossRef]
- Nuti, E.; Cantelmo, A.R.; Gallo, C.; Bruno, A.; Bassani, B.; Camodeca, C.; Tuccinardi, T.; Vera, L.; Orlandini, E.; Nencetti, S.; et al. N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity. J. Med. Chem. 2015, 58, 7224–7240. [Google Scholar] [CrossRef]
- Murray, J.B.; Davidson, J.; Chen, I.; Davis, B.; Dokurno, P.; Graham, C.J.; Harris, R.; Jordan, A.; Matassova, N.; Pedder, C.; et al. Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1. ACS Omega 2019, 4, 8892–8906. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Salman, G.; Pehlivanoglu, S.; Aydin Acar, C.; Yesilot, S. Anticancer Effects of Vitis vinifera L. Mediated Biosynthesized Silver Nanoparticles and Cotreatment with 5 Fluorouracil on HT-29 Cell Line. Biol. Trace Elem. Res. 2021, 200, 3159–3170. [Google Scholar] [CrossRef]
- Sathiyamoorthy, J.; Sudhakar, N. In vitro Cytotoxicity and Apoptotic Assay in HT-29 Cell Line Using Ficus hispida Linn: Leaves Extract. Pharm. Mag 2018, 13 (Suppl. 4), S756–S761. [Google Scholar]
- Kurt-Celep, İ.; Zengin, G.; Sinan, K.I.; Ak, G.; Elbasan, F.; Yıldıztugay, E.; Maggi, F.; Caprioli, G.; Angeloni, S.; Sharmeen, J.B.; et al. Comprehensive evaluation of two Astragalus species (A. campylosema and A. hirsutus) based on biological, toxicological properties and chemical profiling. Food Chem. Toxicol. 2021, 154, 112330. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Herst, P.M.; Tan, A.S. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol Annu Rev. 2005, 11, 127–152. [Google Scholar] [PubMed]
- Kurt-Celep, İ.; Celep, E.; Akyüz, S.; İnan, Y.; Barak, T.H.; Akaydın, G.; Telci, D.; Yesilada, E. Hypericum olympicum L. recovers DNA damage and prevents MMP-9 activation induced by UVB in human dermal fibroblasts. J. Ethnopharmacol 2020, 246, 112202. [Google Scholar] [CrossRef] [PubMed]
- Twarda-Clapa, A.; Olczak, A.; Białkowska, A.M.; Koziołkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef]
- Barak, T.; Bardakcı, H.; Kurt-Celep, İ.; Özdemir, K.; Celep, E. Evaluation of the influence of in vitro human digestion simulation on the chemical composition and bioactivities of Ziziphus jujuba Mill. Acta Aliment. 2022, 51, 105–114. [Google Scholar] [CrossRef]
- İnan, Y.; Akyüz, S.; Kurt-Celep, I.; Celep, E.; Yesilada, E. Influence of In Vitro Human Digestion Simulation on the Phenolics Contents and Biological Activities of the Aqueous Extracts from Turkish Cistus Species. Molecules 2021, 26, 5322. [Google Scholar] [CrossRef]
- Akyüz, S.; Kurt-Celep, İ.; İnan, Y.; Özdemir, O.E.; Celep, E.; Yesilada, E. In vitro evaluation of the bioactivity and bioaccessibility of Hypericum olympicum L. S. Afr. J. Bot. 2021, 142, 316–324. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar]
- Kurt-Celep, İ.; Nihan Kilinc, A.; Griffin, M.; Telci, D. Nitrosylation of tissue transglutaminase enhances fibroblast migration and regulates MMP activation. Matrix Biol. 2022, 105, 1–16. [Google Scholar] [CrossRef]
- Müller, A.; Ganzera, M.; Stuppner, H. Analysis of phenolic glycosides and saponins in Primula elatior and Primula veris (primula root) by liquid chromatography, evaporative light scattering detection and mass spectrometry. J. Chromatogr. A 2006, 1112, 218–223. [Google Scholar] [CrossRef]
- Sinan, K.I.; Zengin, G.; Zheleva-Dimitrova, D.; Gevrenova, R.; Picot-Allain, M.C.N.; Dall’Acqua, S.; Behl, T.; Goh, B.H.; Ying, P.T.S.; Mahomoodally, M.F. Exploring the Chemical Profiles and Biological Values of Two Spondias Species (S. dulcis and S. mombin): Valuable Sources of Bioactive Natural Products. Antioxidants 2021, 10, 1771. [Google Scholar] [CrossRef]
- Fico, G.; Rodondi, G.; Flamini, G.; Passarella, D.; Tomé, F. Comparative phytochemical and morphological analyses of three Italian Primula species. Phytochemistry 2007, 68, 1683–1691. [Google Scholar] [CrossRef]
- He, Y.Q.; Yang, L.; Liu, Y.; Zhang, J.W.; Tang, J.; Su, J.; Li, Y.Y.; Lu, Y.L.; Wang, C.H.; Yang, L.; et al. Characterization of cardamonin metabolism by P450 in different species via HPLC-ESI-ion trap and UPLC-ESI-quadrupole mass spectrometry. Acta Pharm. Sin. 2009, 30, 1462–1470. [Google Scholar] [CrossRef] [Green Version]
- Calis, I.; Sticher, O. Triterpene saponins from plants of the flora of Turkey. Sapon. Used Tradit. Mod. Med. 1996, 404, 485–500. [Google Scholar]
- Włodarczyk, M.; Pasikowski, P.; Osiewała, K.; Frankiewicz, A.; Dryś, A.; Gleńsk, M. In search of high-yielding and single-compound-yielding plants: New sources of pharmaceutically important saponins from the Primulaceae family. Biomolecules 2020, 10, 376. [Google Scholar] [CrossRef] [Green Version]
