Integrated Transcriptome and 16S rDNA Analyses Reveal That Transport Stress Induces Oxidative Stress and Immune and Metabolic Disorders in the Intestine of Hybrid Yellow Catfish (Tachysurus fulvidraco♀ × Pseudobagrus vachellii♂)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish and Experimental Design
2.2. Sample Collection
2.3. Intestine Biochemical Analyses
2.4. Histological Analyses of intestine
2.5. Intestinal Transcriptomic Sequencing and Analysis
2.5.1. RNA Isolation and Transcriptome Sequencing
2.5.2. Assembly and Annotation of Transcripts, Identification of Differentially Expressed Genes
2.5.3. qRT-PCR Analyses
2.6. Intestinal Microbiome Sequencing and Analysis
2.7. Intestinal Microbiota and DEGs Correlation Analyses
2.8. Statistical Analyses
3. Results
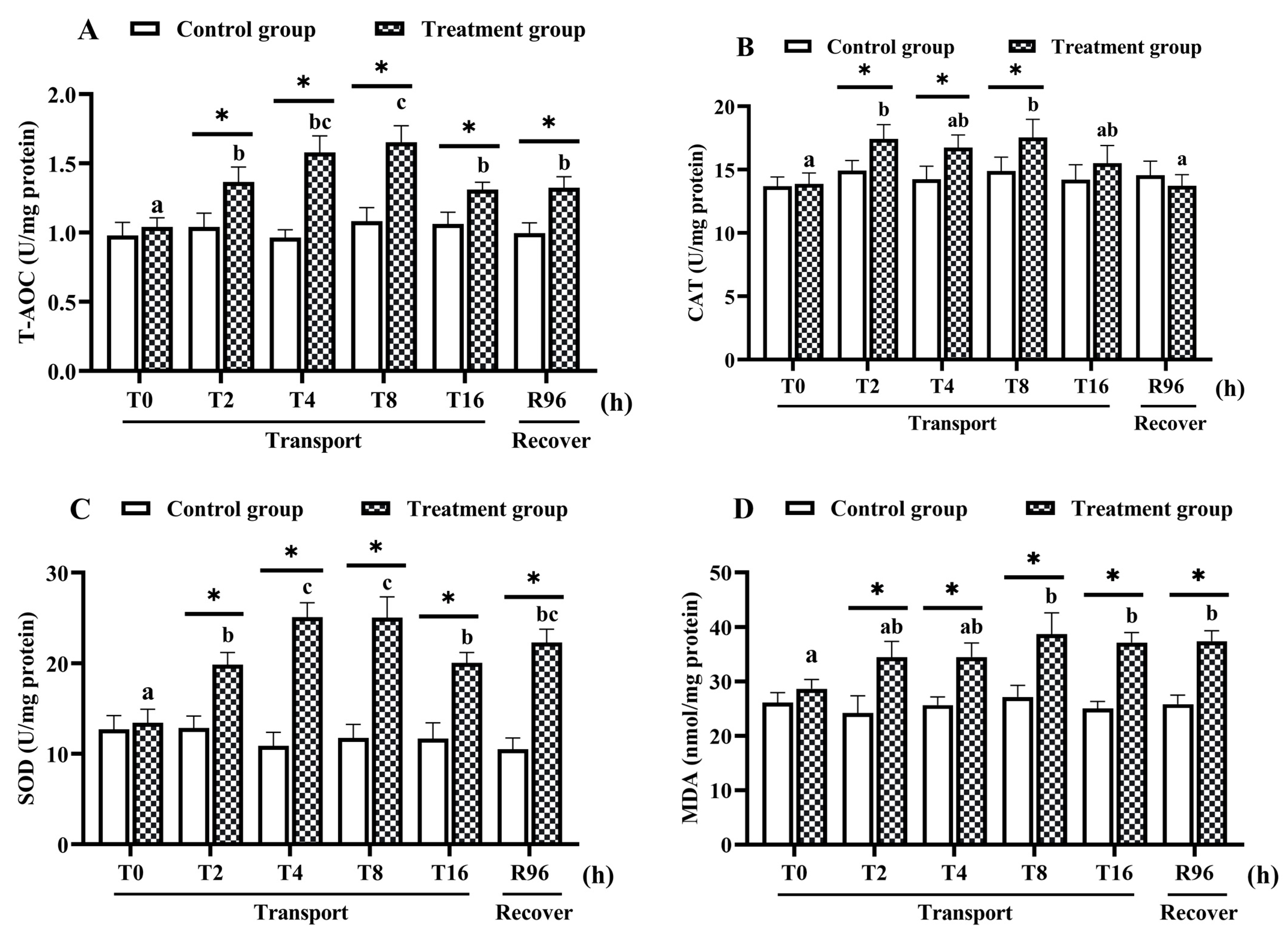
3.1. Intestine Biochemical Parameters
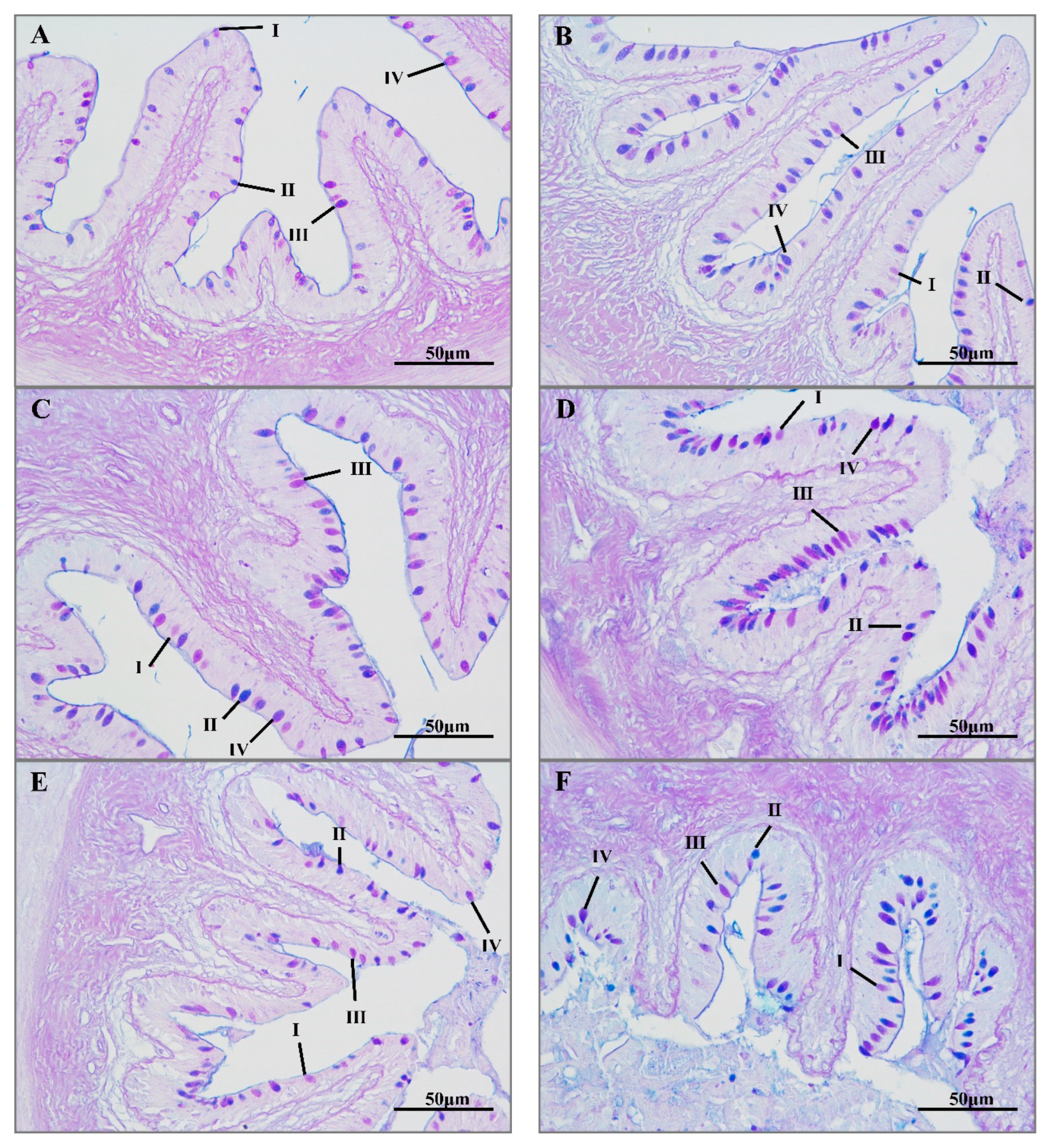
3.2. Histological Analyses
3.3. Intestinal Transcriptome Analysis
3.3.1. Transcriptome Sequencing, Assembly, and Statistics
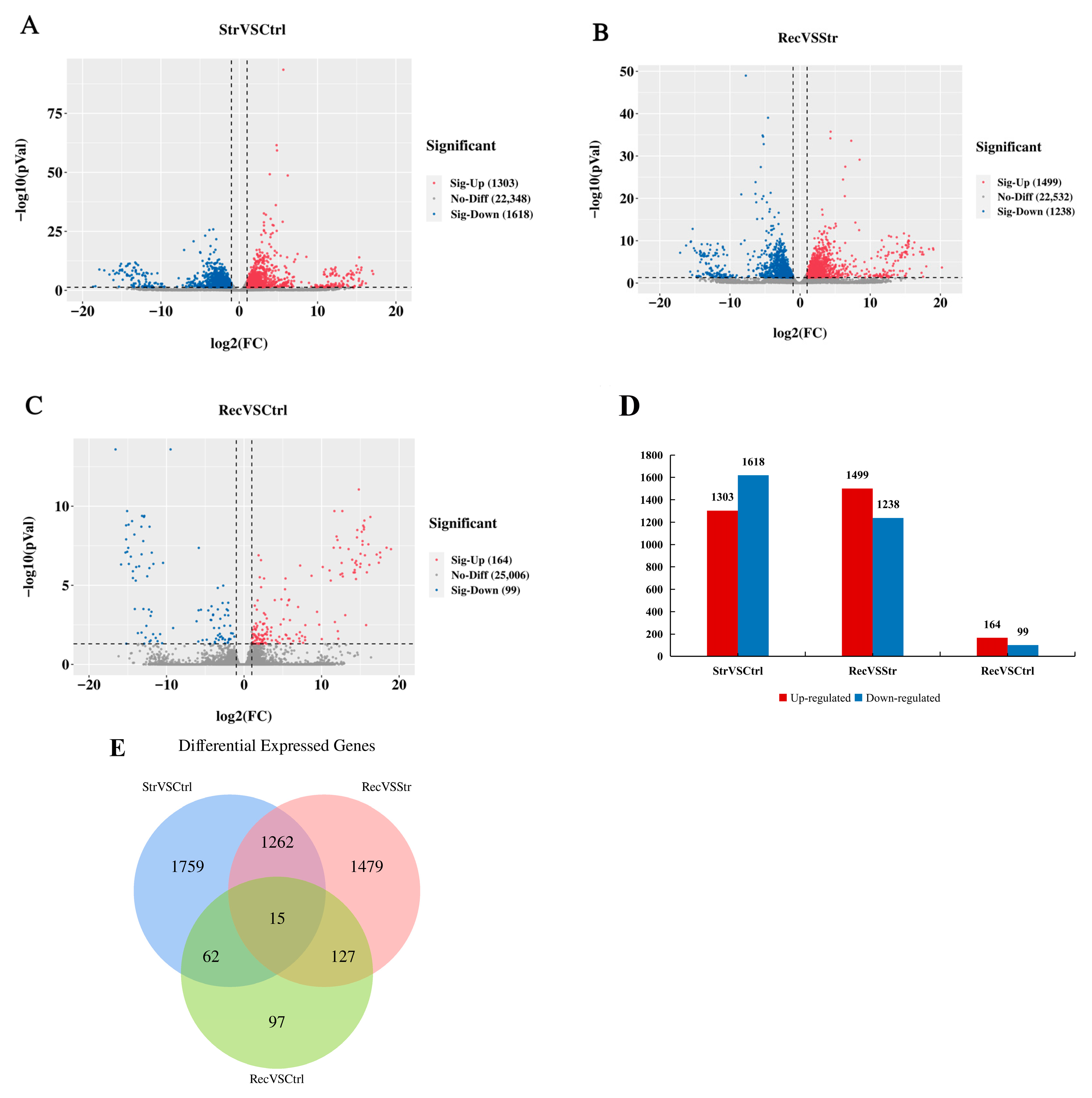
3.3.2. Selection of DEGs
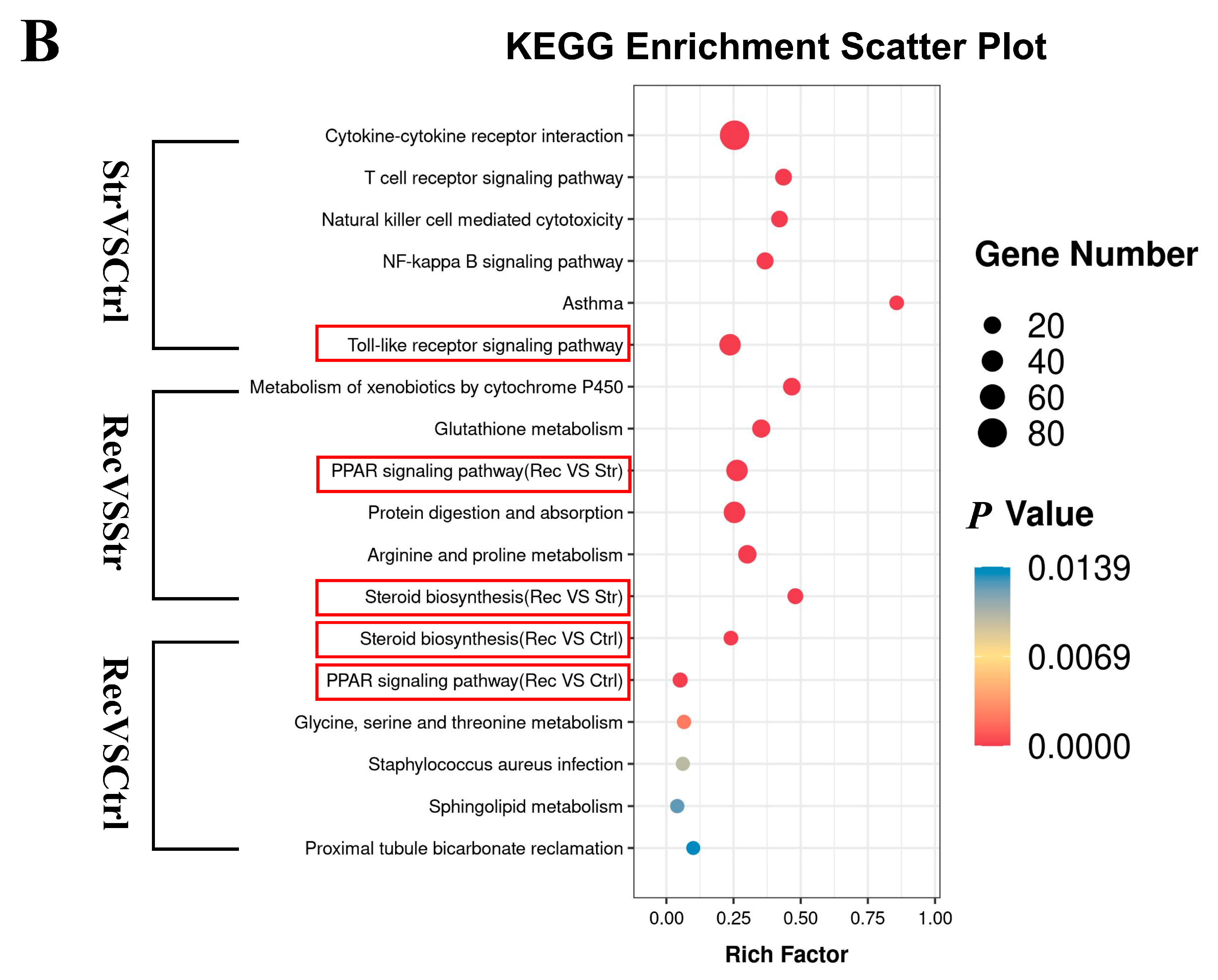
3.3.3. GO and KEGG Enrichment Analyses of DEGs
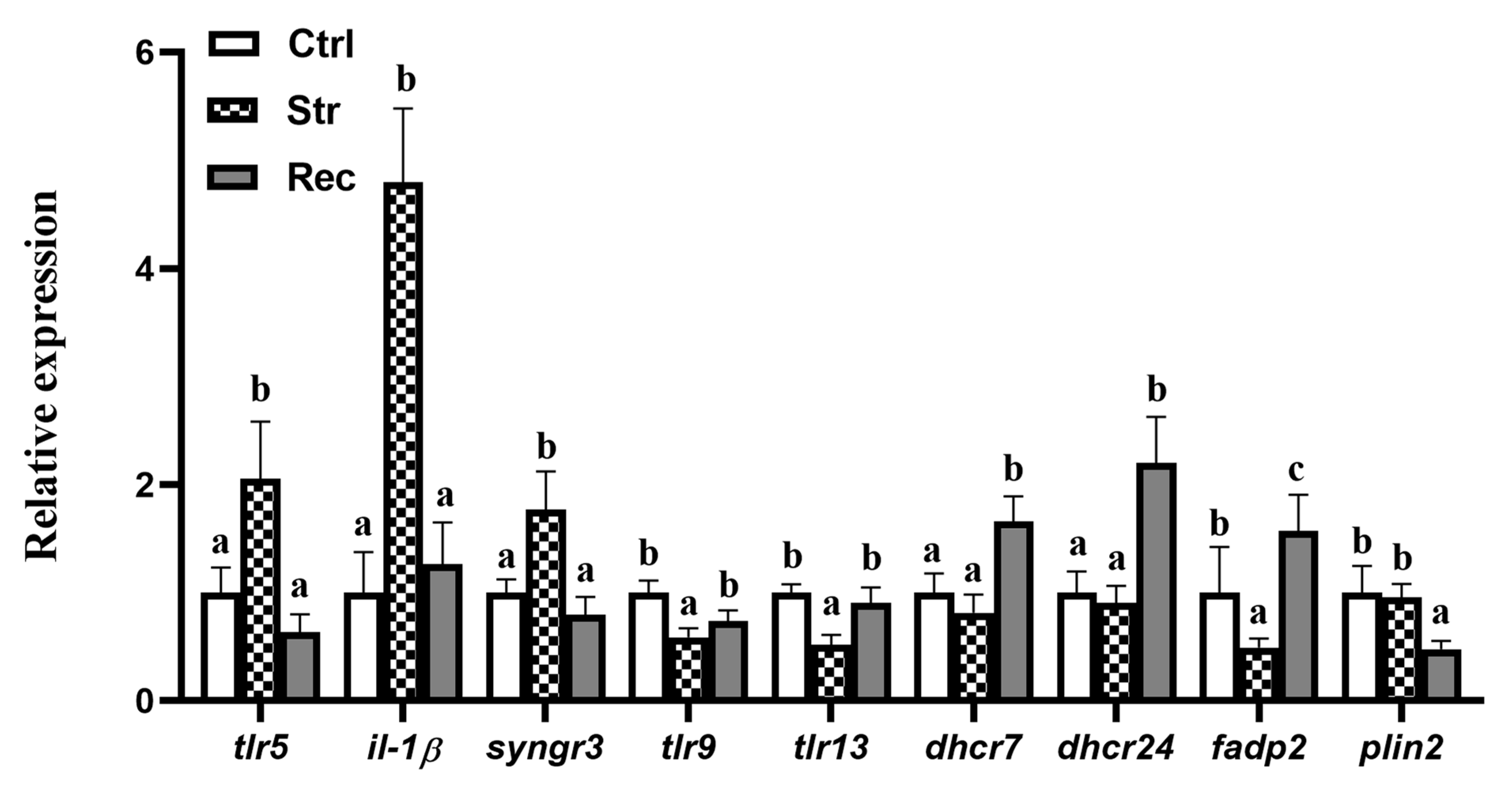
3.3.4. Expression Patterns of DEGs Involved in TLRs Signaling, PPAR Signaling, and Steroid Biosynthesis Pathways under Transport Stress
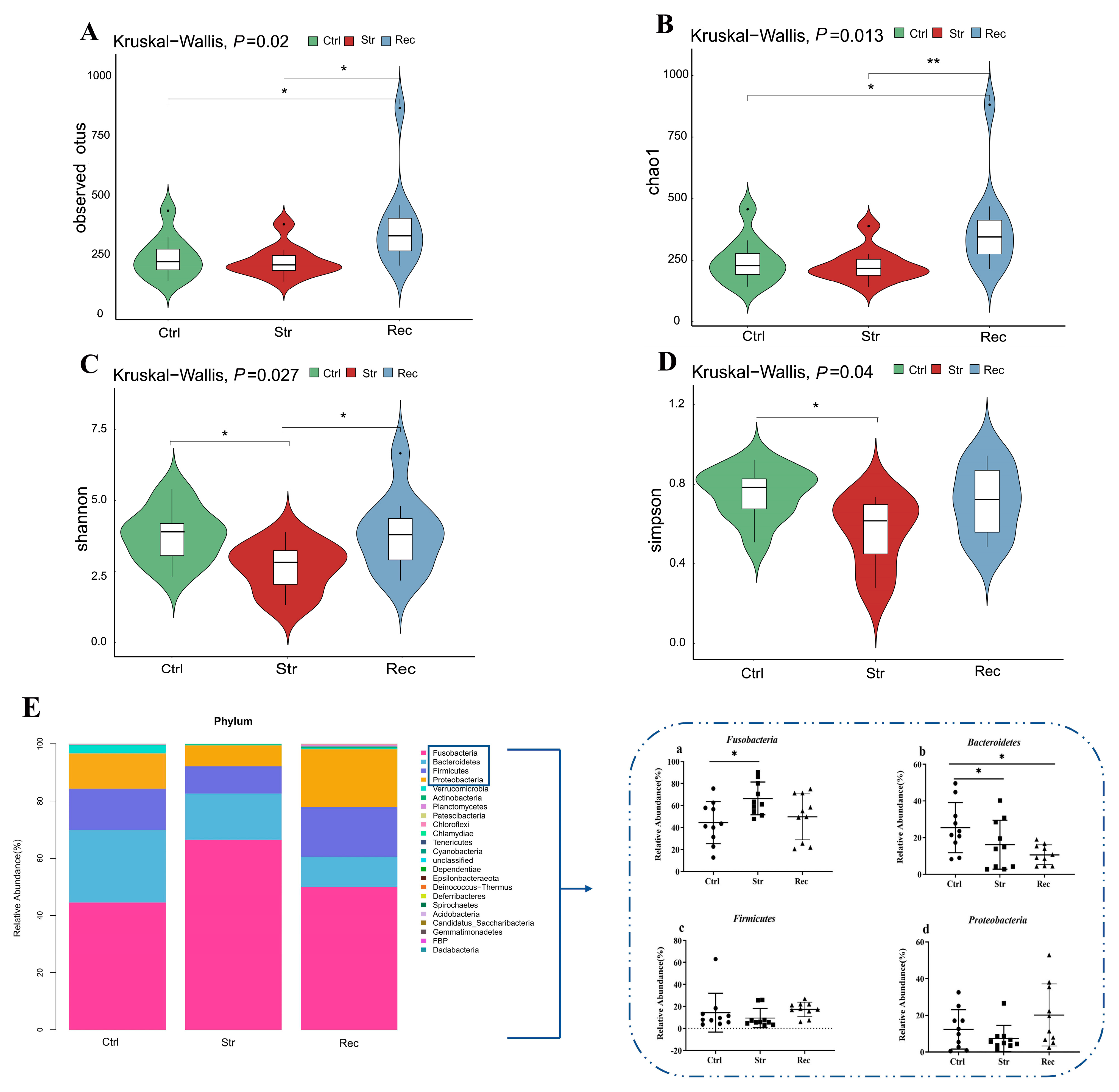
3.4. Intestinal Microbial Composition
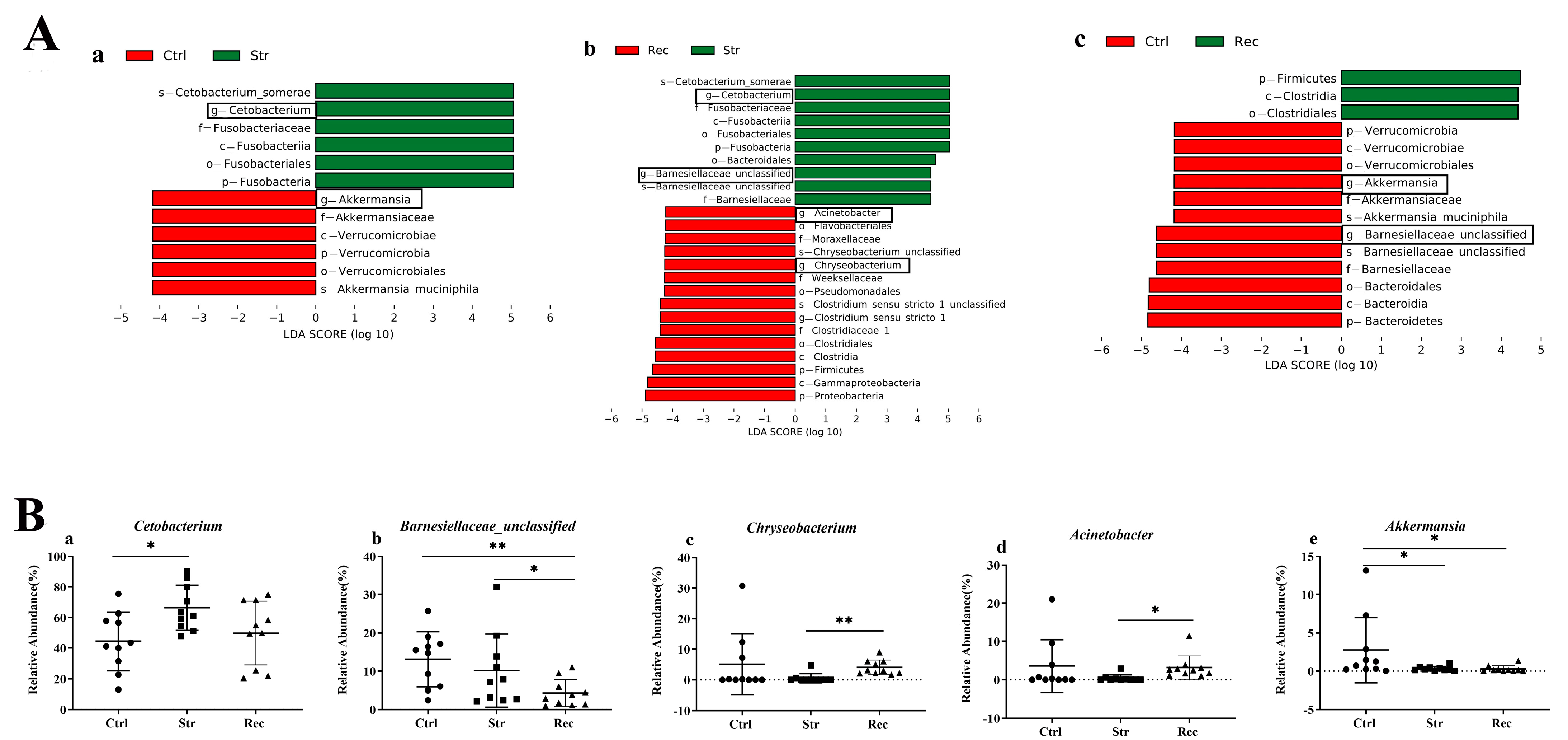
3.5. Relationships between Intestinal Microbiota and DEGs under Transport Stress
4. Discussion
4.1. Changes in the Intestinal Antioxidant System under Transport Stress
4.2. Changes in Mucus Cells in the Intestine under Transport Stress
4.3. Transcriptome Analyses Reveal That Transport Stress Affects Intestinal Immune and Metabolic Function
4.4. 16S rDNA Sequence Analyses Reveal Changes in Intestinal Microecology under Transport Stress
4.5. Intestinal Microbiota and Its Relationship with DEGs under Transport Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harmon, T.S. Methods for reducing stressors and maintaining water quality associated with live fish transport in tanks: A review of the basics. Rev. Aquac. 2009, 1, 58–66. [Google Scholar] [CrossRef]
- Bolasina, S.N. Stress response of juvenile flounder (Paralichthys orbignyanus, Valenciennes 1839), to acute and chronic stressors. Aquaculture 2011, 313, 140–143. [Google Scholar] [CrossRef]
- Wu, S.M.; Chen, J.-R.; Chang, C.-Y.; Tseng, Y.-J.; Pan, B.S. Potential benefit of I-Tiao-Gung (Glycine tomentella) extract to enhance ornamental fish welfare during live transport. Aquaculture 2020, 534, 736304. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Zheng, T.; Song, Z.; Qiang, J.; Tao, Y.; Zhu, H.; Ma, J.; Xu, P. Transport Stress Induces Skin Innate Immunity Response in Hybrid Yellow Catfish (Tachysurus fulvidraco female × P. vachellii male) Through TLR/NLR Signaling Pathways and Regulation of Mucus Secretion. Front. Immunol. 2021, 12, 740359. [Google Scholar] [CrossRef]
