Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains Used in This Study
2.2. Measuring Chronological Lifespan by Flow Citometry
2.3. Oxygen Consumption
2.4. Preparation of Mitochondria-Enriched Fractions
2.5. Citrate Synthase Activity
2.6. Measurement of Complex IV Activity in Mitochondria-Enriched Protein Fractions
2.7. Tolerance to H2O2 in Liquid Cultures—Recording of Growth Curves
2.8. Microscopy and Image Analysis
2.9. Statistics
3. Results
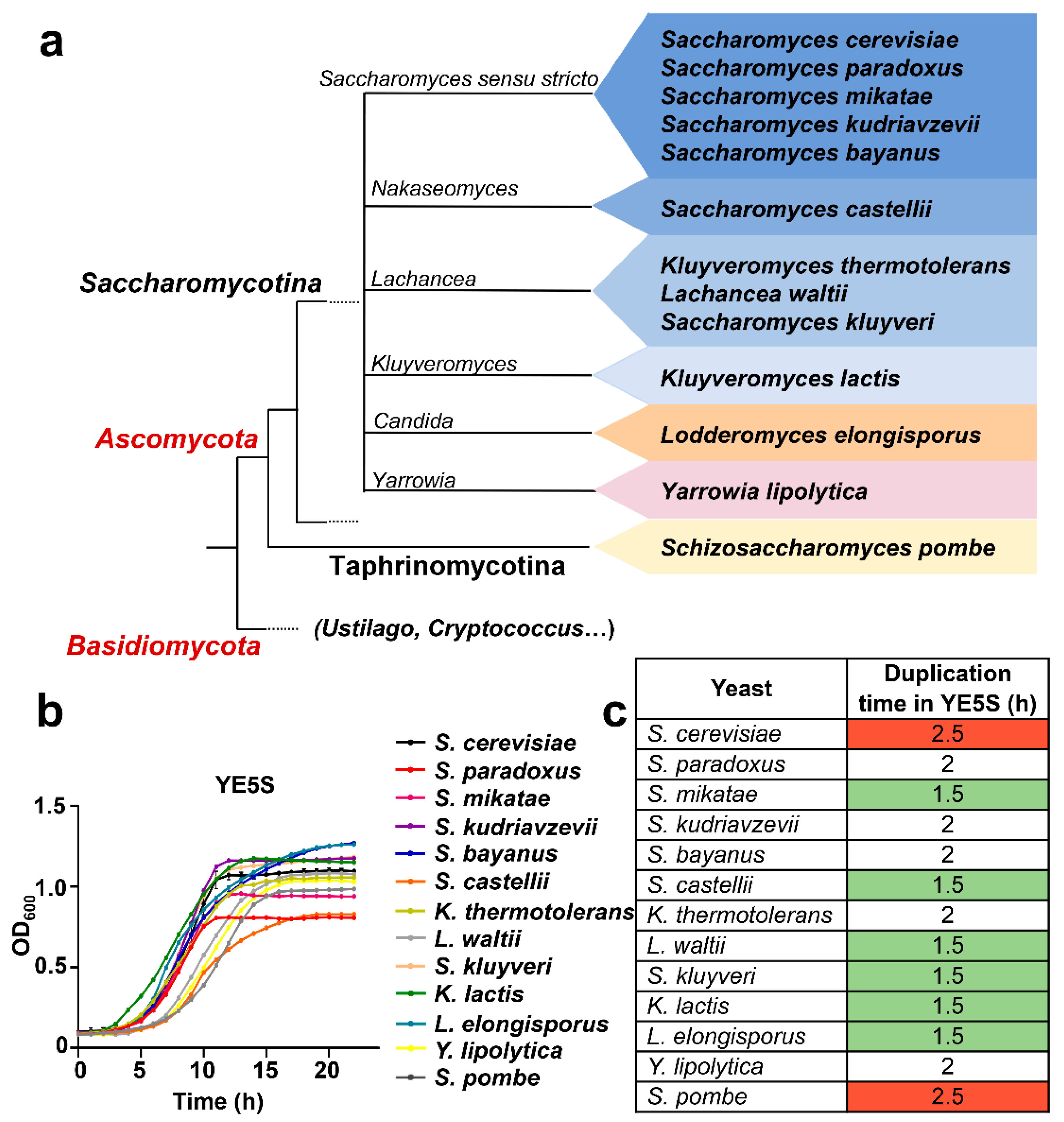
3.1. Growth of 13 Species of Yeast in S. pombe Rich Medium YE5S
3.2. Analysis of CLS in the 13 Yeast Species—Measuring Viability at the Stationary Phase
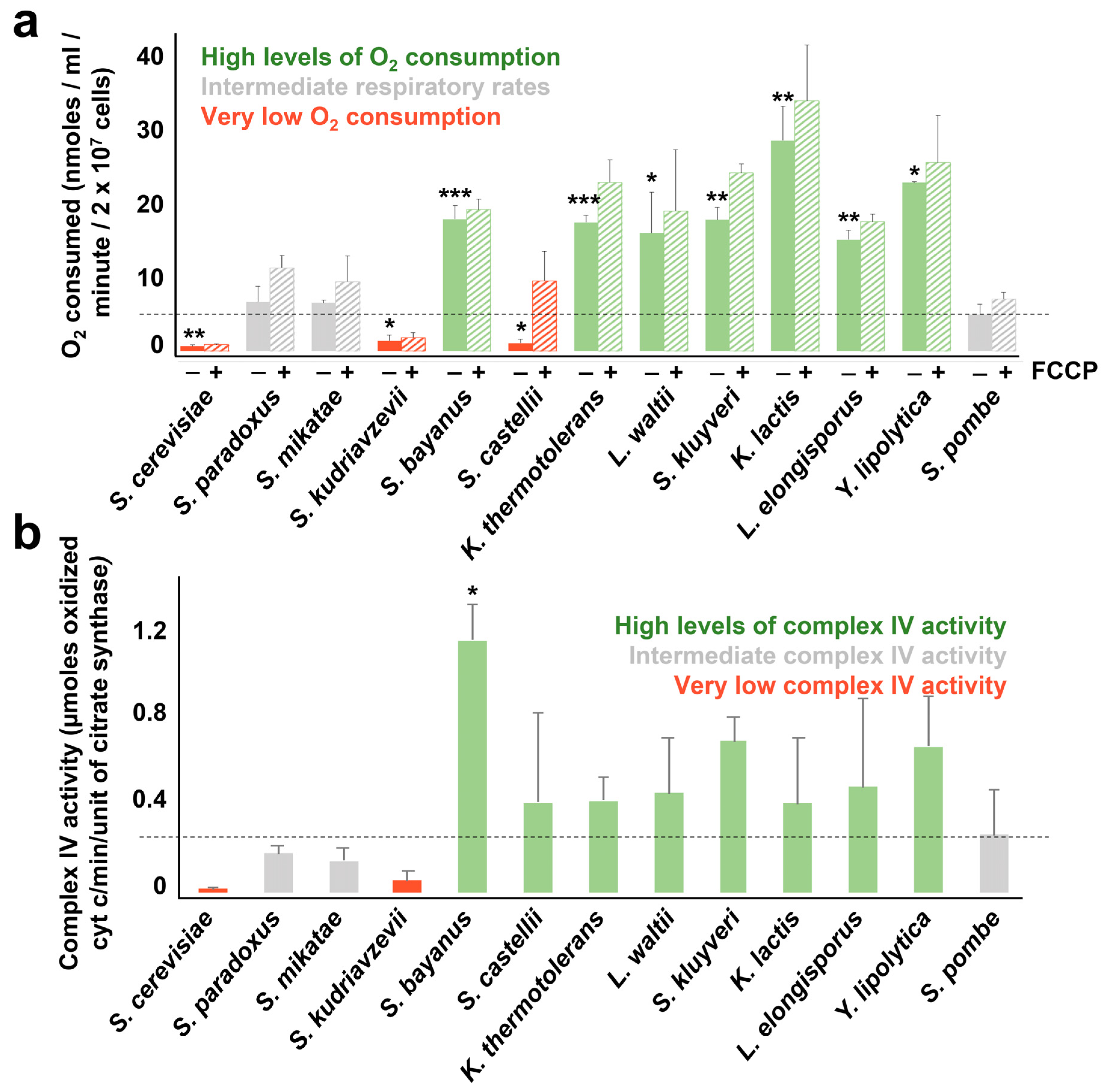
3.3. Measuring Mitochondrial Activity in the Different Yeasts
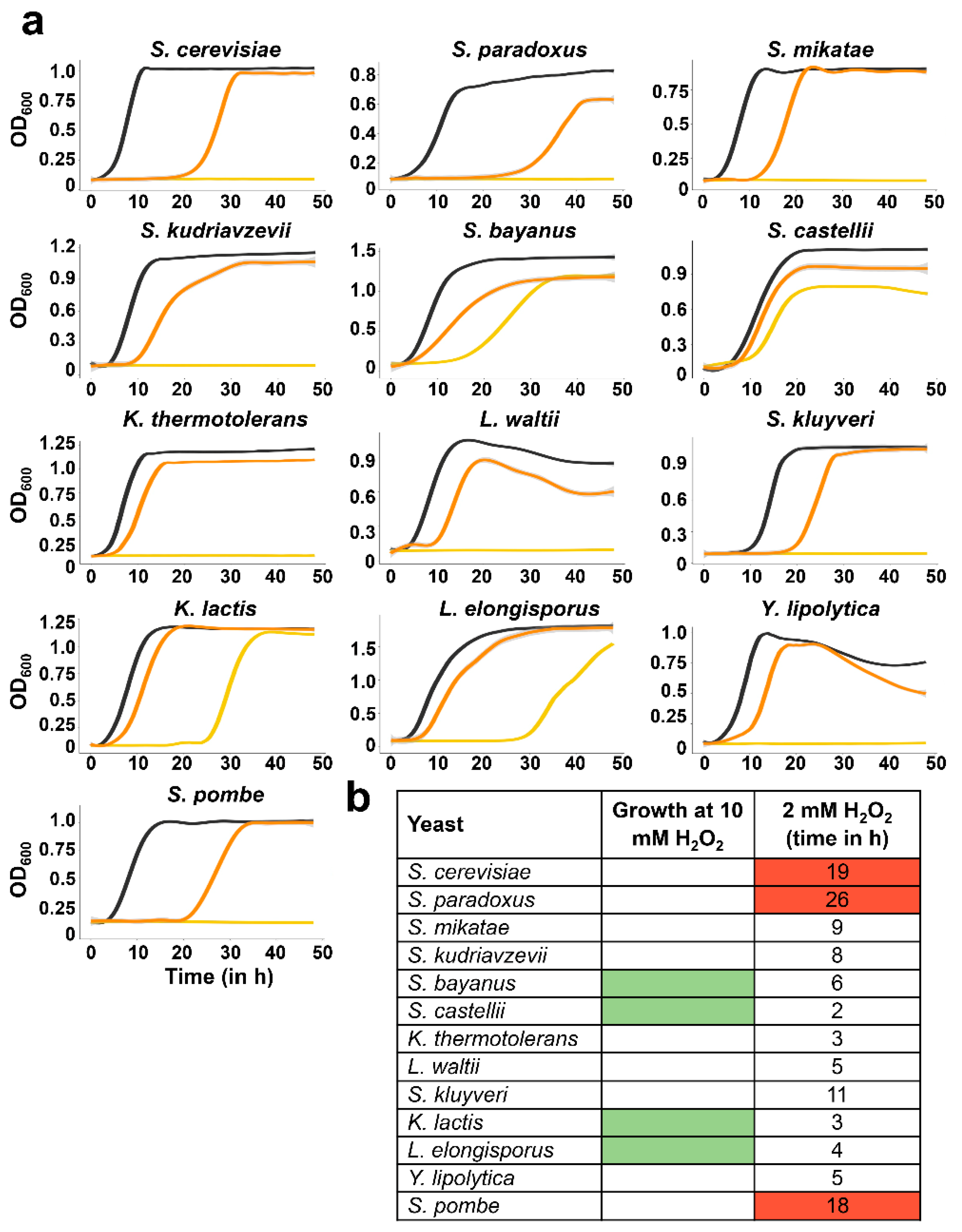
3.4. Measuring Tolerance to Oxidative Stress of the Different Yeasts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Kroemer, G. Hallmarks of Health. Cell 2021, 184, 33–63. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Longo, V.D. Yeast Chronological Lifespan: Longevity Regulatory Genes and Mechanisms. Cells 2022, 11, 1714. [Google Scholar] [CrossRef] [PubMed]
- Legon, L.; Rallis, C. Genome-wide screens in yeast models towards understanding chronological lifespan regulation. Brief. Funct. Genom. 2022, 21, 4–12. [Google Scholar] [CrossRef]
- Zuin, A.; Carmona, M.; Morales-Ivorra, I.; Gabrielli, N.; Vivancos, A.P.; Ayte, J.; Hidalgo, E. Lifespan extension by calorie restriction relies on the Sty1 MAP kinase stress pathway. EMBO J. 2010, 29, 981–991. [Google Scholar] [CrossRef]
- Chen, K.; Shen, W.; Gao, Z.; Luo, C. Stress response capacity analysis during aging and possible new insights into aging studies. Curr. Genet. 2021, 67, 417–420. [Google Scholar] [CrossRef]
- Dawes, I.W.; Perrone, G.G. Stress and ageing in yeast. FEMS Yeast Res. 2020, 20, foz085. [Google Scholar] [CrossRef]
