BAG9 Confers Thermotolerance by Regulating Cellular Redox Homeostasis and the Stability of Heat Shock Proteins in Solanum lycopersicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis and Structural Domain Prediction of BAG Family
2.2. Plant Material, Growth Condition, and Heat Treatment
2.3. Construction of Plant Expression Vector and Tomato Transformation
2.4. Total RNA Extraction and Gene-Expression Analysis
2.5. Gas Exchange and Chlorophyll Fluorescence Measurements
2.6. Analysis of H2O2, O2•− and Malondialdehyde (MDA)
2.7. Antioxidant and Enzyme Activity Assays
2.8. Immunoblotting Assay
2.9. Yeast Two-Hybrid (Y2H) Screen and Assays, and Bimolecular Fluorescence Complementation (BiFC) Assay
2.10. Statistical Analysis
3. Results
3.1. Identification of BAG Homologs in Plants
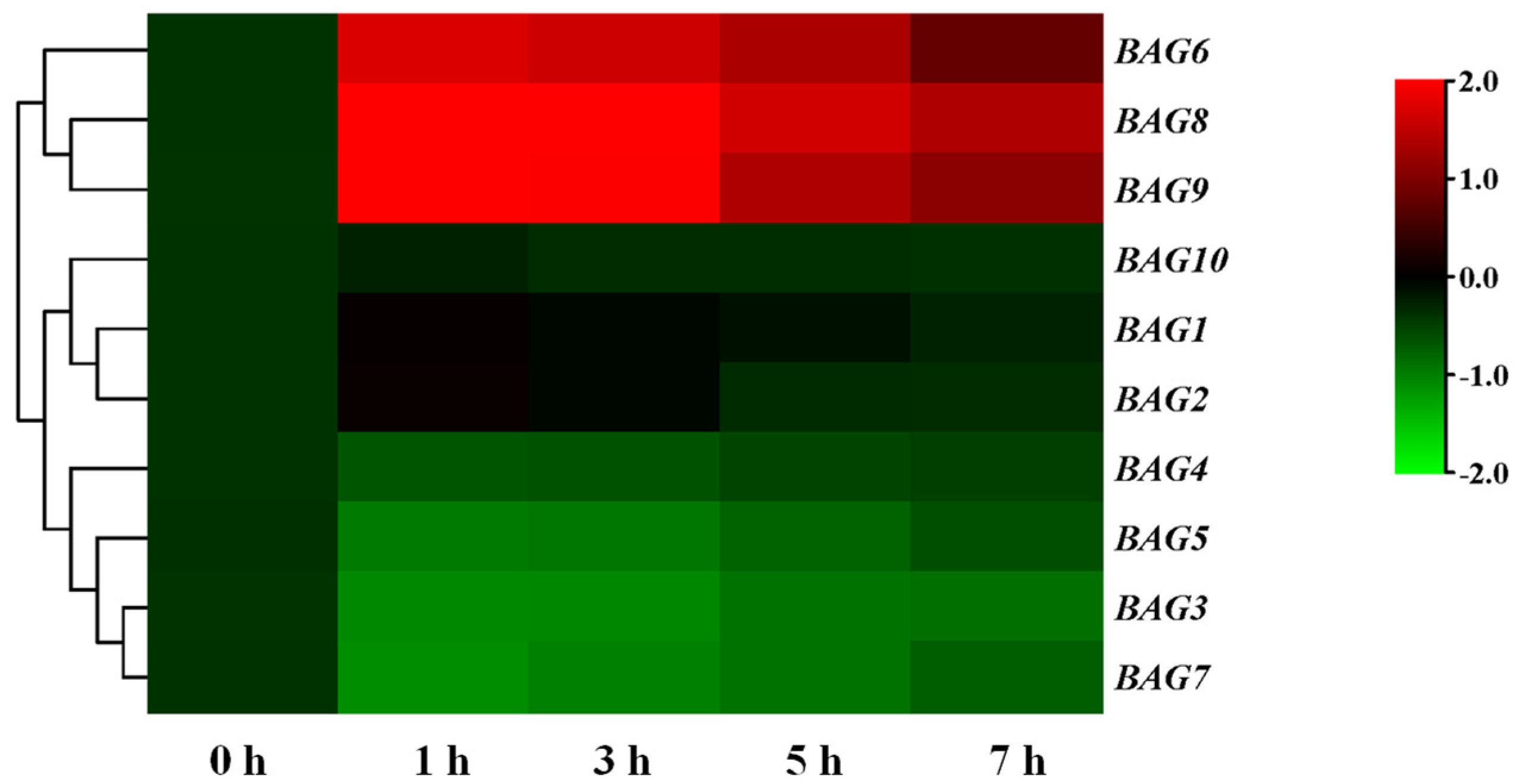
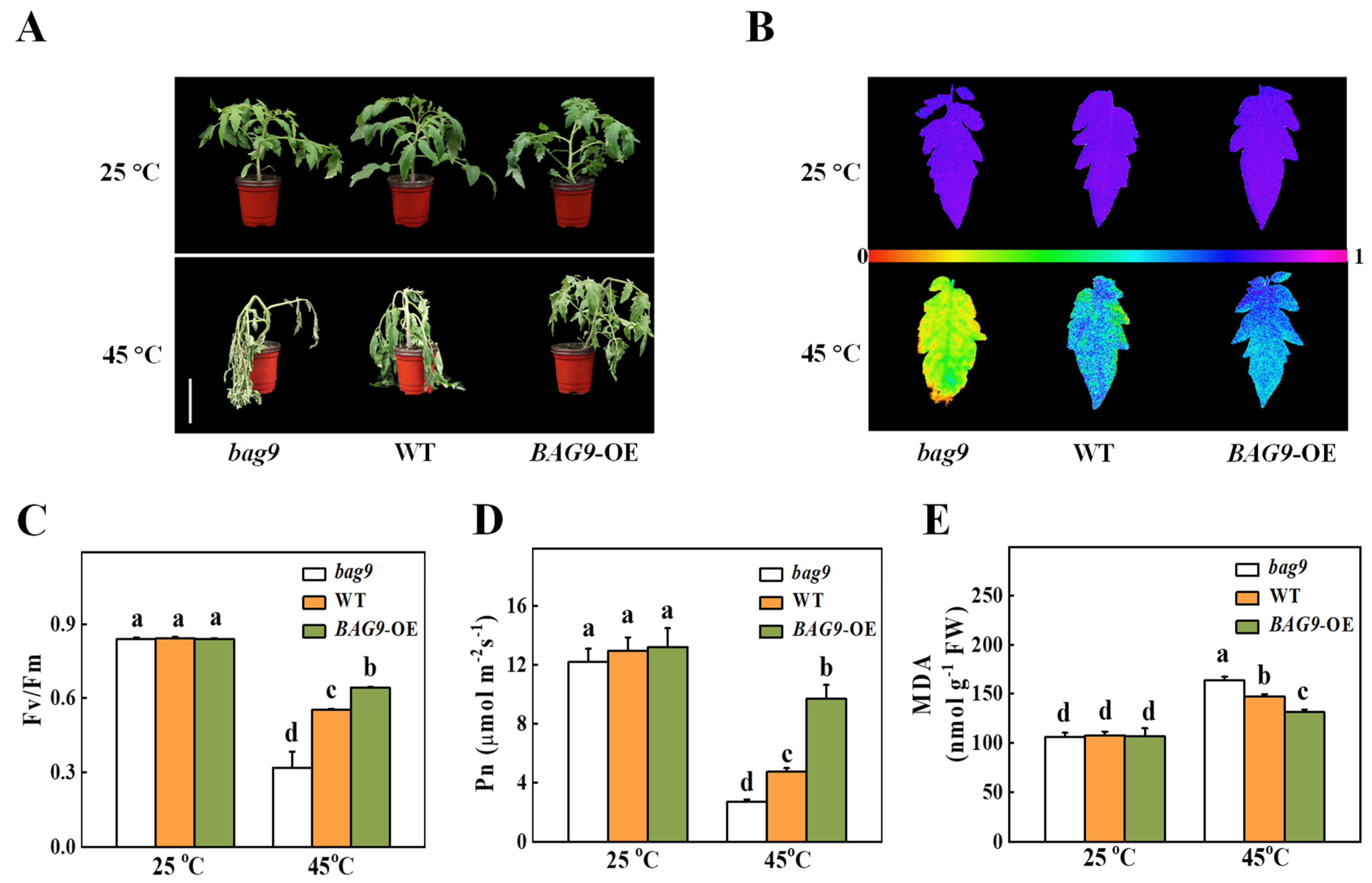
3.2. Involvement of BAG9 in Tomato Thermotolerance
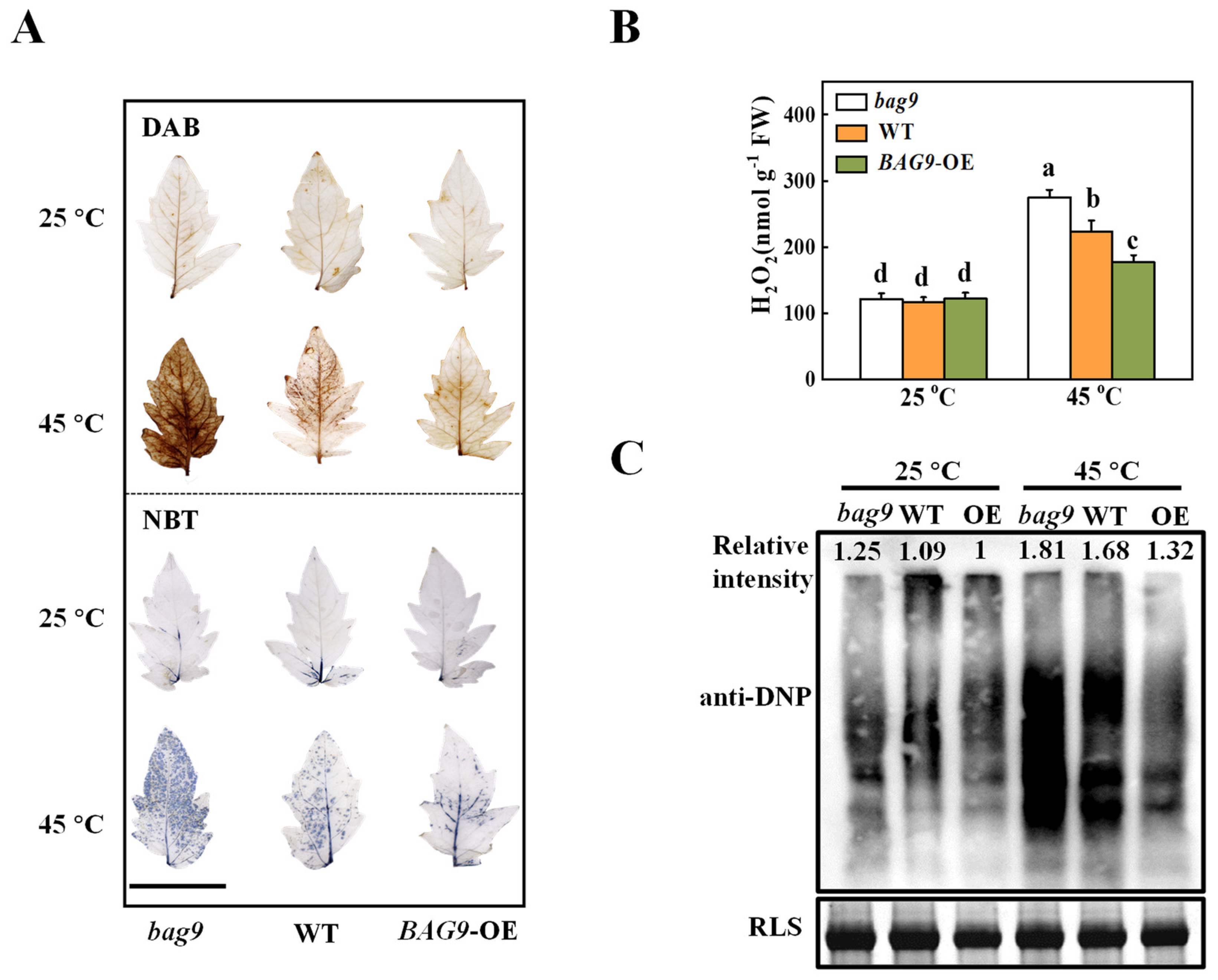
3.3. BAG9 Alleviates Heat-Stress-Induced ROS Accumulation
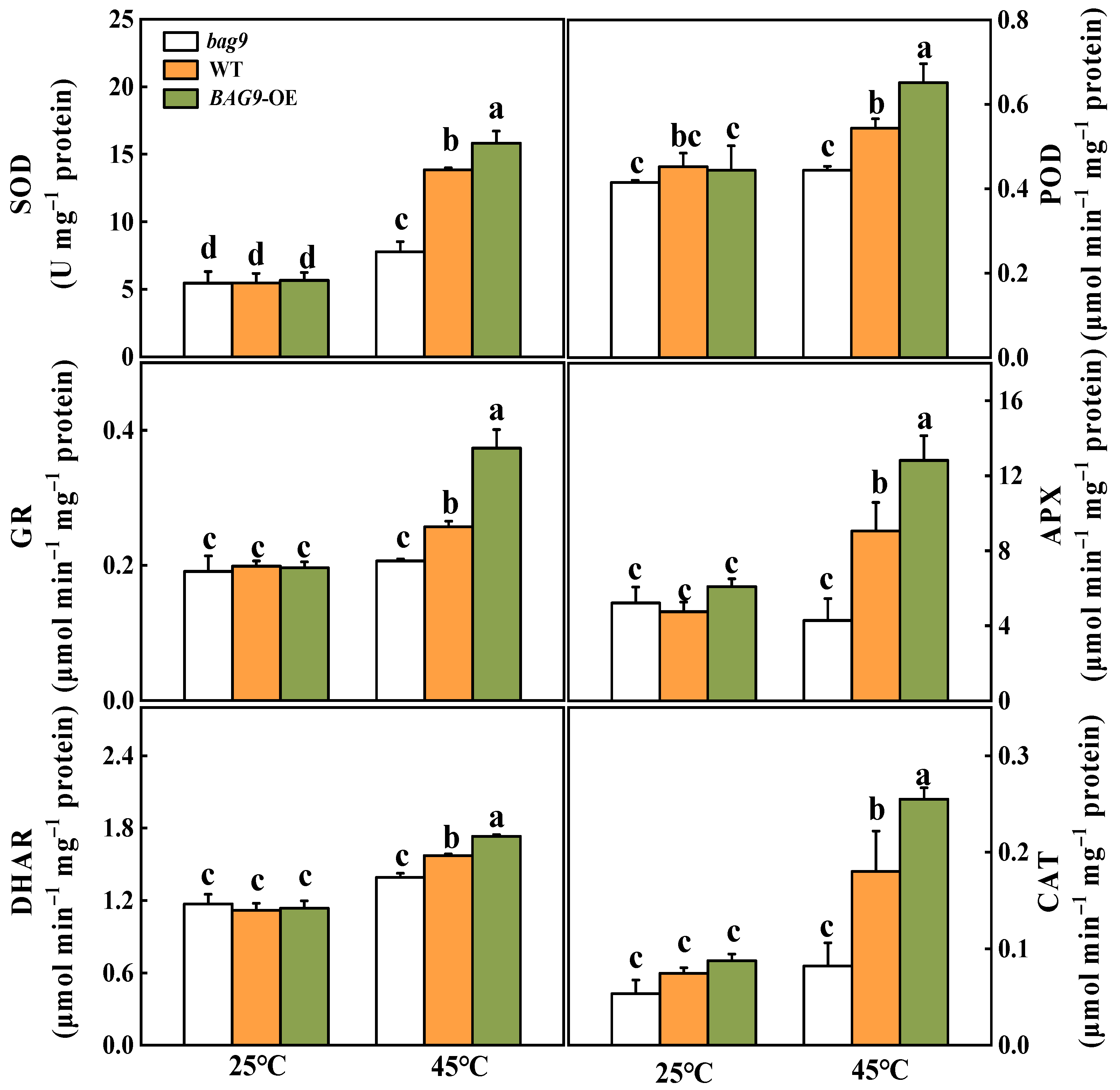
3.4. BAG9 Enhances Antioxidant Capacity under Heat Stress
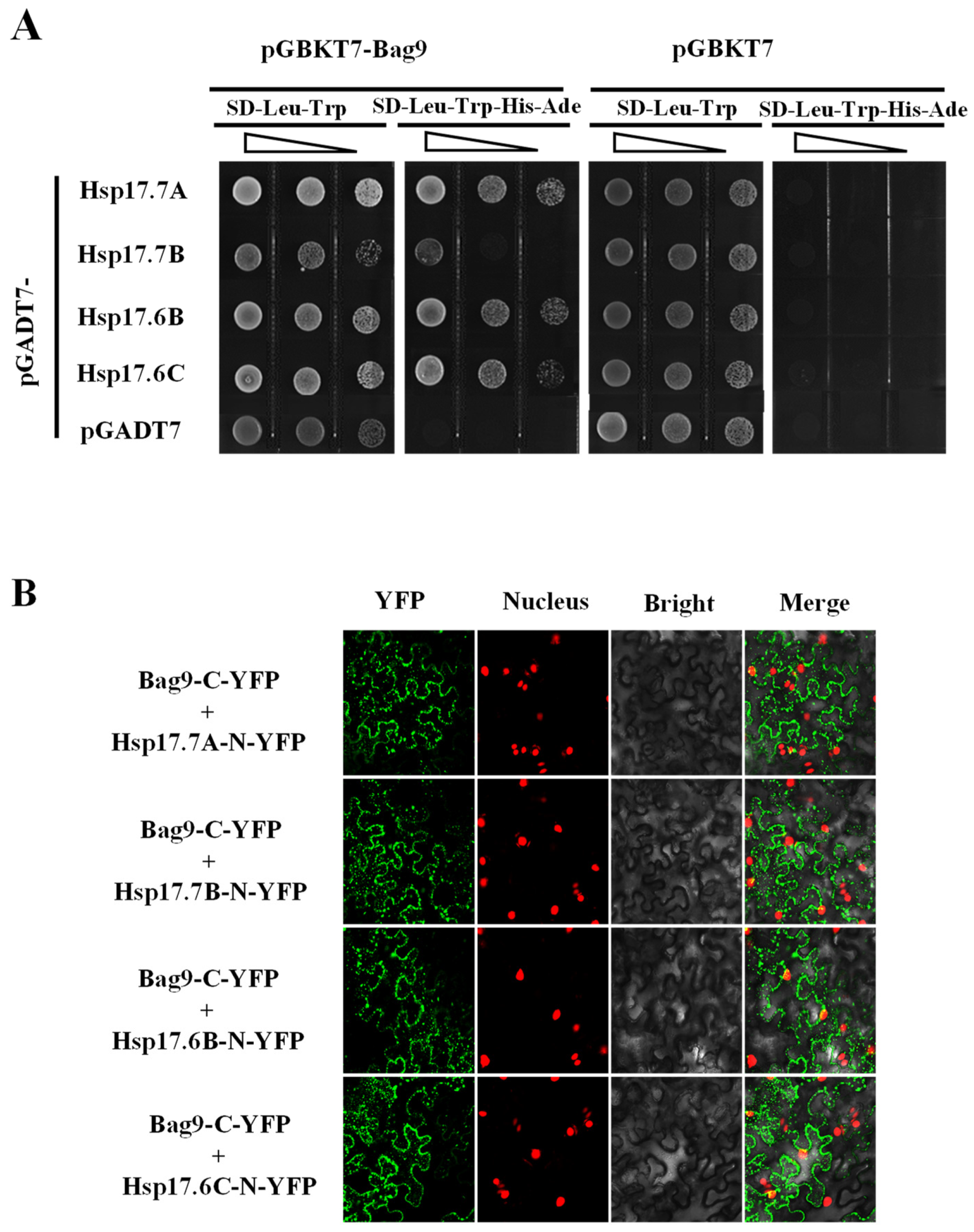
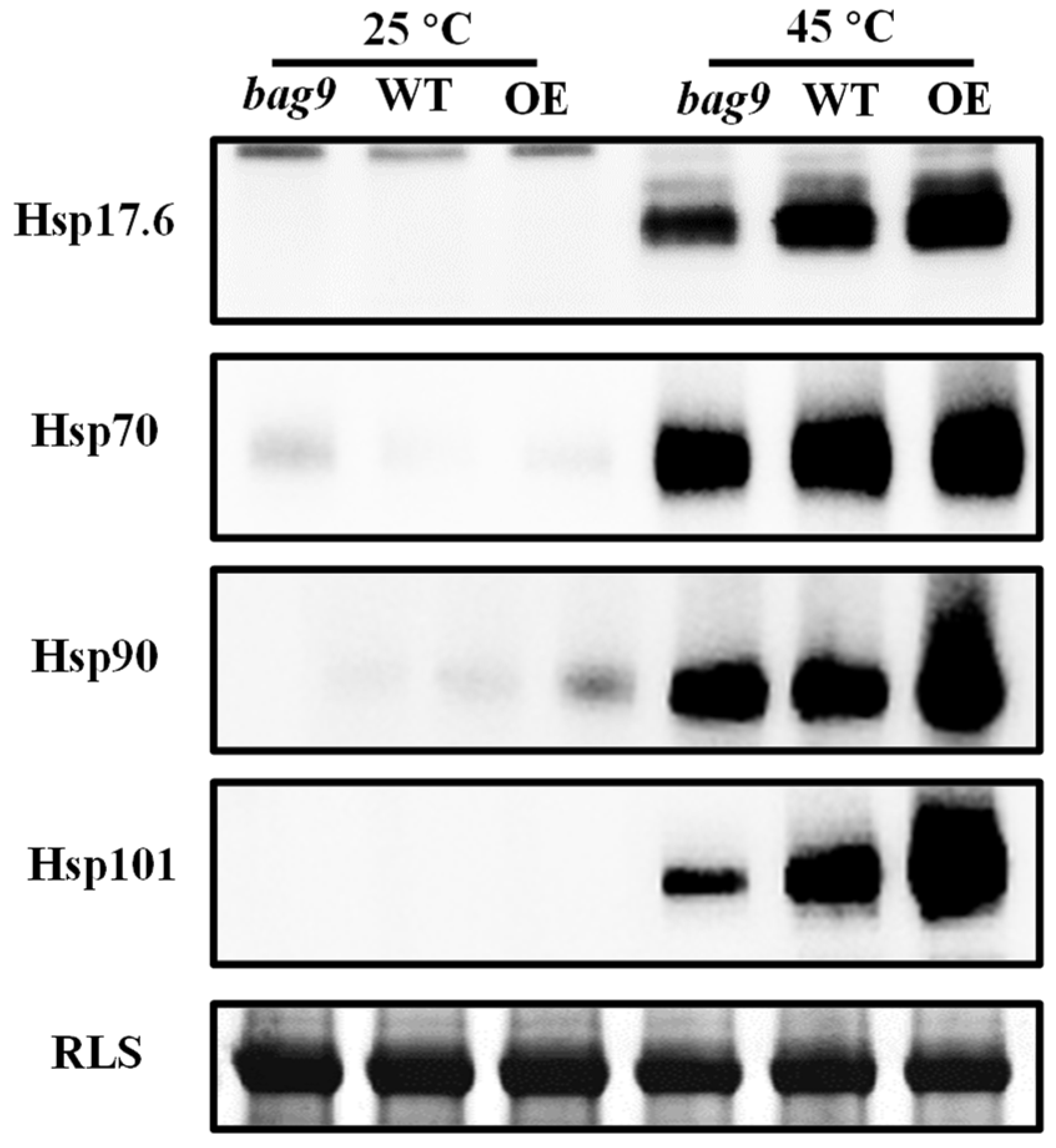
3.5. BAG9 Interacts with Hsp20s and Maintains Hsps Stability under Heat Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mittler, R.; Finka, A.; Goloubinoff, P. How Do Plants Feel the Heat? Trends Biochem. Sci. 2012, 37, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Hassan, F.; Khanday, F.A. Reactive Oxygen and Nitrogen Species: Oxidative Damage and Antioxidative Defense Mechanism in Plants under Abiotic Stress. Plant Abiotic Stress Physiol. 2022, 1, 71. [Google Scholar]
- Narayanan, S.; Tamura, P.J.; Roth, M.; Prasad, P.V.V.; Welti, R. Wheat Leaf Lipids during Heat Stress: I. High Day and Night Temperatures Result in Major Lipid Alterations. Plant Cell Environ. 2016, 39, 787–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharkey, T.D.; Zhang, R. High Temperature Effects on Electron and Proton Circuits of Photosynthesis. J. Integr. Plant Biol. 2010, 52, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Yu, J.; Chen, Z. Role and Regulation of Autophagy in Heat Stress Responses of Tomato Plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, H.; Akter, T. Molecular Chaperones and Acquisition of Thermotolerance in Plants. Handb. Plant Crop Stress 2020, 18, 343–359. [Google Scholar]