- Zheng, T.; Song, Z.; Tao, Y.; Qiang, J.; Ma, J.; Lu, S.; Xu, P. Transport stress induces innate immunity responses through TLR and NLR signaling pathways and increases mucus cell number in gills of hybrid yellow catfish (Tachysurus fulvidraco ♀ × Pseudobagrus vachellii ♂). Fish Shellfish. Immunol. 2022, 127, 166–175. [Google Scholar] [CrossRef]
- Vanderzwalmen, M.; Eaton, L.; Mullen, C.; Henriquez, F.; Carey, P.; Snellgrove, D.; Sloman, K.A. The use of feed and water additives for live fish transport. Rev. Aquac. 2018, 11, 263–278. [Google Scholar] [CrossRef]
- Qiang, J.; Zhang, Z.; Yu, J.; Xu, J.; Liu, H.; Zhang, Z.; Xu, P. Water quality and physiological response of F1hybrid seabream (Pagrus major female × Acanthopagrus schlegelii male) to transport stress at different densities. Aquac. Res. 2017, 49, 767–775. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Yousefi, M.; Hoseinifar, S.H.; Van Doan, H. Cytokines’ gene expression, humoral immune and biochemical responses of common carp (Cyprinus carpio, Linnaeus, 1758) to transportation density and recovery in brackish water. Aquaculture 2019, 504, 13–21. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, L.; Xing, K.; Sun, X.; Sun, J. Short-time Transport Stress Affected Biochemical Parameters in Takifugu Rubripes. In Proceedings of the 5th Annual International Conference on Material Engineering and Application (ICMEA), Wuhan, China, 14 September 2018. [Google Scholar]
- Refaey, M.M.; Tian, X.; Tang, R.; Li, D. Changes in physiological responses, muscular composition and flesh quality of channel catfish Ictalurus punctatus suffering from transport stress. Aquaculture 2017, 478, 9–15. [Google Scholar] [CrossRef]
- Honryo, T.; Oakada, T.; Kawahara, M.; Kurata, M.; Agawa, Y.; Sawada, Y.; Miyashita, S.; Takii, K.; Ishibashi, Y. Estimated time for recovery from transportation stress and starvation in juvenile Pacific bluefin tuna Thunnus orientalis. Aquaculture 2018, 484, 175–183. [Google Scholar] [CrossRef]
- Fang, W.; Chen, Q.; Li, J.; Liu, Y.; Zhao, Z.; Shen, Y.; Mai, K.; Ai, Q. Endoplasmic Reticulum Stress Disturbs Lipid Homeostasis and Augments Inflammation in the Intestine and Isolated Intestinal Cells of Large Yellow Croaker (Larimichthys crocea). Front. Immunol. 2021, 12, 3294. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Xu, G.; Li, Y.; Nie, Z.; Xu, P. Molecular cloning and differential expression of the glucocorticoid receptorgene in the estuarine tapertail anchovy Coilia nasus. Genet. Mol. Res. 2017, 16, 16029125. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wei, H.K.; Xiang, Q.-H.; Wang, J.; Zhou, Y.-F.; Peng, J. Protective effect of quercetin on pig intestinal integrity after transport stress is associated with regulation oxidative status and inflammation. J. Veter-Med. Sci. 2016, 78, 1487–1494. [Google Scholar] [CrossRef]