- Vega, M.; Castillo, D.; de Cubas, L.; Wang, Y.; Huang, Y.; Hidalgo, E.; Cabrera, M. Antagonistic effects of mitochondrial matrix and intermembrane space proteases on yeast aging. BMC Biol. 2022, 20, 160. [Google Scholar] [CrossRef]
- Vazquez-Calvo, C.; Suhm, T.; Buttner, S.; Ott, M. The basic machineries for mitochondrial protein quality control. Mitochondrion 2020, 50, 121–131. [Google Scholar] [CrossRef]
- Malina, C.; Larsson, C.; Nielsen, J. Yeast mitochondria: An overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 2018, 18, foy040. [Google Scholar] [CrossRef]
- Baccolo, G.; Stamerra, G.; Coppola, D.P.; Orlandi, I.; Vai, M. Mitochondrial Metabolism and Aging in Yeast. Int. Rev. Cell Mol. Biol. 2018, 340, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.A.; Louis, E.J. Genome Diversity and Evolution in the Budding Yeasts (Saccharomycotina). Genetics 2017, 206, 717–750. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006, 22, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Leupold, U. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970, 4, 169–177. [Google Scholar]
- Zuin, A.; Vivancos, A.P.; Sanso, M.; Takatsume, Y.; Ayte, J.; Inoue, Y.; Hidalgo, E. The glycolytic metabolite methylglyoxal activates Pap1 and Sty1 stress responses in Schizosaccharomyces pombe. J. Biol. Chem. 2005, 280, 36708–36713. [Google Scholar] [CrossRef]
- Rothstein, R.J. One-step gene disruption in yeast. Methods Enzymol. 1983, 101, 202–211. [Google Scholar] [CrossRef]
- Blevins, W.R.; Ruiz-Orera, J.; Messeguer, X.; Blasco-Moreno, B.; Villanueva-Canas, J.L.; Espinar, L.; Diez, J.; Carey, L.B.; Alba, M.M. Uncovering de novo gene birth in yeast using deep transcriptomics. Nat. Commun. 2021, 12, 604. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar]
- Tsankov, A.M.; Thompson, D.A.; Socha, A.; Regev, A.; Rando, O.J. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010, 8, e1000414. [Google Scholar] [CrossRef]
- Ocampo, A.; Barrientos, A. Quick and reliable assessment of chronological life span in yeast cell populations by flow cytometry. Mech. Ageing Dev. 2011, 132, 315–323. [Google Scholar] [CrossRef]