- Çalis, I.; Yürüker, A.; Rüegger, H.; Wright, A.D.; Sticher, O. Triterpene saponins from Primula veris subsp. macrocalyx and Primula elatior subsp. meyeri. J. Nat. Prod. 1992, 55, 1299–1306. [Google Scholar] [CrossRef]
- Siems, K.; Jaensch, M.; Jakupovic, J. Structures of the two saponins isolated from commercially available root extract of Primula sp. Planta Med. 1998, 64, 272–274. [Google Scholar] [CrossRef]
- Bibi Sadeer, N.; Montesano, D.; Albrizio, S.; Zengin, G.; Mahomoodally, M.F. The versatility of antioxidant assays in food science and safety—Chemistry, applications, strengths, and limitations. Antioxidants 2020, 9, 709. [Google Scholar] [CrossRef]
- Dehghan, H.; Sarrafi, Y.; Salehi, P. Antioxidant and antidiabetic activities of 11 herbal plants from Hyrcania region, Iran. J. Food Drug Anal. 2016, 24, 179–188. [Google Scholar] [CrossRef]
- Alinezhad, H.; Zare, M.; Nabavi, S.F.; Naqinezhad, A.; Nabavi, S.M. Antioxidant, antihemolytic, and inhibitory activities of endemic Primula heterochroma against Fe2+-induced lipid peroxidation and oxidative stress in rat brain in vitro. Pharm. Biol. 2012, 50, 1391–1396. [Google Scholar] [CrossRef]
- Tarapatskyy, M.; Gumienna, A.; Sowa, P.; Kapusta, I.; Puchalski, C. Bioactive phenolic compounds from Primula veris L.: Influence of the extraction conditions and purification. Molecules 2021, 26, 997. [Google Scholar] [CrossRef]
- Lohvina, H.; Sándor, M.; Wink, M. Effect of Ethanol Solvents on Total Phenolic Content and Antioxidant Properties of Seed Extracts of Fenugreek (Trigonella foenum-graecum L.) varieties and determination of phenolic Composition by HPLC-ESI-MS. Diversity 2021, 14, 7. [Google Scholar] [CrossRef]
- Nakilcioğlu-Taş, E.; Ötleş, S. Influence of extraction solvents on the polyphenol contents, compositions, and antioxidant capacities of fig (Ficus carica L.) seeds. An. Acad. Bras. Ciências 2021, 93, e20190526. [Google Scholar] [CrossRef]
- Gedük, A.Ş.; Atsız, S. LC-MS/MS phenolic composition of peach (Prunus persica L. Batsch) extracts and an evaluation of their antidiabetic, antioxidant, and antibacterial activities. S. Afr. J. Bot. 2022, 147, 636–645. [Google Scholar] [CrossRef]
- Noroozisharaf, A.; Samizadeh Lahiji, H.; Hatamzadeh, A.; Bakhshi, D. Phytochemical attributes of endemic endangered primrose (Primula heterochroma Stapf.) accessions grown in Iran. Physiol. Mol. Biol. Plants 2015, 21, 573–581. [Google Scholar] [CrossRef] [Green Version]
- Orhan, D.D.; Özçelik, B.; Hoşbaş, S.; Vural, M. Assessment of antioxidant, antibacterial, antimycobacterial, and antifungal activities of some plants used as folk remedies in Turkey against dermatophytes and yeast-like fungi. Turk. J. Biol. 2012, 36, 672–686. [Google Scholar] [CrossRef]
- Demasi, S.; Caser, M.; Donno, D.; Enri, S.R.; Lonati, M.; Scariot, V. Exploring wild edible flowers as a source of bioactive compounds: New perspectives in horticulture. Folia Hortic. 2021, 33, 27–48. [Google Scholar] [CrossRef]
- Demir, S.; Turan, İ.; Aliyazicioğlu, R.; Aliyazicioğlu, Y. Primula vulgaris Yaprak Ekstraktının Antioksidan ve Sitotoksik Özelliklerinin Değerlendirilmesi. KSÜ Doğa Bilimleri Derg. 2017, 20, 361–367. [Google Scholar] [CrossRef] [Green Version]
- Demir, S.; Turan, İ.; Aliyazicioğlu, Y. Antioxidant properties of Primula vulgaris flower extract and its cytotoxic effect on human cancer cell lines. Kahramanmaraş Sütçü İmam Üniversitesi Tarım Ve Doğa Derg. 2019, 22, 78–84. [Google Scholar]
- Ozkan, M.; Aliyazicioğlu, R.; Demir, S.; Misir, S.; Turan, I.; Yildirmiş, S.; Aliyazicioğlu, Y. Phenolic characterisation and antioxidant activity of Primula vulgaris and its antigenotoxic effect on fibroblast cells. Jundishapur J. Nat. Pharm. Prod. 2017, 12, e40073. [Google Scholar]
- Xu, J.-G.; Hu, Q.-P.; Liu, Y. Antioxidant and DNA-protective activities of chlorogenic acid isomers. J. Agric. Food Chem. 2012, 60, 11625–11630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, Y.; Zhu, Y.; Xu, Z. Assessment of the correlations between reducing power, scavenging DPPH activity and anti-lipid-oxidation capability of phenolic antioxidants. LWT Food Sci. Technol. 2015, 63, 569–574. [Google Scholar] [CrossRef]
- Masek, A.; Chrzescijanska, E.; Latos, M. Determination of antioxidant activity of caffeic acid and p-coumaric acid by using electrochemical and spectrophotometric assays. Int. J. Electrochem. Sci 2016, 11, 10644–10658. [Google Scholar] [CrossRef]