- Poltronieri, C.; Negrato, E.; Bertotto, D.; Majolini, D.; Simontacchi, C.; Radaelli, G. Immunohistochemical localization of constitutive and inducible heat shock protein 70 in carp (Cyprinus carpio) and trout (Oncorhynchus mykiss) exposed to transport stress. Eur. J. Histochem. 2008, 52, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mei, J.; Cao, J.; Xie, J. Effects of Melissa officinalis L. Essential Oil in Comparison with Anaesthetics on Gill Tissue Damage, Liver Metabolism and Immune Parameters in Sea Bass (Lateolabrax maculatus) during Simulated Live Transport. Biology 2021, 11, 11. [Google Scholar] [CrossRef]
- Chandhini, S.; Kumar, V.J.R. Transcriptomics in aquaculture: Current status and applications. Rev. Aquac. 2018, 11, 1379–1397. [Google Scholar] [CrossRef]
- Dhanasiri, A.K.S.; Fernandes, J.M.O.; Kiron, V. Liver Transcriptome Changes in Zebrafish during Acclimation to Transport-Associated Stress. PLoS ONE 2013, 8, e65028. [Google Scholar] [CrossRef]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Llewellyn, M.S.; Boutin, S.; Hoseinifar, S.H.; Derome, N. Teleost microbiomes: The state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 2014, 5, 207. [Google Scholar] [CrossRef] [Green Version]
- Sumithra, T.; Gayathri, S.; Sharma, S.K.; Ebeneezar, S.; Anikuttan, K.; Sajina, K.; Narasimapallavan, G.I.; Reshma, K.; Vishnu, R.; Tamilmani, G.; et al. Metagenomic signatures of transportation stress in the early life stages of cobia (Rachycentron canadum) to aid in mitigation strategies. Aquaculture 2022, 559, 738407. [Google Scholar] [CrossRef]
- Zhu, H.; Qiang, J.; He, J.; Tao, Y.; Bao, J.; Xu, P. Physiological parameters and gut microbiome associated with different dietary lipid levels in hybrid yellow catfish (Tachysurus fulvidraco female × Pseudobagrus vachellii male). Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 37, 100777. [Google Scholar] [CrossRef]
- Zheng, T.; Jia, R.; Cao, L.; Du, J.; Gu, Z.; He, Q.; Xu, P.; Yin, G. Effects of chronic glyphosate exposure on antioxdative status, metabolism and immune response in tilapia (GIFT, Oreochromis niloticus). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 239, 108878. [Google Scholar] [CrossRef] [PubMed]
- da Silva, W.F.; Simões, M.J.; Gutierre, R.C.; Egami, M.I.; Santos, A.A.; Antoniazzi, M.M.; Sasso, G.R.; Ranzani-Paiva, M.J.T. Special dyeing, histochemistry, immunohistochemistry and ultrastructure: A study of mast cells/eosinophilic granules cells (MCs/EGC) from Centropomus parallelus intestine. Fish Shellfish Immunol. 2017, 60, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Ngamkala, S.; Futami, K.; Endo, M.; Maita, M.; Katagiri, T. Immunological effects of glucan and Lactobacillus rhamnosus GG, a probiotic bacterium, on Nile tilapia Oreochromis niloticus intestine with oral Aeromonas challenges. Fish. Sci. 2010, 76, 833–840. [Google Scholar] [CrossRef]