- Gupta, S.C.; Sharma, A.; Mishra, M.; Mishra, R.K.; Chowdhuri, D.K. Heat Shock Proteins in Toxicology: How Close and How Far? Life Sci. 2010, 86, 377–384. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular Chaperones in Protein Folding and Proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Wu, J.; Gao, T.; Hu, J.; Zhao, L.; Yu, C.; Ma, F. Research Advances in Function and Regulation Mechanisms of Plant Small Heat Shock Proteins (SHSPs) under Environmental Stresses. Sci. Total Environ. 2022, 825, 154054. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and Their Function in Protein Homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Petla, B.P.; Kamble, N.U.; Singh, A.; Rao, V.; Salvi, P.; Ghosh, S.; Majee, M. Differentially Expressed Seed Aging Responsive Heat Shock Protein OsHSP18.2 Implicates in Seed Vigor, Longevity and Improves Germination and Seedling Establishment under Abiotic Stress. Front. Plant Sci. 2015, 6, 713. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of Plant Heat-Shock Proteins and Molecular Chaperones in the Abiotic Stress Response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Ishikawa, A.; Tanaka, H.; Nakai, M.; Asahi, T. Deletion of a Chaperonin 60β Gene Leads to Cell Death in the Arabidopsis Lesion Initiation 1 Mutant. Plant Cell Physiol. 2003, 44, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Breiman, A. Plant Hsp90 and Its Co-Chaperones. Curr. Protein Pept. Sci. 2014, 15, 232–244. [Google Scholar] [CrossRef]
- McLoughlin, F.; Basha, E.; Fowler, M.E.; Kim, M.; Bordowitz, J.; Katiyar-Agarwal, S.; Vierling, E. Class I and II Small Heat Shock Proteins Together with HSP101 Protect Protein Translation Factors during Heat Stress. Plant Physiol. 2016, 172, 536. [Google Scholar] [CrossRef] [Green Version]