- Sánchez-Marzo, N.; Pérez-Sánchez, A.; Ruiz-Torres, V.; Martínez-Tébar, A.; Castillo, J.; Herranz-López, M.; Barrajón-Catalán, E. Antioxidant and Photoprotective Activity of Apigenin and its Potassium Salt Derivative in Human Keratinocytes and Absorption in Caco-2 Cell Monolayers. Int. J. Mol. Sci. 2019, 20, 2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Abreu, M.; Gonzalez-Pizarro, R.; Espinoza, L.C.; Rodríguez-Lagunas, M.J.; Espina, M.; García, M.L.; Calpena, A.C. Thiazolidinedione as an alternative to facilitate oral administration in geriatric patients with Alzheimer’s disease. Eur. J. Pharm. Sci. 2019, 129, 173–180. [Google Scholar] [CrossRef]
- Pashaei, H.; Rouhani, A.; Nejabat, M.; Hadizadeh, F.; Mirzaei, S.; Nadri, H.; Maleki, M.F.; Ghodsi, R. Synthesis and molecular dynamic simulation studies of novel N-(1-benzylpiperidin-4-yl) quinoline-4-carboxamides as potential acetylcholinesterase inhibitors. J. Mol. Struct. 2021, 1244, 130919. [Google Scholar] [CrossRef]
- Greig, N.H.; Lahiri, D.K.; Sambamurti, K. Butyrylcholinesterase: An Important New Target in Alzheimer’s Disease Therapy. Int. Psychogeriatr. 2002, 14, 77–91. [Google Scholar] [CrossRef]
- Ebrahimpour, S.; Zakeri, M.; Esmaeili, A. Crosstalk between obesity, diabetes, and alzheimer’s disease: Introducing quercetin as an effective triple herbal medicine. Ageing Res. Rev. 2020, 62, 101095. [Google Scholar] [CrossRef]
- Huang, H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef] [Green Version]
- Montero, J.; Letai, A. Why do BCL-2 inhibitors work and where should we use them in the clinic? Cell Death Differ. 2017, 25, 56–64. [Google Scholar] [CrossRef]
- Demir, S.; Turan, I.; Aliyazicioglu, R.; Yaman, S.O.; Aliyazicioglu, Y. Primula vulgaris extract induces cell cycle arrest and apoptosis in human cervix cancer cells. J. Pharm. Anal. 2018, 8, 307–311. [Google Scholar] [CrossRef]
- Yao, M.; Li, R.; Yang, Z.; Ding, Y.; Zhang, W.; Li, W.; Liu, M.; Zhao, C.; Wang, Y.; Tang, H.; et al. PP9, a steroidal saponin, induces G2/M arrest and apoptosis in human colorectal cancer cells by inhibiting the PI3K/Akt/GSK3β pathway. Chem. Biol. Interact. 2020, 331, 109246. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, Y.; Zheng, H.; Fan, L.; Mei, Q.; Tang, Y.; Duan, X.; Li, Y. Paris saponin VII extracted from trillium tschonoskii suppresses proliferation and induces apoptosis of human colorectal cancer cells. J. Ethnopharmacol. 2019, 239, 111903. [Google Scholar] [CrossRef]
- Saharanavard, S.; Mojab, F.; Boluki Naseri, P.; Behzad, S.; Parsa Khankandi, H. Chemical Compounds Isolated from Aerial Part of Primula auriculata L. Iran. J. Pharm. Sci. 2018, 14, 63–70. [Google Scholar]
- Elekofehinti, O.O.; Iwaloye, O.; Olawale, F.; Ariyo, E.O. Saponins in Cancer Treatment: Current Progress and Future Prospects. Pathophysiology 2021, 28, 250–272. [Google Scholar] [CrossRef]
- Azizian-Farsani, F.; Abedpoor, N.; Hasan Sheikhha, M.; Gure, A.O.; Nasr-Esfahani, M.H.; Ghaedi, K. Receptor for Advanced Glycation End Products Acts as a Fuel to Colorectal Cancer Development. Front. Oncol. 2020, 10, 552283. [Google Scholar] [CrossRef]
- Schröter, D.; Höhn, A. Role of Advanced Glycation End Products in Carcinogenesis and their Therapeutic Implications. Curr. Pharm. Des. 2018, 24, 5245–5251. [Google Scholar] [CrossRef]
- Hafsa, J.; Hammi, K.M.; Khedher, M.R.B.; Smach, M.A.; Charfeddine, B.; Limem, K.; Majdoub, H. Inhibition of protein glycation, antioxidant and antiproliferative activities of Carpobrotus edulis extracts. Biomed. Pharmacother. 2016, 84, 1496–1503. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, H.-y.; Baba, M.; Okada, Y.; Okuyama, T.; Wu, L.-j.; Zhan, L.-b. Extracts and compounds with anti-diabetic complications and anti-cancer activity from Castanea mollissina Blume (Chinese chestnut). BMC Complement. Altern. Med. 2014, 14, 422. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Liu, J.; Dong, L.; Wang, X.; Zhang, X. Novel advances in inhibiting advanced glycation end product formation using natural compounds. Biomed. Pharmacother. 2021, 140, 111750. [Google Scholar] [CrossRef] [PubMed]
- Özbolat, S.N.; Ayna, A. Chrysin Suppresses HT-29 Cell Death Induced by Diclofenac through Apoptosis and Oxidative Damage. Nutr. Cancer 2021, 73, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Hut, E.-F.; Radulescu, M.; Pilut, N.; Macasoi, I.; Berceanu, D.; Coricovac, D.; Pinzaru, I.; Cretu, O.; Dehelean, C. Two Antibiotics, Ampicillin and Tetracycline, Exert Different Effects in HT-29 Colorectal Adenocarcinoma Cells in Terms of Cell Viability and Migration Capacity. Curr. Oncol. 2021, 28, 2466–2480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, G.; Zhuang, X.; Guo, M. Inhibition of Growth of Colon Tumors and Proliferation of HT-29 Cells by Warburgia ugandensis Extract through Mediating G0G1 Cell Cycle Arrest, Cell Apoptosis, and Intracellular ROS Generation. Oxidative Med. Cell. Longev. 2021, 2021, 8807676. [Google Scholar] [CrossRef]