- Biswal, A.; Srivastava, P.P.; Krishna, G.; Paul, T.; Pal, P.; Gupta, S.; Varghese, T.; Jayant, M. An Integrated biomarker approach for explaining the potency of exogenous glucose on transportation induced stress in Labeo rohita fingerlings. Sci. Rep. 2021, 11, 5713. [Google Scholar] [CrossRef]
- Le Guernic, A.; Sanchez, W.; Palluel, O.; Bado-Nilles, A.; Turies, C.; Chadili, E.; Cavalié, I.; Adam-Guillermin, C.; Porcher, J.-M.; Geffard, A.; et al. In situ experiments to assess effects of constraints linked to caging on ecotoxicity biomarkers of the three-spined stickleback (Gasterosteus aculeatus L.). Fish Physiol. Biochem. 2015, 42, 643–657. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Gupta, S.K.; Yousefi, M.; Kulikov, E.V.; Drukovsky, S.G.; Petrov, A.K.; Mirghaed, A.T.; Hoseinifar, S.H.; Van Doan, H. Mitigation of transportation stress in common carp, Cyprinus carpio, by dietary administration of turmeric. Aquaculture 2021, 546, 737380. [Google Scholar] [CrossRef]
- Ren, Y.; Men, X.; Yu, Y.; Li, B.; Zhou, Y.; Zhao, C. Effects of transportation stress on antioxidation, immunity capacity and hypoxia tolerance of rainbow trout (Oncorhynchus mykiss). Aquac. Rep. 2021, 22, 100940. [Google Scholar] [CrossRef]
- Du, S.N.N.; Khajali, F.; Dawson, N.J.; Scott, G.R. Hybridization increases mitochondrial production of reactive oxygen species in sunfish. Evolution 2017, 71, 1643–1652. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Majidiyan, N.; Mirghaed, A.T.; Hoseinifar, S.H.; Van Doan, H. Dietary glycine supplementation alleviates transportation-induced stress in common carp, Cyprinus carpio. Aquaculture 2022, 551, 737959. [Google Scholar] [CrossRef]
- Bosi, G.; DePasquale, J.A.; Rossetti, E.; Dezfuli, B.S. Differential mucins secretion by intestinal mucous cells of Chelon ramada in response to an enteric helminth Neoechinorhynchus agilis (Acanthocephala). Acta Histochem. 2019, 122, 151488. [Google Scholar] [CrossRef] [PubMed]
- Al-Zaidan, A.S.; Endo, M.; Maita, M.; Gonçalves, A.T.; Futami, K.; Katagiri, T. A toxicity bioassay study concerning the effect of un-ionized ammonia on the mucus cells response originating from the gills of zebrafish Danio rerio. Fish. Sci. 2012, 79, 129–142. [Google Scholar] [CrossRef]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar] [CrossRef]
- Zhao, A.; Qin, H.; Sun, M.; Tang, M.; Mei, J.; Ma, K.; Fu, X. Chemical conversion of human epidermal stem cells into intestinal goblet cells for modeling mucus-microbe interaction and therapy. Sci. Adv. 2021, 7, eabb2213. [Google Scholar] [CrossRef]
- Gill, R.; Tsung, A.; Billiar, T.R. Linking oxidative stress to inflammation: Toll-like receptors. Free Radic. Biol. Med. 2010, 48, 1121–1132. [Google Scholar] [CrossRef]
- Feng, T.; Cong, Y.; Alexander, K.; Elson, C.O. Regulation of Toll-like Receptor 5 Gene Expression and Function on Mucosal Dendritic Cells. PLoS ONE 2012, 7, e35918. [Google Scholar] [CrossRef]