- Zuin, A.; Gabrielli, N.; Calvo, I.A.; Garcia-Santamarina, S.; Hoe, K.L.; Kim, D.U.; Park, H.O.; Hayles, J.; Ayte, J.; Hidalgo, E. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS ONE 2008, 3, e2842. [Google Scholar] [CrossRef] [PubMed]
- Chiron, S.; Gaisne, M.; Guillou, E.; Belenguer, P.; Clark-Walker, G.D.; Bonnefoy, N. Studying mitochondria in an attractive model: Schizosaccharomyces pombe. Methods Mol. Biol. 2007, 372, 91–105. [Google Scholar] [CrossRef]
- Yarian, C.S.; Toroser, D.; Sohal, R.S. Aconitase is the main functional target of aging in the citric acid cycle of kidney mitochondria from mice. Mech. Ageing Dev. 2006, 127, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, C.; Dujardin, G. Preparation of respiratory chain complexes from Saccharomyces cerevisiae wild-type and mutant mitochondria: Activity measurement and subunit composition analysis. Methods Mol. Biol. 2008, 432, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Calvo, I.A.; Gabrielli, N.; Iglesias-Baena, I.; Garcia-Santamarina, S.; Hoe, K.L.; Kim, D.U.; Sanso, M.; Zuin, A.; Perez, P.; Ayte, J.; et al. Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS ONE 2009, 4, e6619. [Google Scholar] [CrossRef]
- Garcia, P.; Encinar Del Dedo, J.; Ayte, J.; Hidalgo, E. Genome-wide Screening of Regulators of Catalase Expression: Role of a Transcription Complex and Histone and Trna Modification Complexes on Adaptation to Stress. J. Biol. Chem. 2016, 291, 790–799. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Dujon, B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010, 11, 512–524. [Google Scholar] [CrossRef]
- Alfa, C.; Fantes, P.; Hyams, J.; McLeod, M.; Warbrick, E. Experiments with Fission Yeast: A Laboratory Course Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1993. [Google Scholar]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef]
