BACH1 Expression Is Promoted by Tank Binding Kinase 1 (TBK1) in Pancreatic Cancer Cells to Increase Iron and Reduce the Expression of E-Cadherin
Abstract
:1. Introduction
2. Methods
2.1. Reagents
2.2. Cells and Cell Culture
2.3. Western Blotting
2.4. Antibodies
2.5. siRNAs
2.6. Quantitative Real-Time PCR
2.7. Immunofluorescent Staining
2.8. Detection of Labile Iron and Cell Death
2.9. RNA Sequence
2.10. The Analysis and the Visualization of Public Data
2.11. Statistics
3. Results
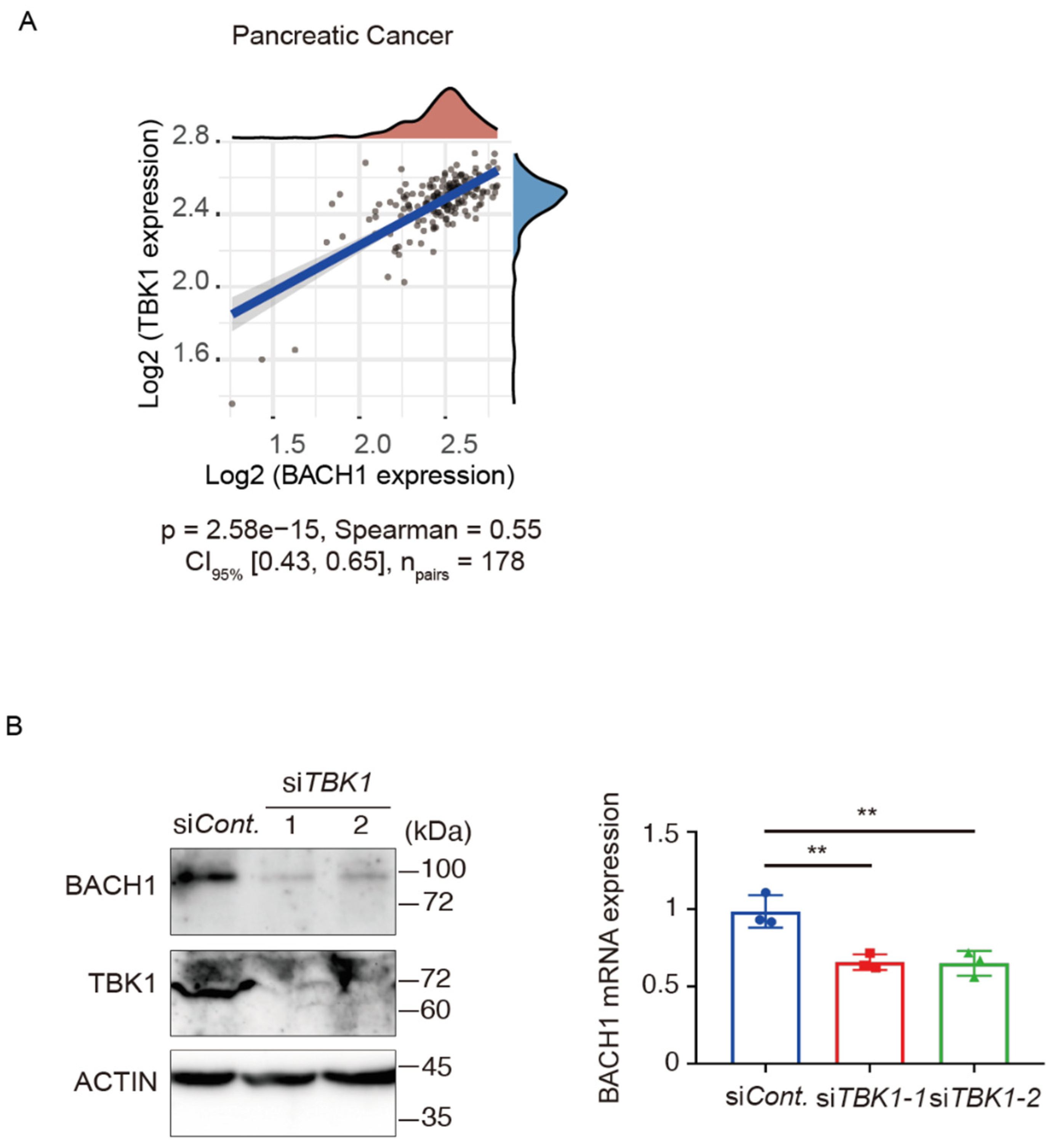
3.1. TBK1 Promotes BACH1 Expression and Accumulation
3.2. TBK1-BACH1 Pathway Regulates Iron Homeostasis and Cell Migration
3.3. BACH1 Increases the Iron Content by Reducing the Expression of Ferritin
3.4. Iron Availability Regulates BACH1 and E-Cadherin Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robbins, E.; Pedrson, H. Iron: Its intracellular localization and possible role in cell division. Proc. Natl. Acad. Sci. USA 1970, 66, 1244–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudland, P.S.; Durbin, H.; Clingan, D.; de Asua, L.J. Iron salts and transferrin are specifically required for cell division of cultured 3T6 cells. Biochem. Biophys. Res. Commun. 1977, 75, 556–562. [Google Scholar] [CrossRef]
- White, M.F.; Dillingham, M.S. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C. Essential functions of iron-requiring proteins in DNA replication, repair and cell cycle control. Protein. Cell 2014, 5, 750–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuss, J.O.; Tsai, C.L.; Ishida, J.P.; Tainer, J.A. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta 2015, 1853, 1253–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig, S.; Ramos-Alonso, L.; Romero, A.M.; Martinez-Pastor, M.T. The elemental role of iron in DNA synthesis and repair. Metallomics 2017, 9, 1483–1500. [Google Scholar] [CrossRef] [Green Version]
- Read, A.D.; Bentley, R.E.; Archer, S.L.; Dunham-Snary, K.J. Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox. Biol. 2021, 47, 102164. [Google Scholar] [CrossRef]
- Kiley, P.J.; Beinert, H. Oxygen sensing by the global regulator, FNR: The role of the iron-sulfur cluster. FEMS Microbiol. Rev. 1998, 22, 341–352. [Google Scholar] [CrossRef]
- Kobayashi, K.; Fujikawa, M.; Kozawa, T. Oxidative stress sensing by the iron-sulfur cluster in the transcription factor, SoxR. J. Inorg. Biochem. 2014, 133, 87–91. [Google Scholar] [CrossRef]
- Barton, J.K.; Silva, R.M.B.; O’Brien, E. Redox Chemistry in the Genome: Emergence of the [4Fe4S] Cofactor in Repair and Replication. Annu. Rev. Biochem. 2019, 88, 163–190. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, C.; Wu, Q.; An, P.; Huang, L.; Wang, J.; Chen, C.; Chen, X.; Zhang, F.; Ma, L.; et al. Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat. Commun. 2019, 10, 2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, C.; Yang, C.G.; He, C. A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 2009, 42, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, S.; Comer, J.M.; Konduri, P.C.; Shah, A.; Wang, T.; Lewis, A.; Shoffner, G.; Guo, F.; Zhang, L. Heme promotes transcriptional and demethylase activities of Gis1, a member of the histone demethylase JMJD2/KDM4 family. Nucleic Acids Res. 2018, 46, 215–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Sheng, X.; Chang, Z.; Wu, Q.; Wang, S.; Xuan, Z.; Li, D.; Wu, Y.; Shang, Y.; Kong, X.; et al. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014, 7, 180–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Chen, X. P53 tumor suppressor and iron homeostasis. FEBS J. 2019, 286, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Fenton, H.J.H. LXXIII.—Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 1894, 65, 899–911. [Google Scholar] [CrossRef] [Green Version]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Conrad, M.; Pratt, D.A. The chemical basis of ferroptosis. Nat. Chem. Biol. 2019, 15, 1137–1147. [Google Scholar] [CrossRef]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin protein nanocages use ion channels, catalytic sites, and nucleation channels to manage iron/oxygen chemistry. Curr. Opin. Chem. Biol. 2011, 15, 304–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torti, S.V.; Torti, F.M. Iron and cancer: More ore to be mined. Nat. Rev. Cancer 2013, 13, 342–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.F.; Zhang, J.; Su, Y.; Shen, Y.Y.; Jiang, D.X.; Hou, Y.Y.; Geng, M.Y.; Ding, J.; Chen, Y. G9a regulates breast cancer growth by modulating iron homeostasis through the repression of ferroxidase hephaestin. Nat. Commun. 2017, 8, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yu, L.; Ding, J.; Chen, Y. Iron Metabolism in Cancer. Int. J. Mol. Sci. 2018, 20, 95. [Google Scholar] [CrossRef] [Green Version]
- El Hout, M.; Dos Santos, L.; Hamai, A.; Mehrpour, M. A promising new approach to cancer therapy: Targeting iron metabolism in cancer stem cells. Semin. Cancer. Biol. 2018, 53, 125–138. [Google Scholar] [CrossRef]
- Prior, R.; Reifenberger, G.; Wechsler, W. Transferrin receptor expression in tumours of the human nervous system: Relation to tumour type, grading and tumour growth fraction. Virchows Arch. A Pathol. Anat. Histopathol. 1990, 416, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Das Gupta, A.; Shah, V.I. Correlation of transferrin receptor expression with histologic grade and immunophenotype in chronic lymphocytic leukemia and non-Hodgkin’s lymphoma. Hematol. Pathol. 1990, 4, 37–41. [Google Scholar] [PubMed]
- Habeshaw, J.A.; Lister, T.A.; Stansfeld, A.G.; Greaves, M.F. Correlation of transferrin receptor expression with histological class and outcome in non-Hodgkin lymphoma. Lancet 1983, 1, 498–501. [Google Scholar] [CrossRef]
- Seymour, G.J.; Walsh, M.D.; Lavin, M.F.; Strutton, G.; Gardiner, R.A. Transferrin receptor expression by human bladder transitional cell carcinomas. Urol. Res. 1987, 15, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.M.; Lee, J.; Finley, L.W.; Schmidt, P.J.; Fleming, M.D.; Haigis, M.C. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene 2015, 34, 2115–2124. [Google Scholar] [CrossRef] [Green Version]