- Shibata, T.; Takemura, N.; Motoi, Y.; Goto, Y.; Karuppuchamy, T.; Izawa, K.; Li, X.; Akashi-Takamura, S.; Tanimura, N.; Kunisawa, J.; et al. PRAT4A-dependent expression of cell surface TLR5 on neutrophils, classical monocytes and dendritic cells. Int. Immunol. 2012, 24, 613–623. [Google Scholar] [CrossRef]
- Yang, W.; Bai, X.; Luan, X.; Min, J.; Tian, X.; Li, H.; Li, H.; Sun, W.; Liu, W.; Fan, W.; et al. Delicate regulation of IL-1β-mediated inflammation by cyclophilin A. Cell Rep. 2022, 38, 110513. [Google Scholar] [CrossRef]
- Šribar, D.; Grabowski, M.; Murgueitio, M.S.; Bermudez, M.; Weindl, G.; Wolber, G. Identification and characterization of a novel chemotype for human TLR8 inhibitors. Eur. J. Med. Chem. 2019, 179, 744–752. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, S.; Huang, J.; Xiao, Y.; Fu, Y.; Liu, W.; Yin, H. Photoactivation of Innate Immunity Receptor TLR8 in Live Mammalian Cells by Genetic Encoding of Photocaged Tyrosine. ChemBioChem 2021, 23, e202100344. [Google Scholar] [CrossRef] [PubMed]
- Largo-Barrientos, P.; Uytterhoeven, V.; de Wit, J.; De Strooper, B.; Verstreken, P. Synaptic tau and synaptogyrin-3 are promising targets to tackle tauopathies. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2021, 17, e054187. [Google Scholar] [CrossRef]
- Haytural, H.; Mermelekas, G.; Emre, C.; Nigam, S.M.; Carroll, S.L.; Winblad, B.; Bogdanovic, N.; Barthet, G.; Granholm, A.-C.; Orre, L.M.; et al. The Proteome of the Dentate Terminal Zone of the Perforant Path Indicates Presynaptic Impairment in Alzheimer Disease. Mol. Cell. Proteom. 2020, 19, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Emami, N.K.; Jung, U.; Voy, B.; Dridi, S. Radical Response: Effects of Heat Stress-Induced Oxidative Stress on Lipid Metabolism in the Avian Liver. Antioxidants 2020, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, X.; Cui, H.; Liu, R.; Zhao, G.; Wen, J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genom. 2019, 20, 863. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Zahmatkesh, M.; Heidari, M.; Hassanzadeh, G.-R.; Karimian, M.; Sarrafnejad, A.; Zarrindast, M.-R. Hippocampal DHCR24 down regulation in a rat model of streptozotocin-induced cognitive decline. Neurosci. Lett. 2015, 587, 107–112. [Google Scholar] [CrossRef]
- Kuehnle, K.; Crameri, A.; Kaälin, R.E.; Luciani, P.; Benvenuti, S.; Peri, A.; Ratti, F.; Rodolfo, M.; Kulic, L.; Heppner, F.; et al. Prosurvival Effect of DHCR24/Seladin-1 in Acute and Chronic Responses to Oxidative Stress. Mol. Cell. Biol. 2008, 28, 539–550. [Google Scholar] [CrossRef]
- Di Stasi, D.; Vallacchi, V.; Campi, V.; Ranzani, T.; Daniotti, M.; Chiodini, E.; Fiorentini, S.; Greeve, I.; Prinetti, A.; Rivoltini, L.; et al. DHCR24 gene expression is upregulated in melanoma metastases and associated to resistance to oxidative stress-induced apoptosis. Int. J. Cancer 2005, 115, 224–230. [Google Scholar] [CrossRef]