- Pendergrass, W.; Wolf, N.; Poot, M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry. Part A J. Int. Soc. Anal. Cytol. 2004, 61, 162–169. [Google Scholar] [CrossRef]
- Keij, J.F.; Bell-Prince, C.; Steinkamp, J.A. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry 2000, 39, 203–210. [Google Scholar] [CrossRef]
- Buckman, J.F.; Hernandez, H.; Kress, G.J.; Votyakova, T.V.; Pal, S.; Reynolds, I.J. MitoTracker labeling in primary neuronal and astrocytic cultures: Influence of mitochondrial membrane potential and oxidants. J. Neurosci. Methods 2001, 104, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Kilgore, J.A.; Dolman, N.J. A Review of Reagents for Fluorescence Microscopy of Cellular Compartments and Structures, Part II: Reagents for Non-Vesicular Organelles. Curr. Protoc. 2023, 3, e752. [Google Scholar] [CrossRef] [PubMed]
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Wolfe, K.H. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 2007, 3, 129. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Kuhl, H.; Ralser, M.; Werber, M.; Lehrach, H.; Breitenbach, M.; Timmermann, B. The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: Insights into ancestry and physiology of a laboratory mutt. Open Biol. 2012, 2, 120093. [Google Scholar] [CrossRef]
- Zadrag-Tecza, R.; Kwolek-Mirek, M.; Bartosz, G.; Bilinski, T. Cell volume as a factor limiting the replicative lifespan of the yeast Saccharomyces cerevisiae. Biogerontology 2009, 10, 481–488. [Google Scholar] [CrossRef]
- Crabtree, H.G. Observations on the carbohydrate metabolism of tumours. Biochem J 1929, 23, 536–545. [Google Scholar] [CrossRef]
- De Deken, R.H. The Crabtree effects and its relation to the petite mutation. J. Gen. Microbiol. 1966, 44, 157–165. [Google Scholar] [CrossRef]
- Gonzalez-Siso, M.I.; Freire-Picos, M.A.; Ramil, E.; Gonzalez-Dominguez, M.; Rodriguez Torres, A.; Cerdan, M.E. Respirofermentative metabolism in Kluyveromyces lactis: Insights and perspectives. Enzym. Microb. Technol. 2000, 26, 699–705. [Google Scholar] [CrossRef]