- Awang, M.A.; Najihah, N.N.; Daud, N.M.; Ismail, N.I.M.; Cheng, P.G.; Ismail, M.F.; Ramaiya, S.D. Antioxidant and cytotoxicity activity of Cordyceps militaris extracts against human colorectal cancer cell line. J. Appl. Pharm. Sci. 2021, 11, 105–109. [Google Scholar]
- Rothwarf, D.M.; Karin, M. The NF-κB Activation Pathway: A Paradigm in Information Transfer from Membrane to Nucleus. Sci. STKE 1999, 1999, re1. [Google Scholar] [CrossRef]
- Li, M.; You, L.; Xue, J.; Lu, Y. Ionizing Radiation-Induced Cellular Senescence in Normal, Non-transformed Cells and the Involved DNA Damage Response: A Mini Review. Front. Pharmacol. 2018, 9, 522. [Google Scholar] [CrossRef]
- Khan, H.; Ullah, H.; Castilho, P.C.M.F.; Gomila, A.S.; D’Onofrio, G.; Filosa, R.; Wang, F.; Nabavi, S.M.; Daglia, M.; Silva, A.S.; et al. Targeting NF-κB signaling pathway in cancer by dietary polyphenols. Crit. Rev. Food Sci. Nutr. 2020, 60, 2790–2800. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [Green Version]
- Mollace, A.; Coluccio, M.L.; Donato, G.; Mollace, V.; Malara, N. Cross-talks in colon cancer between RAGE/AGEs axis and inflammation/immunotherapy. Oncotarget 2021, 12, 1281. [Google Scholar] [CrossRef]
- El-Far, A.H.; Sroga, G.; Al Jaouni, S.K.; Mousa, S.A. Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 3613. [Google Scholar] [CrossRef]
- Waghela, B.N.; Vaidya, F.U.; Ranjan, K.; Chhipa, A.S.; Tiwari, B.S.; Pathak, C. AGE-RAGE synergy influences programmed cell death signaling to promote cancer. Mol. Cell. Biochem. 2021, 476, 585–598. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef]
- Ledoux, A.C.; Perkins, N.D. NF-κB and the cell cycle. Biochem. Soc. Trans. 2014, 42, 76–81. [Google Scholar] [CrossRef]
- Mermelshtein, A.; Gerson, A.; Walfisch, S.; Delgado, B.; Shechter-Maor, G.; Delgado, J.; Fich, A.; Gheber, L. Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas. Br. J. Cancer 2005, 93, 338–345. [Google Scholar] [CrossRef] [Green Version]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [Green Version]
- Vos, M.C.; van der Wurff, A.A.M.; van Kuppevelt, T.H.; Massuger, L.F.A.G. The role of MMP-14 in ovarian cancer: A systematic review. J. Ovarian Res. 2021, 14, 101. [Google Scholar] [CrossRef]
- Liao, H.-Y.; Da, C.-M.; Liao, B.; Zhang, H.-H. Roles of matrix metalloproteinase-7 (MMP-7) in cancer. Clin. Biochem. 2021, 92, 9–18. [Google Scholar] [CrossRef]
- Wiśniowski, T.; Bryda, J.; Kurzepa, J.; Wątroba, S. The role of matrix metalloproteinases in pathogenesis of human bladder cancer. Acta Biochim. Pol. 2021, 68, 547–555. [Google Scholar]
- Barabás, L.; Hritz, I.; István, G.; Tulassay, Z.; Herszényi, L. The Behavior of MMP-2, MMP-7, MMP-9, and Their Inhibitors TIMP-1 and TIMP-2 in Adenoma-Colorectal Cancer Sequence. Dig. Dis. 2021, 39, 217–224. [Google Scholar] [CrossRef]
- Deng, R.; Wu, H.; Ran, H.; Kong, X.; Hu, L.; Wang, X.; Su, Q. Glucose-derived AGEs promote migration and invasion of colorectal cancer by up-regulating Sp1 expression. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1065–1074. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Li, X.; Chen, Y.; Fang, J.; Ge, Z. High-mobility group Box 1: A novel inducer of the epithelial–mesenchymal transition in colorectal carcinoma. Cancer Lett. 2015, 357, 527–534. [Google Scholar] [CrossRef]


| Plant Parts | Solvents | Yield (%) | TPC (mg GAE/g Dry Extract) | TFC (mg RE/g Dry Extract) | PBD (mmol TE/g Dry Extract) |
|---|---|---|---|---|---|
| Aerial parts | EA | 1.73 | 32.36 ± 0.43 g | 9.64 ± 0.17 d | 1.84 ± 0.11 b |
| EtOH | 4.15 | 45.77 ± 0.30 e | 22.84 ± 0.42 c | 1.76 ± 0.08 b | |
| EtOH/Water | 16.67 | 56.81 ± 0.23 c | 63.92 ± 0.41 a | 1.49 ± 0.16 c,d | |
| Infusion | 12.97 | 52.88 ± 0.66 d | 57.71 ± 0.56 b | 1.24 ± 0.01 d | |
| Rhizomes | EA | 1.61 | 36.19 ± 0.26 f | 3.72 ± 0.10 f | 1.45 ± 0.11 c,d |
| EtOH | 7.64 | 96.66 ± 0.61 a | 9.64 ± 0.67 d | 2.59 ± 0.13 a | |
| EtOH/Water | 14.87 | 70.51 ± 0.34 b | 7.72 ± 0.03 e | 1.92 ± 0.02 b | |
| Infusion | 16.06 | 46.08 ± 0.17 e | 6.85 ± 0.13 e | 1.67 ± 0.04 b,c |
| No. | Identified/Tentatively Annotated Compound | Molecular Formula | Exact Mass [M-H] | Fragmentation Pattern in (-) ESI-MS/MS | tR (Min) | Δ ppm | Distribution | Level of Identification (CAWG) |
|---|---|---|---|---|---|---|---|---|
| Phenolic acids and glycosides | ||||||||
| 1. | protocatechuic acid | C7H6O4 | 153.0181 | 153.0181 (17.85), 109.0280 (100) | 2.01 | −7.790 | 1,2,3,4,5,6,7,8 | 1 |
| 2. | caffeic acid | C9H8O4 | 179.0341 | 179.0338 (22.92), 135.0437 (100), 107.0488 (2.38) | 3.54 | −4.926 | 1,2,3,4,7,8 | 1 |