- Lee, G.T.; Ha, Y.-S.; Jung, Y.S.; Moon, S.-K.; Kang, H.W.; Lee, O.-J.; Joung, J.Y.; Choi, Y.H.; Yun, S.-J.; Kim, W.-J.; et al. DHCR24 is an Independent Predictor of Progression in Patients with Non-Muscle-Invasive Urothelial Carcinoma, and Its Functional Role is Involved in the Aggressive Properties of Urothelial Carcinoma Cells. Ann. Surg. Oncol. 2014, 21, 538–545. [Google Scholar] [CrossRef]
- Lee, J.N.; Bae, S.-H.; Paik, Y.-K. Structure and alternative splicing of the rat 7-dehydrocholesterol reductase gene. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1576, 148–156. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, J.N.; Paik, Y.-K. Cholesterol Biosynthesis from lanosterol—A concerted role for Sp1 and NF-Y-binding sites for sterol-mediated regulation of rat 7-dehydrocholesterol reductase gene expression. J. Biol. Chem. 2001, 276, 18153–18160. [Google Scholar] [CrossRef] [PubMed]
- Klapper, M.; Böhme, M.; Nitz, I.; Döring, F. Type 2 diabetes-associated fatty acid binding protein 2 promoter haplotypes are differentially regulated by GATA factors. Hum. Mutat. 2007, 29, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Tsai, T.H.; Li, L.; Saha, P.; Chan, L.; Chang, B.H.-J. PLIN2 is a Key Regulator of the Unfolded Protein Response and Endoplasmic Reticulum Stress Resolution in Pancreatic β Cells. Sci. Rep. 2017, 7, 40855. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, L.-F.; Zhang, R.-P.; Zhang, W.-T. Clinical significance of FABP2 expression in newborns with necrotizing enterocolitis. World J. Pediatr. 2015, 12, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, F.; Wang, J.; Wang, Y.; Fang, Y. Retracted: Intestinal fatty acid-binding protein mediates atherosclerotic progress through increasing intestinal inflammation and permeability. J. Cell. Mol. Med. 2020, 24, 5205–5212. [Google Scholar] [CrossRef]
- Son, S.-H.; Goo, Y.-H.; Choi, M.; Saha, P.K.; Oka, K.; Chan, L.C.B.; Paul, A. Enhanced atheroprotection and lesion remodelling by targeting the foam cell and increasing plasma cholesterol acceptors. Cardiovasc. Res. 2015, 109, 294–304. [Google Scholar] [CrossRef]
- Guerreiro, I.; Enes, P.; Rodiles, A.; Merrifield, D.; Oliva-Teles, A. Effects of rearing temperature and dietary short-chain fructooligosaccharides supplementation on allochthonous gut microbiota, digestive enzymes activities and intestine health of turbot (Scophthalmus maximus L.) juveniles. Aquac. Nutr. 2015, 22, 631–642. [Google Scholar] [CrossRef]
- Li, W.; Liu, B.; Liu, Z.; Yin, Y.; Xu, G.; Han, M.; Xie, L. Effect of dietary histamine on intestinal morphology, inflammatory status, and gut microbiota in yellow catfish (Pelteobagrus fulvidraco). Fish Shellfish Immunol. 2021, 117, 95–103. [Google Scholar] [CrossRef]
- Gur, C.; Mandelboim, O.; Bachrach, G. “Messieurs, c’est les microbes qui auront le dernier mot”: Gentlemen, it is the microbes who have the last word (Louis Pasteur)—Fusobacterium nucleatumprotect tumors from killing by immune cells. OncoImmunology 2015, 4, e1038690. [Google Scholar] [CrossRef]