- Leung, L.; Radulovich, N.; Zhu, C.Q.; Organ, S.; Bandarchi, B.; Pintilie, M.; To, C.; Panchal, D.; Tsao, M.S. Lipocalin2 promotes invasion, tumorigenicity and gemcitabine resistance in pancreatic ductal adenocarcinoma. PLoS ONE 2012, 7, e46677. [Google Scholar] [CrossRef] [PubMed]
- Kakhlon, O.; Gruenbaum, Y.; Cabantchik, Z.I. Repression of ferritin expression modulates cell responsiveness to H-ras-induced growth. Biochem. Soc. Trans. 2002, 30, 777–780. [Google Scholar] [CrossRef]
- Kakhlon, O.; Gruenbaum, Y.; Cabantchik, Z.I. Ferritin expression modulates cell cycle dynamics and cell responsiveness to H-ras-induced growth via expansion of the labile iron pool. Biochem. J. 2002, 363, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.P.; Elliott, R.L.; Head, J.F. Manipulation of iron transporter genes results in the suppression of human and mouse mammary adenocarcinomas. Anticancer Res. 2010, 30, 759–765. [Google Scholar] [PubMed]
- Xue, D.; Zhou, C.X.; Shi, Y.B.; Lu, H.; He, X.Z. Decreased expression of ferroportin in prostate cancer. Oncol. Lett. 2015, 10, 913–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyake, T.; Itoh, K.; Motohashi, H.; Hayashi, N.; Hoshino, H.; Nishizawa, M.; Yamamoto, M.; Igarashi, K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell Biol. 1996, 16, 6083–6095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hintze, K.J.; Katoh, Y.; Igarashi, K.; Theil, E.C. Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, beta-globin, and NADP(H) quinone (oxido) reductase1. J. Biol. Chem. 2007, 282, 34365–34371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, H.; Matsumoto, M.; Shindo, T.; Saigusa, D.; Kato, H.; Suzuki, K.; Sato, M.; Ishii, Y.; Shimokawa, H.; Igarashi, K. Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J. Biol. Chem. 2020, 295, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Marro, S.; Chiabrando, D.; Messana, E.; Stolte, J.; Turco, E.; Tolosano, E.; Muckenthaler, M.U. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position -7007 of the FPN1 promoter. Haematologica 2010, 95, 1261–1268. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, K.; Sun, J.; Taketani, S.; Nakajima, O.; Nishitani, C.; Sassa, S.; Hayashi, N.; Yamamoto, M.; Shibahara, S.; Fujita, H.; et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001, 20, 2835–2843. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Tian, Q.; Steuerwald, N.M.; Schrum, L.W.; Bonkovsky, H.L. The let-7 microRNA enhances heme oxygenase-1 by suppressing Bach1 and attenuates oxidant injury in human hepatocytes. Biochim. Biophys. Acta 2012, 1819, 1113–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igarashi, K.; Nishizawa, H.; Saiki, Y.; Matsumoto, M. The transcription factor BACH1 at the crossroads of cancer biology: From epithelial-mesenchymal transition to ferroptosis. J. Biol. Chem. 2021, 297, 101032. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, H.; Lei, R.; Chong, R.A.; Wei, Y.; Lu, X.; Tagkopoulos, I.; Kung, S.Y.; Yang, Q.; Hu, G.; et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J. Biol. Chem. 2012, 287, 33533–33544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Yesilkanal, A.E.; Wynne, J.P.; Frankenberger, C.; Liu, J.; Yan, J.; Elbaz, M.; Rabe, D.C.; Rustandy, F.D.; Tiwari, P.; et al. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature 2019, 568, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Matsumoto, M.; Saiki, Y.; Alam, M.; Nishizawa, H.; Rokugo, M.; Brydun, A.; Yamada, S.; Kaneko, M.K.; Funayama, R.; et al. BACH1 Promotes Pancreatic Cancer Metastasis by Repressing Epithelial Genes and Enhancing Epithelial-Mesenchymal Transition. Cancer Res. 2020, 80, 1279–1292. [Google Scholar] [CrossRef] [Green Version]