| 3. | neochlorogenic (3-caffeoylquinic) acid | C16H18O9 | 353.0873 | 353.0873 (39.89), 191.0551 (100), 179.0338 (65.22), 135.0438 (67.15) | 2.37 | −1.488 | 1,2,3,4,5,6,7,8 | 1 |
| 4. | chlorogenic (5-caffeoylquinic) acid | C16H18O9 | 353.0880 | 353.0880 (4.84), 191.0552 (100), 179.0334 (0.93), 161.0233 (2.30), 135.0436 (0.85) | 3.19 | 0.665 | 1,2,3,4,5,6,7,8 | 1 |
| 5. | primeverin/primulaverin | C20H28O13 | 475.1468 | 521.1525 (11.02), 475.1468 (4.90), 443.1199 (12.31), 293.0880 (100), 233.0674 (1.33), 181.0500 (10.28), 166.0261 (30.25), 149.0444 (9.37), 131.0332 (7.67) | 4.70 | 2.328 | 2,3,5,6,7,8 | 2 |
| Flavones and flavonols | ||||||||
| 6. | 4′-hydroxyflavone | C15H10O3 | 237.0555 | 237.0552 (100), 209.0603 (7.98), 117.0331 (65.16) | 9.42 | −1.044 | 1,2,3,4,5,6,7,8 | 1 |
| 7. | 7,4′-dihydroxyflavone | C15H10O4 | 253.0504 | 253.0504 (100), 225.0538 (1.05), 209.0609 (0.99), 133.0281 (51.31), 117.0330 (1.00) | 7.94 | −0.799 | 1,2,3,4,5,6,7 | 2 |
| 8. | 7,3′,4′-trihydroxyflavone | C15H10O5 | 269.0458 | 269.0458 (100), 241.8075 (0.35), 225.0551 (2.66), 201.0185 (13.80), 173.0233 (3.58), 153.0177 (0.50), 135.0073 (37.58) (1,3A), 133.0281 (25.40) (1,3B), 91.0173 (0,4A) | 8.17 | 0.867 | 1,2,3 | 2 |
| 9. | apigenin | C15H10O5 | 269.0454 | 269.0454 (100), 225.0555 (2.26), 204.9892 (0.99), 151.0022 (6.20), 149.0231 (5.05), 117.0330 (19.33), 107.0123 (4.96) | 8.60 | −0.396 | 1,2,3 | 1 |
| 10. | acacetin | C16H12O5 | 283.0612 | 283.0612 (100), 268.0378 (67.81), 211.0401 (1.79), 151.0023 (2.29), 133.0284 (0.48), 107.0122 (0.48) | 11.35 | 0.718 | 1,2,3,4 | 2 |
| 11. | luteolin | C15H10O6 | 285.0405 | 285.0404 (100), 257.0452 (0.37), 241.0504 (0.25), 151.0024 (4.61), 133.0281 (23.07), 107.0123 (4.33) | 7.56 | 0.136 | 1,2,3,4,5,6,7 | 1 |
| 12. | kaempferol | C15H10O6 | 285.0405 | 285.0404 (100), 178.9914 (0.49), 161.7458 (0.48), 151.0025 (2.07), 117.0331 (0.92), 107.0122 (0.98) | 8.82 | 0.031 | 1,2,3,4,5,6,7,8 | 1 |
| 13. | quercetin | C15H10O7 | 301.0355 | 301.0353 (100), 273.0403 (2.68), 257.0466 (1.25), 178.9975 (19.30), 161.0230 (0.40), 151.0024 (44.12), 121.0280 (13.69), 107.0123 (14.49) | 7.60 | 0.279 | 1,2,4,5,6,7,8 | 1 |
| 14. | cirsimaritin | C17H14O6 | 313.0719 | 313.0719 (100), 298.0484 (60.56), 283.0252 (2.28), 255.0291 (1.74), 163.0027 (1.02), 149.9946 (14.61), 133.0282 (20.31), 1798.9915 (1.35), 151.0023 (0.69) | 11.00 | 0.539 | 1,3 | 2 |
| 15. | dihydroxy-dimethoxy flavone | C17H14O6 | 313.0714 | 313.0714 (11.33), 298.0483 (100), 283.0245 (17.27), 255.0301 (3.48), 133.0280 (8.28), 116.9938 (1.29) | 11.18 | −1.122 | 1,3,4 | 3 |
| 16. | rhamnetin | C16H12O7 | 315.0507 | 315.0507 (90.55), 301.0298 (7.33), 300.0273 (100), 272.0330 (4.37), 255.0293 (2.53), 227.0333 (2.38), 165.9898 (12.11) | 8.80 | −0.971 | 1,2,3 | 1 |
| 17. | isorhamnetin | C16H12O7 | 315.0514 | 315.0511 (100), 301.0323 (3.08), 300.0275 (47.91), 151.0021 (11.36), 107.0121 (9.85) | 9.10 | 1.156 | 1,3 | 1 |
| 18. | 2′-hydroxyflavone-7-O-hexoside | C21H20O9 | 415.1037 | 415.1036 (13.39), 253.0503 (100), 225.0549 (1.28), 209.0602 (1.08), 133.0280 (32.83), 117.0331 (1.34) | 6.23 | 0.662 | 1 | 3 |
| 19. | kaempferol 3-O-pentoside | C20H18O10 | 417.0830 | 417.0830 (100), 285.0397 (10.33), 284.0326 (41.06), 255.0297 (31.74), 227.0339 (26.28), 211.0378 (2.32) | 6.09 | 0.624 | 1,2,3,4 | 2 |
| 20. | kaempferol 7-O-pentoside | C20H18O10 | 417.0830 | 417.0830 (100), 285.0405 (63.95), 284.0326 (66.41), 255.0298 (44.05), 227.0348 (31.22) | 6.33 | 0.696 | 1 | 2 |
| 21. | kaempferol 7-O-deoxyhexoside | C21H20O10 | 431.0984 | 431.0984 (100), 285.0403 (70.22), 284.0327 (54.75), 255.0298 (6.51), 227.0345 (31.17) | 6.58 | 0.186 | 1,2,3,4 | 2 |
| 22. | apigenin 7-O-glucoside | C21H20O10 | 431.0986 | 431.0982 (16.52), 269.0456 (100), 227.0336 (2.06), 241.0504 (0.89), 150.9850 (0.50), 133.0280 (11.87), 117.5589 (0.59) | 7.69 | 0.464 | 1,2,3,4 | 1 |
| 23. | quercetin 3-O-pentoside | C20H18O11 | 433.0774 | 433.0774 (100), 301.0350 (84.54), 300.0275 (86.50), 271.0251 (39.29), 255.0300 (19.52), 227.0324 (2.46), 178.9983 (1.91), 151.0022 (7.32), 107.0123 (2.35) | 5.74 | −0.565 | 1,2 | 2 |
| 24. | luteolin 7-O-glucoside | C21H20O11 | 447.0932 | 447.0932 (100), 285.0403 (90.49), 284.0326 (37.15), 227.0344 (1.65), 151.0022 (4.77), 133.0283 (3.73), 107.0122 (1.81) | 5.35 | −0.122 | 1,2,3,4 | 1 |
| 25. | luteolin 3′-O-hexoside | C21H20O11 | 447.0936 | 447.0927 (100), 285.0400 (22.81), 284.0326 (56.02), 255.029 (43.74) 227.0344 (40.57), 211.0397 (0.62), 151.0389 (5.68), 133.0283 (3.21), 107.0486 (0.63) | 5.90 | 0.773 | 1,2,5,6,8 | 2 |
| 26. | kaempferol 3-O-glucoside | C21H20O11 | 447.0935 | 447.0935 (100), 285.0393 (16.96), 284.0327 (51.53), 255.0298 (41.70), 227.0345 (40.54), 211.0412 (0.87), 151.0023 (1.10) | 5.63 | 0.571 | 1,2,3,4,5,6,7,8 | 1 |