- Zhang, X.; Wen, K.; Ding, D.; Liu, J.; Lei, Z.; Chen, X.; Ye, G.; Zhang, J.; Shen, H.; Yan, C.; et al. Size-dependent adverse effects of microplastics on intestinal microbiota and metabolic homeostasis in the marine medaka (Oryzias melastigma). Environ. Int. 2021, 151, 106452. [Google Scholar] [CrossRef]
- Gibiino, G.; Lopetuso, L.R.; Scaldaferri, F.; Rizzatti, G.; Binda, C.; Gasbarrini, A. Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Dig. Liver Dis. 2018, 50, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, F. Gut Microbiota Parameters Potentially Useful in Clinical Perspective. Microorganisms 2021, 9, 2402. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Lapébie, P.; Lombard, V.; Drula, E.; Terrapon, N.; Henrissat, B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019, 10, 2043. [Google Scholar] [CrossRef] [PubMed]
- Machate, D.J.; Figueiredo, P.S.; Marcelino, G.; Guimarães, R.D.C.A.; Hiane, P.A.; Bogo, D.; Pinheiro, V.A.Z.; De Oliveira, L.C.S.; Pott, A. Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis. Int. J. Mol. Sci. 2020, 21, 4093. [Google Scholar] [CrossRef] [PubMed]
- Amaretti, A.; Gozzoli, C.; Simone, M.; Raimondi, S.; Righini, L.; Pérez-Brocal, V.; Lopez, R.G.; Moya, A.; Rossi, M. Profiling of Protein Degraders in Cultures of Human Gut Microbiota. Front. Microbiol. 2019, 10, 2614. [Google Scholar] [CrossRef]
- Ofek, T.; Lalzar, M.; Laviad-Shitrit, S.; Izhaki, I.; Halpern, M. Comparative Study of Intestinal Microbiota Composition of Six Edible Fish Species. Front. Microbiol. 2021, 12, 760266. [Google Scholar] [CrossRef]
- Xiao, F.; Liao, L.; Xu, Q.; He, Z.; Xiao, T.; Wang, J.; Huang, J.; Yu, Y.; Wu, B.; Yan, Q. Host–microbiota interactions and responses to grass carp reovirus infection in Ctenopharyngodon idellus. Environ. Microbiol. 2020, 23, 431–447. [Google Scholar] [CrossRef]
- Liu, T.; Guo, L.; Zhangying, Y.; Ruan, S.; Liu, W.; Zhang, X.; Feng, F. Dietary medium-chain 1-monoglycerides modulates the community and function of cecal microbiota of broilers. J. Sci. Food Agric. 2021, 102, 2242–2252. [Google Scholar] [CrossRef]
- Nicholson, S.E.; Merrill, D.; Zhu, C.; Burmeister, D.M.; Zou, Y.; Lai, Z.; Darlington, D.N.; Lewis, A.M.; Newton, L.; Scroggins, S.; et al. Polytrauma independent of therapeutic intervention alters the gastrointestinal microbiome. Am. J. Surg. 2018, 216, 699–705. [Google Scholar] [CrossRef]
- Loch, T.P.; Fujimoto, M.; Woodiga, S.A.; Walker, E.D.; Marsh, T.; Faisal, M. Diversity of Fish-Associated Flavobacteria of Michigan. J. Aquat. Anim. Health 2013, 25, 149–164. [Google Scholar] [CrossRef]
- Loch, T.; Faisal, M. Polyphasic characterization reveals the presence of novel fish-associated Chryseobacterium spp. in the Great Lakes of North America. Dis. Aquat. Org. 2015, 113, 113–125. [Google Scholar] [CrossRef]