- Merret, R.; Carpentier, M.-C.; Favory, J.-J.; Picart, C.; Descombin, J.; Bousquet-Antonelli, C.; Tillard, P.; Lejay, L.; Deragon, J.-M.; Charng, Y. Heat Shock Protein HSP101 Affects the Release of Ribosomal Protein mRNAs for Recovery after Heat Shock. Plant Physiol. 2017, 174, 1216–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.; Chai, K.; Ko, S.; Kuang, L.; Lur, H.; Charng, Y. A Positive Feedback Loop between HSP101 and HSA32 Modulates Long-Term Acquired Thermotolerance Illustrating Diverse Heat Stress Responses in Rice Varieties. Plant Physiol. 2014, 164, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Colvin, T.A.; Gabai, V.L.; Gong, J.; Calderwood, S.K.; Li, H.; Gummuluru, S.; Matchuk, O.N.; Smirnova, S.G.; Orlova, N.V.; Zamulaeva, I.A.; et al. Hsp70-Bag3 Interactions Regulate Cancer-Related Signaling Networks. Cancer Res. 2014, 74, 4731–4740. [Google Scholar] [CrossRef] [Green Version]
- Genevaux, P.; Georgopoulos, C.; Kelley, W.L. The Hsp70 Chaperone Machines of Escherichia Coli: A Paradigm for the Repartition of Chaperone Functions. Mol. Microbiol. 2007, 66, 840–857. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Gymnastics of Molecular Chaperones. Mol. Cell 2010, 39, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.P.; Bukau, B. Hsp70 Chaperones: Cellular Functions and Molecular Mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Takeda, M.; Morimoto, R.I. Bag1–Hsp70 Mediates a Physiological Stress Signalling Pathway That Regulates Raf-1/ERK and Cell Growth. Nat. Cell Biol. 2001, 3, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Kong, F.; Zhang, S.; Meng, X.; Wang, Y.; Meng, Q. A Tomato Chloroplast-Targeted DnaJ Protein Protects Rubisco Activity under Heat Stress. J. Exp. Bot. 2015, 66, 3027–3040. [Google Scholar] [CrossRef] [Green Version]
- Bracher, A.; Verghese, J. The Nucleotide Exchange Factors of Hsp70 Molecular Chaperones. Front. Mol. Biosci. 2015, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Brive, L.; Takayama, S.; Briknarová, K.; Homma, S.; Ishida, S.K.; Reed, J.C.; Ely, K.R. The Carboxyl-Terminal Lobe of Hsc70 ATPase Domain Is Sufficient for Binding to BAG1. Biochem. Biophys. Res. Commun. 2001, 289, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Dickman, M.B. The BAG Proteins: A Ubiquitous Family of Chaperone Regulators. Cell. Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Reed, J.C. Molecular Chaperone Targeting and Regulation by BAG Family Proteins. Nat. Cell Biol. 2001, 3, E237–E241. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Li, Z.; Luo, Q.; Li, L.; Shen, Y.; Men, S. The BAG2 and BAG6 Genes Are Involved in Multiple Abiotic Stress Tolerances in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 5856. [Google Scholar] [CrossRef]