| 27. | luteolin 3′-O-pentoside | C21H20O11 | 447.0936 | 447.0927 (100), 285.0400 (22.81), 284.0326 (56.02), 255.02977 (43.74), 227.0344 (40.57), 211.0397 (0.62), 151.0389 (5.68), 133.0283 (3.21), 107.0486 (0.63) | 5.90 | 0.773 | 1,2,5,6,8 | 2 |
| 28. | myricetin 3-O-pentoside | C20H18O12 | 449.0726 | 449.0716 (100), 317.0298 (28.80), 316.0223 (87.23), 287.0207 (14.49), 271.0246 (24.62), 178.9969 (2.77), 151.0019 (3.07) | 5.01 | 0.113 | 1,2,3,4 | 2 |
| 29. | 7,4′-dihydroxyflavone O-acetylhexoside | C23H22O10 | 457.1147 | 457.1142 (24.71), 397.0938 (7.06), 295.0595 (0.88), 253.0504 (100), 225.0548 (1.59), 209.0607 (1.28), 117.0326 (1.09), 133.0281 (37.57) | 7.06 | 1.597 | 1,2,3,4,5,6,7,8 | 3/4 |
| 30. | myricitrin (myricetin 3-O-rhamnoside) | C21H20O12 | 463.0886 | 463.0884 (100), 317.0295 (22.04), 316.0224 (84.26), 287.0198 (15.13), 271.0248 (28.78), 178.9973 (2.76), 151.0027 (5.24), 107.0123 (1.22) | 5.10 | 3.113 | 1,2,3,4,5,6,7,8 | 1 |
| 31. | isoquercitrin (quercetin 3-O-glucoside) | C21H20O12 | 463.0888 | 463.0883 (100), 301.0349 (41.27), 300.0274 (71.68), 271.0248 (3.70), 151.0026 (6.10), 107.0121 (2.54) | 5.27 | 1.319 | 1,2,3,4,5,6,7,8 | 1 |
| 32. | 7,4′-dihydroxyflavone O-hexoside-O-ethylmaleate | C27H28O13 | 559.1457 | 559.1469 (0.85), 457.1139 (4.62), 415.1035 (47.92), 253.0504 (100), 225.0548 (1.51), 209.0604 (1.70), 133.0281 (29.38), 117.0328 (0.59) | 7.07 | −0.043 | 1,2,3,4,5,6,7,8 | 4 |
| 33. | acaciin (acacetin 7-O-rutinoside) | C28H32O14 | 591.1730 | 591.1726 (16.76), 284.0646 (8.43), 283.0612 (100), 269.0413 (3.92), 268.0377 (43.72), 267.0296 (0.21), 240.0430 (1.02), 239.0346 (0.65), 151.0025 (0.51), 107.0125 (0.18) | 7.58 | 1.051 | 1,2,3,5 | 1 |
| 34. | kaempferol 4′-O-rutinoside | C27H30O15 | 593.1516 | 593.1516 (85.52), 285.0406 (100), 284.0329 (12.77) | 5.21 | 0.601 | 1,2,3,4,6,8 | 2 |
| 35. | kaempferol 3-O-rutinoside | C27H30O15 | 593.1514 | 593.1517 (100), 285.0400 (26.30), 284.0327 (54.97), 255.0298 (34.80), 227.0346 (22.41), 211.0393 (1.14), 151.0023 (2.39), 107.0123 (1.20) | 5.40 | 0.399 | 1,2,3,4,6,8 | 1 |
| 36. | kaempferol 7-O-rutinoside | C27H30O15 | 593.1515 | 593.1523 (100), 285.0406 (57.00), 284.0323 (34.01), 255.0307 (32.37), 227.0347 (10.38) | 5.63 | 0.500 | 1,2,3,4,5,6,7,8 | 2 |
| 37. | rutin (quercetin 3-O-rutinoside) | C27H30O16 | 609.1467 | 609.1468 (100), 301.0349 (31.23), 300.0276 (76.39), 271.0249 (40.61), 255.0297 (17.68), 178.9976 (3.20), 151.0024 (5.81), 107.0123 (2.36) | 5.00 | 1.120 | 1,2,3,4,5,6,7,8 | 1 |
| 38. | pectolinarin (pectolinarigenin 7-O-rutinoside) | C29H34O15 | 621.1831 | 621.1830 (11.33), 313.0720 (100), 298.0484 (19.01), 284.0285 (2.59), 283.0249 (33.40), 255.0300 (6.17), 227.0341 (1.49), 163.0025 (2.87), 117.0329 (1.36) | 7.66 | 0.993 | 1,2,3,4,5,6,7 | 1 |
| 39. | myricetin 3-O-rutinoside | C27H30O17 | 625.1418 | 625.1418 (100), 317.0292 (14.25), 316.0224 (72.60), 287.0196 (18.02), 271.0247 (22.26), 178.9972 (1.95), 151.0024 (6.58), 107.0116 (1.20) | 4.45 | 1.324 | 1,2,3,4,5,6,8 | 2 |
| 40. | kaempferol 3-O-hex-deoxyhex-deoxyhex | C33H40O19 | 739.2093 | 739.2108 (86.61), 285.0405 (100), 284.0326 (53.84), 255.0297 (56.20), 227.0346 (39.82), 211.0399 (3.86), 151.0027 (2.49), 135.0073 (1.92), 107.0125 (3.60) | 5.31 | 0.254 | 1,2,3,4,5,6,7,8 | 2 |
| 41. | quercetin 3-O-hex-deoxyhex-hex | C33H40O20 | 755.2040 | 755.2049 (100), 301.0348 (46.03), 300.0275 (78.15), 271.0248 (54.98), 255.0298 (25.98), 227.0339 (4.48), 151.0028 (10.89), 107.0120 (2.11) | 4.24 | 0.539 | 1,2,3,4 | 2 |
| 42. | kaempferol 3-O-hex-hex-hex | C33H40O21 | 771.1992 | 771.1996 (51.83), 285.0405 (100), 255.0295 (18.63), 227.0348 (12.85), 211.0397 (3.88), 151.0021 (1.07), 107.0122 (3.19) | 4.71 | 0.297 | 1,2,3,4,5,6,7,8 | 2 |
| 43. | quercetin 3-O-hex-hex-hex | C33H40O22 | 787.1943 | 787.1937 (83.60), 301.0351 (100), 300.0278 (79.80), 271.0247 (56.82), 255.0295 (20.25), 151.0024 (18.60), 107.0123 (3.69) | 4.24 | 0.539 | 1,2,3,4,5,7 | 2 |
| Chalcones and dihydochalcones | ||||||||
| 44. | hydroxychalcone | C15H12O2 | 223.0760 | 223.0759 (100), 205.0648 (0.38), 202.9935 (2.10), 195.0807 (49.88), 182.9862 (2.59), 145.0281 (1.59), 119.0487 (0.58), 117.0331 (16.14), 93.0330 (8.74) | 11.93 | −1.896 | 1,4 | 3 |
| 45. | dihydroxychalcone | C15H12O3 | 239.0709 | 239.0710 (28.19), 195.0808 (1.34), 121.0282 (1.08), 119.0487 (100), 117.0332 (3.36), 93.0330 (33.11) | 13.19 | −1.872 | 1,2,3,4,5,6,7,8 | 3 |
| 46. | dihydroxydihydrochalcone | C15H14O3 | 241.0868 | 241.0867 (20.44), 197.0960 (1.02), 147.0438 (7.62), 135.0437 (100), 121.0276 (0.18), 119.0486 (0.34), 117.0337 (0.34), 93.0330 (44.52) | 12.87 | −0.820 | 1,2,3,4,5,6,7 | 3 |