- Li, J.; Cao, J.; Wang, X.; Liu, N.; Wang, W.; Luo, Y. Acinetobacter pittii, an emerging new multi-drug resistant fish pathogen isolated from diseased blunt snout bream (Megalobrama amblycephala Yih) in China. Appl. Microbiol. Biotechnol. 2017, 101, 6459–6471. [Google Scholar] [CrossRef] [PubMed]
- Malick, R.C.; Bera, A.K.; Chowdhury, H.; Bhattacharya, M.; Abdulla, T.; Swain, H.S.; Baitha, R.; Kumar, V.; Das, B.K. Identification and pathogenicity study of emerging fish pathogens Acinetobacter junii and Acinetobacter pittii recovered from a disease outbreak in Labeo catla (Hamilton, 1822) and Hypophthalmichthys molitrix (Valenciennes, 1844) of freshwater wetland in West Bengal, India. Aquac. Res. 2020, 51, 2410–2420. [Google Scholar] [CrossRef]
- Hieu, D.Q.; Hang, B.T.B.; Lokesh, J.; Garigliany, M.-M.; Huong, D.T.T.; Yen, D.T.; Liem, P.T.; Tam, B.M.; Hai, D.M.; Son, V.N.; et al. Salinity significantly affects intestinal microbiota and gene expression in striped catfish juveniles. Appl. Microbiol. Biotechnol. 2022, 106, 3245–3264. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Ngugi, D.K.; Stingl, U. Phylogenetic Diversity, Distribution, and Cophylogeny of Giant Bacteria (Epulopiscium) with their Surgeonfish Hosts in the Red Sea. Front. Microbiol. 2016, 7, 285. [Google Scholar] [CrossRef]
- Parata, L.; Nielsen, S.; Xing, X.; Thomas, T.; Egan, S.; Verges, A. Age, gut location and diet impact the gut microbiome of a tropical herbivorous surgeonfish. FEMS Microbiol. Ecol. 2019, 96, fiz179. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Y.; Wen, Y.; Ji, W.; Zhou, Y.; Ji, Y.; Liu, X.; Wang, W.; Asim, M.; Liang, X.; et al. Analysis of the transcriptomic profilings of Mandarin fish (Siniperca chuatsi) infected with Flavobacterium columnare with an emphasis on immune responses. Fish Shellfish. Immunol. 2015, 43, 111–119. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, T.; Tao, Y.; Lu, S.; Qiang, J.; Xu, P. Integrated Transcriptome and 16S rDNA Analyses Reveal That Transport Stress Induces Oxidative Stress and Immune and Metabolic Disorders in the Intestine of Hybrid Yellow Catfish (Tachysurus fulvidraco♀ × Pseudobagrus vachellii♂). Antioxidants 2022, 11, 1737. https://doi.org/10.3390/antiox11091737
Zheng T, Tao Y, Lu S, Qiang J, Xu P. Integrated Transcriptome and 16S rDNA Analyses Reveal That Transport Stress Induces Oxidative Stress and Immune and Metabolic Disorders in the Intestine of Hybrid Yellow Catfish (Tachysurus fulvidraco♀ × Pseudobagrus vachellii♂). Antioxidants. 2022; 11(9):1737. https://doi.org/10.3390/antiox11091737
Chicago/Turabian StyleZheng, Tao, Yifan Tao, Siqi Lu, Jun Qiang, and Pao Xu. 2022. "Integrated Transcriptome and 16S rDNA Analyses Reveal That Transport Stress Induces Oxidative Stress and Immune and Metabolic Disorders in the Intestine of Hybrid Yellow Catfish (Tachysurus fulvidraco♀ × Pseudobagrus vachellii♂)" Antioxidants 11, no. 9: 1737. https://doi.org/10.3390/antiox11091737