- Li, Y.; Williams, B.; Dickman, M. Arabidopsis B-Cell Lymphoma2 (Bcl-2)-Associated Athanogene 7 (BAG7)-Mediated Heat Tolerance Requires Translocation, Sumoylation and Binding to WRKY29. New Phytol. 2017, 214, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.; Kabbage, M.; Britt, R.; Dickman, M.B. AtBAG7, an Arabidopsis Bcl-2-Associated Athanogene, Resides in the Endoplasmic Reticulum and Is Involved in the Unfolded Protein Response. Proc. Natl. Acad. Sci. USA 2010, 107, 6088–6093. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kabbage, M.; Liu, W.; Dickman, M. Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants. Plant Cell 2016, 28, 233–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Li, L.; Kang, H.; Yang, X.; Men, S.; Shen, Y. Chronic Mitochondrial Calcium Elevation Suppresses Leaf Senescence. Biochem. Biophys. Res. Commun. 2017, 487, 672–677. [Google Scholar] [CrossRef]
- He, M.; Wang, Y.; Jahan, M.S.; Liu, W.; Raziq, A.; Sun, J.; Shu, S.; Guo, S. Characterization of SlBAG Genes from Solanum Lycopersicum and Its Function in Response to Dark-Induced Leaf Senescence. Plants 2021, 10, 947. [Google Scholar] [CrossRef]
- Doukhanina, E.V.; Chen, S.; van der Zalm, E.; Godzik, A.; Reed, J.; Dickman, M.B. Identification and Functional Characterization of the BAG Protein Family in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 18793–18801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locascio, A.; Marqués, M.C.; García-Martínez, G.; Corratgé-Faillie, C.; Andrés-Colás, N.; Rubio, L.; Fernández, J.A.; Véry, A.-A.; Mulet, J.M.; Yenush, L. BCL2-Associated Athanogene4 Regulates the KAT1 Potassium Channel and Controls Stomatal Movement. Plant Physiol. 2019, 181, 1277–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R.; Davis, S.J. Ambient Thermometers in Plants: From Physiological Outputs towards Mechanisms of Thermal Sensing. Curr. Biol. 2010, 20, R1086–R1092. [Google Scholar] [CrossRef] [Green Version]