| 47. | trihydroxychalcone | C15H12O4 | 255.0663 | 255.0661 (25.50), 211.0757 (100), 183.0809 (0.79), 169.0648 (3.71), 143.0490 (4.45), 119.0486 (1.08), 93.0329 (1.86) | 9.63 | −0.008 | 1,3,4,7,8 | 3 |
| 48. | trihydroxydihydrochalcone | C15H14O4 | 257.0818 | 257.0818 (100), 213.0914 (94.48), 171.0802 (1.46), 151.0391 (0.27), 121.0280 (10.62), 117.5003 (0.22), 107.0487 (43.67), 93.0330 (4.79) | 9.71 | −0.670 | 2,3,4,5,6,7,8 | 3 |
| 49. | tetrahydroxychalcone | C15H12O5 | 271.0616 | 271.0609 (100), 151.0024 (65.21), 119.0490 (43.91), 107.0127 (18.87), 93.0330 (6.17) | 8.57 | 1.414 | 2,3,7,8 | 3 |
| 50. | tetrahydroxydihydrochalcone | C15H14O5 | 273.0769 | 273.0769 (100), 229.0866 (55.68), 121.0280 (17.11), 137.0231 (7.82), 107.0487 (44.35), 93.0329 (1.50) | 8.43 | 0.305 | 2,3,4,7 | 3 |
| Saponins | ||||||||
| 51. | primulasaponin isomer I(primula acid I) | C54H88O23 | 1103.5644 | 1103.5649 (100), 957.5015 (0.8), 923.5041 (2.8), 795.4615 (0.3), 553.3922 (0.5), 455.3522 (0.9), 437.3407 (0.3), 407.3314 (1.0) | 10.17 | 0.524 | 1,2,3,4,5,6,7,8 | 2 |
| 52. | primulasaponin(primula acid I) isomer II | C54H88O23 | 1103.5644 | 1103.5647 (100), 923.5104 (2.8), 795.4513 (0.5), 455.3532 (1.9) | 11.99 | 0.306 | 1,2,3,4,5,6,7,8 | 2 |
| 53. | priverosaponin B isomer I | C54H88O24 | 1119.5593 | 1119.5597 (100), 973.5049 (0.6), 957.5066 (0.6), 939.4974 (3.1), 811.4457 (0.4), 775.4327 (0.1), 749.4576 (0.1), 569.3849 (1.0), 473.3637 (0.9), 423.3281 (0.8), 407.2965 (0.3), 391.3006 (0.5) | 8.17 | 0.369 | 1,2,3,4,5,6,7,8 | 2 |
| 54. | priverosaponin B isomer II | C54H88O24 | 1119.5593 | 1119.5599 (100), 939.4963 (3.3), 811.4561 (0.3), 775.4206 (0.1), 569.3799 (0.9), 473.3624 (2.0), 423.2854 (0.3) | 8.94 | 0.593 | 1,2,3,4,5,6,7,8 | 2 |
| 55. | priverosaponin B isomer III | C54H88O24 | 1119.5593 | 1119.5603 (100), 973.4953 (0.3), 939.4949 (3.0), 811.4513 (0.3), 569.3843 (1.0), 473.3639 (1.1), 423.3256 (0.4) | 10.15 | 0.914 | 1,2,3,4,5,6,7,8 | 2 |
| 56. | priverosaponin B isomer IV | C54H88O24 | 1119.5593 | 1119.5601 (100), 939.4949 (3.5), 569.3826 (1.4), 423.3278 (0.7), 405.3184 (0.4) | 10.99 | 0.700 | 1,2,3,4,5,6,7,8 | 2 |
| 57. | primacrosaponin isomer I | C54H88O25 | 1135.5542 | 1135.5552 (100), 955.4901 (3.9) | 8.09 | 0.871 | 1,2,3,4,5,6,7,8 | 2 |
| 58. | primacrosaponin isomer II | C54H88O25 | 1135.5542 | 1135.5551 (100), 955.4885 (3.8), 585.3813 (0.5), 489.3595 (0.6) | 8.42 | 0.756 | 1,2,3,4,5,6,7,8 | 2 |
| 59. | priverosaponin B 22 acetate isomer I | C56H90O25 | 1161.5698 | 1161.5706 (100), 981.5074 (2.9), 611.3977 (1.2), 515.3732 (1.2) | 9.48 | 0.0.619 | 1,2,3,4,5,6,7,8 | 2 |
| 60. | priverosaponin B 22 acetateisomer II | C56H90O25 | 1161.5698 | 1161.5706 (100), 1101.5438 (0.2), 1015.5159 (0.6), 981.5071 (2.8), 853.4595 (0.5), 611.3957 (1.0), 583.4039 (0.5), 515.3780 (1.0), 405.3154 (0.2) | 9.65 | 0.619 | 1,2,4,5,6,7,8 | 2 |
| 61. | priverosaponin B 22 acetateisomer III | C56H90O25 | 1161.5698 | 1161.5703 (100), 1015.5275 (0.6), 981.5071 (3.1), 853.4584 (0.5), 611.3962 (0.6), 513.3572 (1.6) | 9.92 | 0.404 | 1,2,3,4,5,6,7,8 | 2 |
| 62. | priverosaponin B 22 acetateisomer IV | C56H90O25 | 1161.5698 | 1161.5701 (100), 981.5082 (2.8), 853.4612 (0.1), 835.4540 (0.1), 817.4373 (0.1), 673.4022 (0.1), 611.4022 (0.9), 583.4010 (0.5), 515.3792 (0.6), 473.3643 (1.6) | 10.49 | 0.197 | 1,2,3,4,5,6,7,8 | 2 |
| Other | ||||||||
| 63. | azelaic acid | C9H16O4 | 187.0966 | 187.0966 (49.74), 125.0957 (100), 123.0800 (1.46), 97.0642 (6.88) | 6.30 | −5.250 | 1,2,3,4,5,6,7,8 | 2 |
| 64. | quinic acid | C7H12O6 | 191.0553 | 191.0561 (100), 173.0450 (1.72), 127.0387 (4.07), 111.0440 (1.41), 93.0331 (5.06), 85.0279 (18.16) | 3.18 | −5.032 | 1,2,3,4,5,6,7,8 | 2 |
| 65. | dihydroresveratrol 3-O-hexoside | C20H24O8 | 391.1402 | 391.1401 (6.24), 229.0865 (100), 137.0229 (6.22), 122.0359 (45.14), 108.0201 (17.96), 93.0329 (2.61) | 4.38 | 0.995 | 1,2,3,5,6,7,8 | 3 |
| Plant Parts | Solvents | DPPH (mg TE/g Dry Extract) | ABTS (mg TE/g Dry Extract) | CUPRAC (mg TE/g Dry Extract) | FRAP (mg TE/g Dry Extract) | MCA (mg EDTAE/g Dry Extract) |
|---|---|---|---|---|---|---|
| Aerial parts | EA | 5.41 ± 0.23 g | 38.40 ± 0.95 e | 63.15 ± 0.54 g | 26.55 ± 0.11 h | 3.96 ± 0.48 g |
| EtOH | 52.22 ± 1.58 f | 75.51 ± 1.61 e | 136.92 ± 7.41 e | 68.96 ± 0.21 f | 6.66 ± 0.24 f | |
| EtOH/Water | 132.65 ± 2.91 c | 180.87 ± 3.72 c | 172.46 ± 1.26 c | 108.37 ± 0.70 c | 11.75 ± 0.28 d | |
| Infusion | 106.89 ± 3.01 d | 158.77 ± 8.98 c | 148.70 ± 1.75 d | 95.40 ± 0.46 d | 25.21 ± 0.09 a | |
| Rhizomes | EA | 47.85 ± 0.07 f | 79.02 ± 0.26 d | 99.65 ± 1.14 f | 51.08 ± 0.15 g | 13.40 ± 0.60 c |
| EtOH | 415.06 ± 1.36 a | 638.30 ± 21.10 a | 477.77 ± 3.34 a | 301.02 ± 1.00 a | 3.90 ± 0.09 g | |