- Lignitto, L.; LeBoeuf, S.E.; Homer, H.; Jiang, S.; Askenazi, M.; Karakousi, T.R.; Pass, H.I.; Bhutkar, A.J.; Tsirigos, A.; Ueberheide, B.; et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 2019, 178, 316–329.e318. [Google Scholar] [CrossRef] [PubMed]
- Wiel, C.; Le Gal, K.; Ibrahim, M.X.; Jahangir, C.A.; Kashif, M.; Yao, H.; Ziegler, D.V.; Xu, X.; Ghosh, T.; Mondal, T.; et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 2019, 178, 330–345.e322. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.F.; Meloche, S.; Servant, M.J. The IKK-related kinases: From innate immunity to oncogenesis. Cell Res. 2008, 18, 889–899. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Wang, L.; Berman, M.; Kong, Y.Y.; Dorf, M.E. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 2011, 35, 426–440. [Google Scholar] [CrossRef] [Green Version]
- Sonia, S.; Benjamin, R.; Nathalie, G.; Guo, P.Z.; Rongtuan, L.; John, H. Triggering the interferon antiviral response through an IKK-related pathway. Science 2003, 300, 1148–1151. [Google Scholar]
- Zhao, P.; Wong, K.I.; Sun, X.; Reilly, S.M.; Uhm, M.; Liao, Z.; Skorobogatko, Y.; Saltiel, A.R. TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell 2018, 172, 731–743.e712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yum, S.; Li, M.; Fang, Y.; Chen, Z.J. TBK1 recruitment to STING activates both IRF3 and NF-kappaB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. USA 2021, 118, e2100225118. [Google Scholar] [CrossRef]
- Balka, K.R.; Louis, C.; Saunders, T.L.; Smith, A.M.; Calleja, D.J.; D’Silva, D.B.; Moghaddas, F.; Tailler, M.; Lawlor, K.E.; Zhan, Y.; et al. TBK1 and IKKε Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells. Cell Rep. 2020, 31, 107492. [Google Scholar] [CrossRef] [PubMed]
- Basit, A.; Cho, M.G.; Kim, E.Y.; Kwon, D.; Kang, S.J.; Lee, J.H. The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp. Mol. Med. 2020, 52, 643–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, K.A.; McWhirter, S.M.; Faia, K.L.; Rowe, D.C.; Latz, E.; Golenbock, D.T.; Coyle, A.J.; Liao, S.M.; Maniatis, T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003, 4, 491–496. [Google Scholar] [CrossRef]
- Chien, Y.; Kim, S.; Bumeister, R.; Loo, Y.M.; Kwon, S.W.; Johnson, C.L.; Balakireva, M.G.; Romeo, Y.; Kopelovich, L.; Gale, M., Jr.; et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006, 127, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.H.; Torres, M.; Ram, R.; Formstecher, E.; Roland, C.; Cheng, T.; Brekken, R.; Wurz, R.; Tasker, A.; Polverino, T.; et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell 2011, 41, 458–470. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Cao, Y.; Yang, X.; Zheng, Z.; Guan, K.; Wang, Q.; Tai, Y.; Zhang, Y.; Ma, S.; Cao, Y.; et al. Elevated expression of TANK-binding kinase 1 enhances tamoxifen resistance in breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E601–E610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.M.; Ou, Y.H.; McMillan, E.A.; Vaden, R.M.; Zaman, A.; Bodemann, B.O.; Makkar, G.; Posner, B.A.; White, M.A. TBK1 provides context-selective support of the activated AKT/mTOR pathway in lung cancer. Cancer Res. 2017, 77, 5077–5094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparrer, K.M.J.; Gableske, S.; Zurenski, M.A.; Parker, Z.M.; Full, F.; Baumgart, G.J.; Kato, J.; Pacheco-Rodriguez, G.; Liang, C.; Pornillos, O.; et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2017, 2, 1543–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, J.N.S.; Wang, C.; Bunker, E.; Hao, L.; Maric, D.; Schiavo, G.; Randow, F.; Youle, R.J. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell 2019, 74, 347–362.e346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozawa, T.; Sano, S.; Minowa-Nozawa, A.; Toh, H.; Nakajima, S.; Murase, K.; Aikawa, C.; Nakagawa, I. TBC1D9 regulates TBK1 activation through Ca(2+) signaling in selective autophagy. Nat. Commun. 2020, 11, 770. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xie, H.; Liu, X.; Potjewyd, F.; James, L.I.; Wilkerson, E.M.; Herring, L.E.; Xie, L.; Chen, X.; Cabrera, J.C.; et al. TBK1 is a synthetic lethal target in cancer with VHL loss. Cancer Discov. 2020, 10, 460–475. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, J.M.; Dowdle, W.E.; DeJesus, R.; Wang, Z.; Bergman, P.; Kobylarz, M.; Lindeman, A.; Xavier, R.J.; McAllister, G.; Nyfeler, B.; et al. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep. 2017, 20, 2341–2356. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, M.; Kondo, K.; Shiraki, T.; Brydun, A.; Funayama, R.; Nakayama, K.; Yaegashi, N.; Katagiri, H.; Igarashi, K. Genomewide approaches for BACH1 target genes in mouse embryonic fibroblasts showed BACH1-Pparg pathway in adipogenesis. Genes Cells 2016, 21, 553–567. [Google Scholar] [CrossRef] [Green Version]
- Taheem, D.K.; Foyt, D.A.; Loaiza, S.; Ferreira, S.A.; Ilic, D.; Auner, H.W.; Grigoriadis, A.E.; Jell, G.; Gentleman, E. Differential Regulation of Human Bone Marrow Mesenchymal Stromal Cell Chondrogenesis by Hypoxia Inducible Factor-1alpha Hydroxylase Inhibitors. Stem. Cells 2018, 36, 1380–1392. [Google Scholar] [CrossRef] [Green Version]
- Leermakers, P.A.; Remels, A.H.V.; Zonneveld, M.I.; Rouschop, K.M.A.; Schols, A.; Gosker, H.R. Iron deficiency-induced loss of skeletal muscle mitochondrial proteins and respiratory capacity; the role of mitophagy and secretion of mitochondria-containing vesicles. FASEB J. 2020, 34, 6703–6717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallander, M.L.; Leibold, E.A.; Eisenstein, R.S. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 2006, 1763, 668–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muckenthaler, M.U.; Galy, B.; Hentze, M.W. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008, 28, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Watanabe-Matsui, M. Wearing red for signaling: The heme-bach axis in heme metabolism, oxidative stress response and iron immunology. Tohoku J. Exp. Med. 2014, 232, 229–253. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Tashiro, S.; Hira, S.; Sun, J.; Yamazaki, C.; Zenke, Y.; Ikeda-Saito, M.; Yoshida, M.; Igarashi, K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004, 23, 2544–2553. [Google Scholar] [CrossRef] [Green Version]
- Zenke-Kawasaki, Y.; Dohi, Y.; Katoh, Y.; Ikura, T.; Ikura, M.; Asahara, T.; Tokunaga, F.; Iwai, K.; Igarashi, K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell Biol. 2007, 27, 6962–6971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, K.J.; Sharp, P.A. Iron elevates mesenchymal and metastatic biomarkers in HepG2 cells. Sci. Rep. 2020, 10, 21926. [Google Scholar] [CrossRef]
- Gomes, A.C.; Moreira, A.C.; Mesquita, G.; Gomes, M.S. Modulation of Iron Metabolism in Response to Infection: Twists for All Tastes. Pharmaceuticals 2018, 11, 84. [Google Scholar] [CrossRef] [Green Version]
- Ratledge, C. Iron metabolism and infection. Food Nutr. Bull. 2007, 28, S515–S523. [Google Scholar] [CrossRef]
- Drakesmith, H.; Prentice, A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, D.; Yue, F.; Zheng, M.; Kovacevic, Z.; Richardson, D.R. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1). J. Biol. Chem. 2012, 287, 17016–17028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, H.; Liu, L.; Liu, Z.; Qin, H.; Liao, Z.; Xia, P.; Yang, Y.; Li, B.; Gao, F.; Cai, J. Blocking TBK1 alleviated radiation-induced pulmonary fibrosis and epithelial-mesenchymal transition through Akt-Erk inactivation. Exp. Mol. Med. 2019, 51, 1–17. [Google Scholar] [CrossRef]
- Liu, W.; Huang, Y.J.; Liu, C.; Yang, Y.Y.; Liu, H.; Cui, J.G.; Cheng, Y.; Gao, F.; Cai, J.M.; Li, B.L. Inhibition of TBK1 attenuates radiation-induced epithelial-mesenchymal transition of A549 human lung cancer cells via activation of GSK-3beta and repression of ZEB1. Lab. Investig. 2014, 94, 362–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.M.; Jung, Y.; Lee, J.M.; Kim, W.; Cho, J.K.; Jeong, J.; Kim, S.J. Loss of TBK1 induces epithelial-mesenchymal transition in the breast cancer cells by ERalpha downregulation. Cancer Res. 2013, 73, 6679–6689. [Google Scholar] [CrossRef] [Green Version]
- Cruz, V.H.; Arner, E.N.; Du, W.; Bremauntz, A.E.; Brekken, R.A. Axl-mediated activation of TBK1 drives epithelial plasticity in pancreatic cancer. JCI Insight 2019, 5, e126117. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Kitamuro, T.; Takahashi, K.; Ogawa, K.; Udono-Fujimori, R.; Takeda, K.; Furuyama, K.; Nakayama, M.; Sun, J.; Fujita, H.; Hida, W.; et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 2003, 278, 9125–9133. [Google Scholar] [CrossRef] [Green Version]
- Ponka, P.; Schulman, H.M. Acquisition of iron from transferrin regulates reticulocyte heme synthesis. J. Biol. Chem. 1985, 260, 14717–14721. [Google Scholar] [CrossRef]






| siRNA Name | Accession Name | Sequence (5′-3′) |
|---|---|---|
| siBACH1-1 | NCBI Gene ID 571 | GGUCAAAGGACUUUCACAACAUUAA |