- Freel, K.C.; Friedrich, A.; Schacherer, J. Mitochondrial genome evolution in yeasts: An all-encompassing view. FEMS Yeast Res. 2015, 15, fov023. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.J.; Zarse, K.; Voigt, A.; Urban, N.; Birringer, M.; Ristow, M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007, 6, 280–293. [Google Scholar] [CrossRef] [PubMed]


| Yeast Strains | Phenotypes | Mitochondrial Activity c | |||
|---|---|---|---|---|---|
| Longevity by FACS a | H2O2 Tolerance b | O2 Consumption | MitoTracker Staining | CIV Activity | |
| Group 1 | |||||
| K. lactis | |||||
| S. bayanus | |||||
| L. elongisporus | |||||
| Group 2 | |||||
| S. kluyveri | |||||
| K. thermotolerans | |||||
| L. waltii | |||||
| Y. lipolytica | |||||
| Group 3 | |||||
| S. mikatae | |||||
| S. paradoxus | |||||
| S. pombe | |||||
| Group 4 | |||||
| S. kudriavzevii | |||||
| S. castellii | |||||
| S. cerevisiae | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gröger, A.; Martínez-Albo, I.; Albà, M.M.; Ayté, J.; Vega, M.; Hidalgo, E. Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species. Antioxidants 2023, 12, 1810. https://doi.org/10.3390/antiox12101810
Gröger A, Martínez-Albo I, Albà MM, Ayté J, Vega M, Hidalgo E. Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species. Antioxidants. 2023; 12(10):1810. https://doi.org/10.3390/antiox12101810
Chicago/Turabian StyleGröger, Anna, Ilune Martínez-Albo, M. Mar Albà, José Ayté, Montserrat Vega, and Elena Hidalgo. 2023. "Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species" Antioxidants 12, no. 10: 1810. https://doi.org/10.3390/antiox12101810
APA StyleGröger, A., Martínez-Albo, I., Albà, M. M., Ayté, J., Vega, M., & Hidalgo, E. (2023). Comparing Mitochondrial Activity, Oxidative Stress Tolerance, and Longevity of Thirteen Ascomycota Yeast Species. Antioxidants, 12(10), 1810. https://doi.org/10.3390/antiox12101810