| EtOH/Water | 297.12 ± 1.43 b | 457.89 ± 7.97 b | 327.21 ± 2.55 b | 209.70 ± 2.13 b | 8.06 ± 0.58 e | |
| Infusion | 97.18 ± 1.19 e | 166.73 ± 5.06 c | 142.74 ± 3.66 d,e | 81.50 ± 0.25 e | 21.95 ± 0.15 b |
| Plant Parts | Solvents | AChE (mg GALAE/g Dry Extract) | BChE (mg GALAE/g Dry Extract) | Tyrosinase (mg KAE/g Dry Extract) | Amylase (mmol ACAE/g Dry Extract) | Glucosidase (mmol ACAE/g Dry Extract) |
|---|---|---|---|---|---|---|
| Aerial parts | EA | 1.63 ± 0.21 c | 3.54 ± 0.23 a | 66.73 ± 0.81 d | 0.67 ± 0.01 a | 1.14 ± 0.01 b,c |
| EtOH | 2.16 ± 0.05 b | 1.03 ± 0.24 b,c | 67.94 ± 0.19 c,d | 0.47 ± 0.01 c | 1.13 ± 0.02 c | |
| EtOH/Water | 2.22 ± 0.04 b | 0.48 ± 0.06 c,d | 73.02 ± 1.01 a,b | 0.44 ± 0.01 d | 1.16 ± 0.01 a | |
| Infusion | 1.13 ± 0.08 d | na | 27.92 ± 3.40 e | 0.11 ± 0.01 g | 1.12 ± 0.01 c | |
| Rhizomes | EA | 2.31 ± 0.07 b | 3.45 ± 0.44 a | 70.98 ± 0.17 b,c | 0.54 ± 0.02 b | 1.15 ± 0.01 a,b |
| EtOH | 2.63 ± 0.01 a | 3.51 ± 0.06 a | 76.36 ± 0.36 a | 0.42 ± 0.01 d | 1.16 ± 0.01 a | |
| EtOH/Water | 2.61 ± 0.01 a | 3.39 ± 0.08 a | 73.64 ± 0.20 a,b | 0.37 ± 0.01 e | 1.16 ± 0.01 a | |
| Infusion | 1.17 ± 0.04 d | 1.49 ± 0.14 b | 16.58 ± 0.90 f | 0.18 ± 0.01 f | 1.15 ± 0.01 a,b |
| Compound | AChE | BChE | Tyrosinase | Amylase | Glucosidase | MMP-9 | BCL-2 |
|---|---|---|---|---|---|---|---|
| Kcal/mol | |||||||
| protocatechuic acid | −7.14 | −6.41 | −6.44 | −6.35 | −8.12 | −8.28 | −4.64 |
| kaempferol 3-O-rutinoside | −11.21 | −11.92 | −6.17 | −9.95 | −10.65 | −11.35 | −10.04 |
| rutin (quercetin 3-O-glucoside) | −13.89 | −11.63 | −6.23 | −9.88 | −10.33 | −9.62 | −8.8 |
| 4′-hydroxyflavone | −4.58 | −4.38 | −5.41 | −5.33 | −4.87 | −3.57 | −5.12 |
| 4-hydroxychalcone | −8.87 | −6.89 | −5.78 | −6.88 | −6.90 | −6.09 | −6.22 |
| myricitrin (myricetin 3-O-rhamnoside) | −11.35 | −7.70 | −6.21 | −6.71 | −12.01 | −10.00 | −5.81 |
| primulasaponin | - | - | - | - | - | −8.79 | −10.41 |
| azelaic acid | −6.25 | −4.66 | −6.56 | −4.78 | −6.82 | −9.00 | −2.17 |
| dihydroresveratrol 3-O-hexoside | −12.85 | −8.19 | −5.70 | −6.87 | −9.67 | −8.02 | −5.00 |
| Compound | GI Absorption | BBB Permeant | CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | Lipinksi Rule | PAINS |
|---|---|---|---|---|---|---|---|---|---|
| protocatechuic acid | High | No | No | No | No | No | No | Yes; 0 violation | 0 alerts |
| kaempferol 3-O-rutinoside | Low | No | No | No | No | No | No | No; 3 violations: MW > 500; NorO > 10; NHorOH > 5 | 0 alerts |
| rutin (quercetin 3-O-glucoside) | Low | No | No | No | No | No | No | No; 3 violations: MW > 500; NorO > 10; NHorOH > 5 | 1 alert: catechol amine |
| 4′-hydroxyflavone | High | Yes | No | No | No | No | No | Yes; 0 violation | 0 alerts |
| hydroxychalcone | High | Yes | No | No | No | No | No | Yes; 0 violation | 1 alert: Michael acceptor |
| myricitrin (myricetin 3-O-rhamnoside) | Low | No | No | No | No | No | No | No; 2 violations: NorO > 10; NHorOH > 5 | 1 alert: catechol amine |
| Primulasaponin | Low | No | No | No | No | No | No | No; 3 violations: MW > 500; NorO > 10; NHorOH > 5 | |
| azelaic acid | High | Yes | No | No | No | No | No | Yes; 0 violation | 0 alerts |
| dihydroresveratrol 3-O-hexoside | Low | No | No | No | No | No | No | No; 1 violation: NorO > 5 | 0 alerts |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurt-Celep, I.; Zheleva-Dimitrova, D.; Gevrenova, R.; Uba, A.I.; Zengin, G.; Yıldıztugay, E.; Picot-Allain, C.M.N.; Lorenzo, J.M.; Mahomoodally, M.F.; Montesano, D. An In-Depth Study on the Metabolite Profile and Biological Properties of Primula auriculata Extracts: A Fascinating Sparkle on the Way from Nature to Functional Applications. Antioxidants 2022, 11, 1377. https://doi.org/10.3390/antiox11071377
Kurt-Celep I, Zheleva-Dimitrova D, Gevrenova R, Uba AI, Zengin G, Yıldıztugay E, Picot-Allain CMN, Lorenzo JM, Mahomoodally MF, Montesano D. An In-Depth Study on the Metabolite Profile and Biological Properties of Primula auriculata Extracts: A Fascinating Sparkle on the Way from Nature to Functional Applications. Antioxidants. 2022; 11(7):1377. https://doi.org/10.3390/antiox11071377
Chicago/Turabian StyleKurt-Celep, Inci, Dimitrina Zheleva-Dimitrova, Reneta Gevrenova, Abdullahi Ibrahim Uba, Gokhan Zengin, Evren Yıldıztugay, Carene Marie Nancy Picot-Allain, José Manuel Lorenzo, Mohamad Fawzi Mahomoodally, and Domenico Montesano. 2022. "An In-Depth Study on the Metabolite Profile and Biological Properties of Primula auriculata Extracts: A Fascinating Sparkle on the Way from Nature to Functional Applications" Antioxidants 11, no. 7: 1377. https://doi.org/10.3390/antiox11071377