| siBACH1-2 | NCBI Gene ID 571 | GGGCACCAGGGAAGAUAGUAGUGUU |
| siTBK1-1 | NCBI Gene ID 29110 | GGACUACCAGAAUCUGAAUUCUUAA |
| siTBK1-2 | NCBI Gene ID 29110 | GCGAGAUGUGGUGGGUGGAAUGAAU |
| siTBK1-3 | NCBI Gene ID 29110 | GGGAACCUCUGAAUACCAUAGGAUU |
| Gene Name | Accession Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| BACH1 | NCBI Gene ID 571 | GTTACTTCCACTCAAGAATCGT | ACATTTGCACACTTCATCCA |
| CDH1 | NCBI Gene ID 999 | TCCTGGCCTCAGAAGACAGA | CCTTGGCCAGTGATGCTGTA |
| FBXO22 | NCBI Gene ID 26263 | ATTGCTTGGTTCGCGTGGTA | GCTCTCTTATGGCCACGACA |
| FTH1 | NCBI Gene ID 2495 | TGAAGCTGCAGAACCAACGAGG | GCACACTCCATTGCATTCAGCC |
| FTL | NCBI Gene ID 2512 | TACGAGCGTCTCCTGAAGATGC | GGTTCAGCTTTTTCTCCAGGGC |
| HMOX1 | NCBI Gene ID 3162 | TTTCAGAAGGGCCAGGTGAC | AGTAGACAGGGGCGAAGACT |
| IRP1 | NCBI Gene ID 48 | TGCTTCCTCAGGTGATTGGCTACA | TAGCTCGGTCAGCAATGGACAACT |
| IRP2 | NCBI Gene ID 3658 | ACCAGAGGTGGTTGGATGTGAGTT | ACTCCTACTTGCCTGAGGTGCTTT |
| MMP7 | NCBI Gene ID 4316 | ATCATGATTGGCTTTGCGCG | CCAGCGTTCATCCTCATCGA |
| RPL13A | NCBI Gene ID 23521 | TCGTACGCTGTGAAGGCATC | GTGGGGCAGCATACCTCG |
| SLC40A1 | NCBI Gene ID 30061 | GATCCTTGGCCGACTACCTG | CACATCCGATCTCCCCAAGT |
| SNAI2 | NCBI Gene ID 6591 | CAACGCCTCCAAAAAGCCAA | ACAGTGATGGGGCTGTATGC |
| TBK1 | NCBI Gene ID 29110 | AGCGGCAGAGTTAGGTGAAA | CCAGTGATCCACCTGGAGAT |
| VIM | NCBI Gene ID 7431 | GGACCAGCTAACCAACGACA | GGGTGTTTTCGGCTTCCTCT |
| Gene | NCBI Reference Sequence |
|---|---|
| BACH1 | NG_029658.2 |
| CDH1 | NG_008021.1 |
| FTH1 | NG_008346.1 |
| FTL | NG_008152.1 |
| SLC40A1 | NG_009027.1 |
| TBK1 | NG_046906.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Matsumoto, M.; Matsui-Watanabe, M.; Ochiai, K.; Callens, B.K.K.; Nguyen, L.C.; Kozuki, Y.; Tanaka, M.; Nishizawa, H.; Igarashi, K. BACH1 Expression Is Promoted by Tank Binding Kinase 1 (TBK1) in Pancreatic Cancer Cells to Increase Iron and Reduce the Expression of E-Cadherin. Antioxidants 2022, 11, 1460. https://doi.org/10.3390/antiox11081460
Liu L, Matsumoto M, Matsui-Watanabe M, Ochiai K, Callens BKK, Nguyen LC, Kozuki Y, Tanaka M, Nishizawa H, Igarashi K. BACH1 Expression Is Promoted by Tank Binding Kinase 1 (TBK1) in Pancreatic Cancer Cells to Increase Iron and Reduce the Expression of E-Cadherin. Antioxidants. 2022; 11(8):1460. https://doi.org/10.3390/antiox11081460
Chicago/Turabian StyleLiu, Liang, Mitsuyo Matsumoto, Miki Matsui-Watanabe, Kyoko Ochiai, Bert K. K. Callens, Long Chi Nguyen, Yushi Kozuki, Miho Tanaka, Hironari Nishizawa, and Kazuhiko Igarashi. 2022. "BACH1 Expression Is Promoted by Tank Binding Kinase 1 (TBK1) in Pancreatic Cancer Cells to Increase Iron and Reduce the Expression of E-Cadherin" Antioxidants 11, no. 8: 1460. https://doi.org/10.3390/antiox11081460
APA StyleLiu, L., Matsumoto, M., Matsui-Watanabe, M., Ochiai, K., Callens, B. K. K., Nguyen, L. C., Kozuki, Y., Tanaka, M., Nishizawa, H., & Igarashi, K. (2022). BACH1 Expression Is Promoted by Tank Binding Kinase 1 (TBK1) in Pancreatic Cancer Cells to Increase Iron and Reduce the Expression of E-Cadherin. Antioxidants, 11(8), 1460. https://doi.org/10.3390/antiox11081460