- Chi, C.; Xu, X.; Wang, M.; Zhang, H.; Fang, P.; Zhou, J.; Xia, X.; Shi, K.; Zhou, Y.; Yu, J. Strigolactones Positively Regulate Abscisic Acid-Dependent Heat and Cold Tolerance in Tomato. Hortic. Res. 2021, 8, 237. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Shi, K.; Xia, X.; Zhou, Y.H.; Yu, J. Hydrogen Peroxide Is Involved in the Cold Acclimation-Induced Chilling Tolerance of Tomato Plants. Plant Physiol. Biochem. 2012, 60, 141–149. [Google Scholar] [CrossRef]
- Hu, Z.; Ma, Q.; Foyer, C.H.; Lei, C.; Choi, H.W.; Zheng, C.; Li, J.; Zuo, J.; Mao, Z.; Mei, Y.; et al. High CO2- and Pathogen-driven Expression of the Carbonic Anhydrase βCA3 Confers Basal Immunity in Tomato. New Phytol. 2021, 229, 2827–2843. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Li, X.; Xia, X.; Zhou, Y.; Shi, K.; Chen, Z.; Yu, J. H2O2 Mediates the Crosstalk of Brassinosteroid and Abscisic Acid in Tomato Responses to Heat and Oxidative Stresses. J. Exp. Bot. 2014, 65, 4371–4383. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. Oxidative Stress and Antioxidative Systems: Recipes for Successful Data Collection and Interpretation: Methods in Oxidative Stress Research. Plant Cell Environ. 2016, 39, 1140–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.-J.; Wang, Y.-J.; Zhou, Y.-H.; Tao, Y.; Mao, W.-H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.-Q. Reactive Oxygen Species Are Involved in Brassinosteroid- Induced Stress Tolerance in Cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Yin, L.-L.; Zhou, J.; Xia, X.-J.; Shi, K.; Yu, J.-Q.; Zhou, Y.-H.; Foyer, C.H. Interactions between 2-Cys Peroxiredoxins and Ascorbate in Autophagosome Formation during the Heat Stress Response in Solanum lycopersicum. J. Exp. Bot. 2016, 67, 1919–1933. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-C.; Chi, C.; Jin, L.-J.; Zhu, J.; Yu, J.-Q.; Zhou, Y.-H. The bZip Transcription Factor HY5 Mediates CRY1a-Induced Anthocyanin Biosynthesis in Tomato. Plant Cell Environ. 2018, 41, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, Z.; Wang, X.; Li, X.; Zhang, Z.; Fan, B.; Zhu, C.; Chen, Z. Dicot-Specific ATG8-Interacting ATI3 Proteins Interact with Conserved UBAC2 Proteins and Play Critical Roles in Plant Stress Responses. Autophagy 2018, 14, 487–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devireddy, A.R.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Chen, J.-G. Role of Reactive Oxygen Species and Hormones in Plant Responses to Temperature Changes. Int. J. Mol. Sci. 2021, 22, 8843. [Google Scholar] [CrossRef]
- Rana, R.M.; Dong, S.; Ali, Z.; Khan, A.I.; Zhang, H.S. Identification and Characterization of the Bcl-2- Associated Athanogene (BAG) Protein Family in Rice. Afr. J. Biotechnol. 2012, 11, 88–98. [Google Scholar] [CrossRef]
- Hu, L.; Chen, J.; Guo, J.; Zhao, Y.; Zhu, L.; Huang, Y. Functional Divergence and Evolutionary Dynamics of BAG Gene Family in Maize (Zea Mays). Int. J. Agric. Biol. 2013, 15, 200–206. [Google Scholar]
- Lee, D.; Kim, S.; Oh, Y.; Choi, B.; Lee, J.; Hwang, I. Arabidopsis BAG1 Functions as a Cofactor in Hsc70-Mediated Proteasomal Degradation of Unimported Plastid Proteins. Mol. Plant 2016, 9, 1428–1431. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Takato, H.; Fujita, K.; Suzuki, S. HSG1, a Grape Bcl-2-Associated Athanogene, Promotes Floral Transition by Activating CONSTANS Expression in Transgenic Arabidopsis Plant. Mol. Biol. Rep. 2012, 39, 4367–4374. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa-Yokoi, A.; Yoshida, E.; Yabuta, Y.; Shigeoka, S. Analysis of the Regulation of Target Genes by an Arabidopsis Heat Shock Transcription Factor, HsfA2. Biosci. Biotechnol. Biochem. 2009, 73, 890–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Qian, L.; Jiang, H.; Ji, Y.; Fang, Y.; Sheng, J.; Xu, X.; Ge, C. Overexpression of a Bcl-2-Associated Athanogene SlBAG9 Negatively Regulates High-Temperature Response in Tomato. Int. J. Biol. Macromol. 2022, 194, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ji, Y.; Sheng, J.; Wang, Y.; Liu, X.; Xiao, P.; Ding, H. Genome-Wide Identification of the Bcl-2 Associated Athanogene (BAG) Gene Family in Solanum Lycopersicum and the Functional Role of SlBAG9 in Response to Osmotic Stress. Antioxidants 2022, 11, 598. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under Stressful Environments: An Overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Nakamoto, H.; Suzuki, N.; Roy, S.K. Constitutive Expression of a Small Heat-Shock Protein Confers Cellular Thermotolerance and Thermal Protection to the Photosynthetic Apparatus in Cyanobacteria. FEBS Lett. 2000, 483, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Inoue, H.; Li, M.; Schnell, D.J. An Essential Role for Chloroplast Heat Shock Protein 90 (Hsp90C) in Protein Import into Chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, N.; Nakamoto, H. HtpG Is Essential for the Thermal Stress Management in Cyanobacteria. FEBS Lett. 1999, 458, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Li, J.; Ding, S.; Cheng, F.; Li, X.; Jiang, Y.; Yu, J.; Foyer, C.H.; Shi, K. The Protein Kinase CPK28 Phosphorylates Ascorbate Peroxidase and Enhances Thermotolerance in Tomato. Plant Physiol. 2021, 186, 1302–1317. [Google Scholar] [CrossRef]
- Hoang, T.M.L.; Moghaddam, L.; Williams, B.; Khanna, H.; Dale, J.; Mundree, S.G. Development of Salinity Tolerance in Rice by Constitutive-Overexpression of Genes Involved in the Regulation of Programmed Cell Death. Front. Plant Sci. 2015, 6, 175. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and Redox Signalling in the Response of Plants to Abiotic Stress: ROS and Redox Signalling in Plants. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Luo, Q.; Yang, W.; Ahammed, G.J.; Ding, S.; Chen, X.; Wang, J.; Xia, X.; Shi, K. A Novel Role of Pipecolic Acid Biosynthetic Pathway in Drought Tolerance through the Antioxidant System in Tomato. Antioxidants 2021, 10, 1923. [Google Scholar] [CrossRef] [PubMed]
- Saed-Moucheshi, A.; Shekoofa, A.; Pessarakli, M. Reactive Oxygen Species (ROS) Generation and Detoxifying in Plants. J. Plant Nutr. 2014, 37, 1573–1585. [Google Scholar] [CrossRef]
- Wu, Y.; Ye, B. Effects of Combined Elevated Temperature and Drought Stress on Anti-Oxidative Enzyme Activities and Reactive Oxygen Species Metabolism of Broussonetia papyrifera Seedlings. Acta Ecol. Sin. 2016, 36, 403–410. [Google Scholar]
- Soengas, P.; Rodríguez, V.M.; Velasco, P.; Cartea, M.E. Effect of Temperature Stress on Antioxidant Defenses in Brassica oleracea. ACS Omega 2018, 3, 5237–5243. [Google Scholar] [CrossRef]
- Zou, M.; Yuan, L.; Zhu, S.; Liu, S.; Ge, J.; Wang, C. Response of Osmotic Adjustment and Ascorbate-Glutathione Cycle to Heat Stress in a Heat-Sensitive and a Heat-Tolerant Genotype of Wucai (Brassica campestris L.). Sci. Hortic. 2016, 211, 87–94. [Google Scholar] [CrossRef]
- Cabrera, M.; Boronat, S.; Marte, L.; Vega, M.; Perez, P.; Ayte, J.; Hidalgo, E. Chaperone-Facilitated Aggregation of Thermo-Sensitive Proteins Shields Them from Degradation during Heat Stress. Cell Rep. 2020, 30, 2430–2443.e4. [Google Scholar] [CrossRef] [Green Version]
- Irfan, M.; Kumar, P.; Ahmad, I.; Datta, A. Unraveling the Role of Tomato Bcl-2-Associated Athanogene (BAG) Proteins during Abiotic Stress Response and Fruit Ripening. Sci. Rep. 2021, 11, 21734. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Li, J.; He, J.; Liu, H.; Zhang, H. A Class I Cytosolic HSP20 of Rice Enhances Heat and Salt Tolerance in Different Organisms. Sci. Rep. 2020, 10, 1383. [Google Scholar] [CrossRef]
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Genome-Wide Identification and Analysis of Biotic and Abiotic Stress Regulation of Small Heat Shock Protein (HSP20) Family Genes in Bread Wheat. J. Plant Physiol. 2017, 211, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Pratt, W.B.; Toft, D.O. Regulation of Signaling Protein Function and Trafficking by the Hsp90/Hsp70-Based Chaperone Machinery. Exp. Biol. Med. 2003, 228, 111–133. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Liu, C.; Yang, C.; Kanwar, M.K.; Shao, S.; Qi, Z.; Zhou, J. BAG9 Confers Thermotolerance by Regulating Cellular Redox Homeostasis and the Stability of Heat Shock Proteins in Solanum lycopersicum. Antioxidants 2022, 11, 1467. https://doi.org/10.3390/antiox11081467
Huang H, Liu C, Yang C, Kanwar MK, Shao S, Qi Z, Zhou J. BAG9 Confers Thermotolerance by Regulating Cellular Redox Homeostasis and the Stability of Heat Shock Proteins in Solanum lycopersicum. Antioxidants. 2022; 11(8):1467. https://doi.org/10.3390/antiox11081467
Chicago/Turabian StyleHuang, Huamin, Chenxu Liu, Chen Yang, Mukesh Kumar Kanwar, Shujun Shao, Zhenyu Qi, and Jie Zhou. 2022. "BAG9 Confers Thermotolerance by Regulating Cellular Redox Homeostasis and the Stability of Heat Shock Proteins in Solanum lycopersicum" Antioxidants 11, no. 8: 1467. https://doi.org/10.3390/antiox11081467





