Characterization of the Metallothionein Gene Family in Avena sativa L. and the Gene Expression during Seed Germination and Heavy Metal Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oat Metallothionein Genome-Wide Identification and Analysis of Putative AsMT Proteins
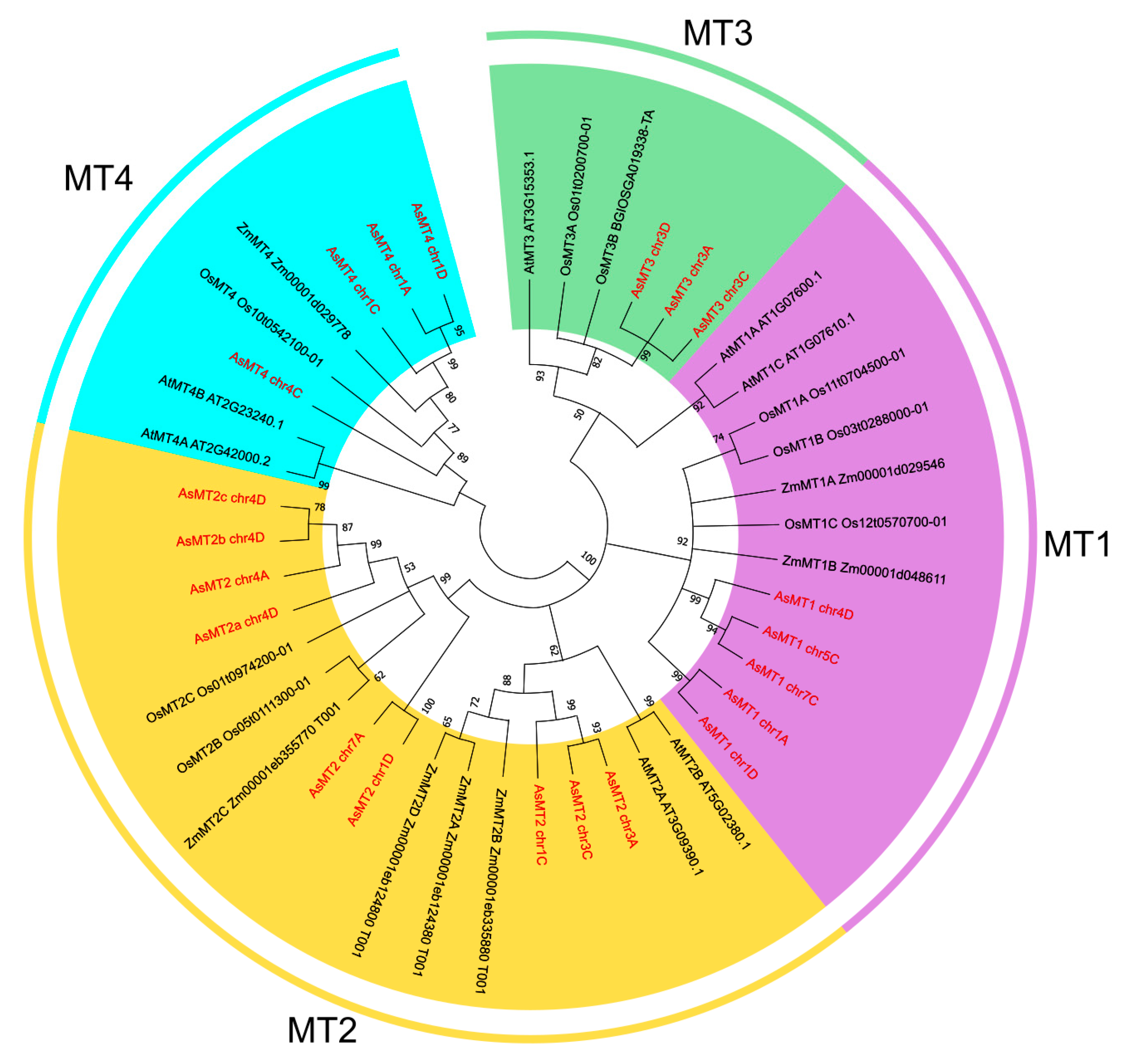
2.2. Phylogenetic Analysis and Conserved Motif Analysis
2.3. Prediction of Cis-Responsive Elements in AsMT Promoters
2.4. Plant Material
2.5. Total RNA Isolation
2.6. Quantitative Real-Time PCR (RT-qPCR) Analysis
2.7. Determination of Photosynthetic Pigments
2.8. Determination of Total Phenolic Content
2.9. Determination of Antioxidant Capacity
2.9.1. ABTS Assay
2.9.2. Ferric Reducing Antioxidant Power (FRAP) Assay
2.10. Statistical Analysis
3. Results
3.1. Identification and Chromosome Distribution of A. sativa Metallothionein (AsMT) Genes
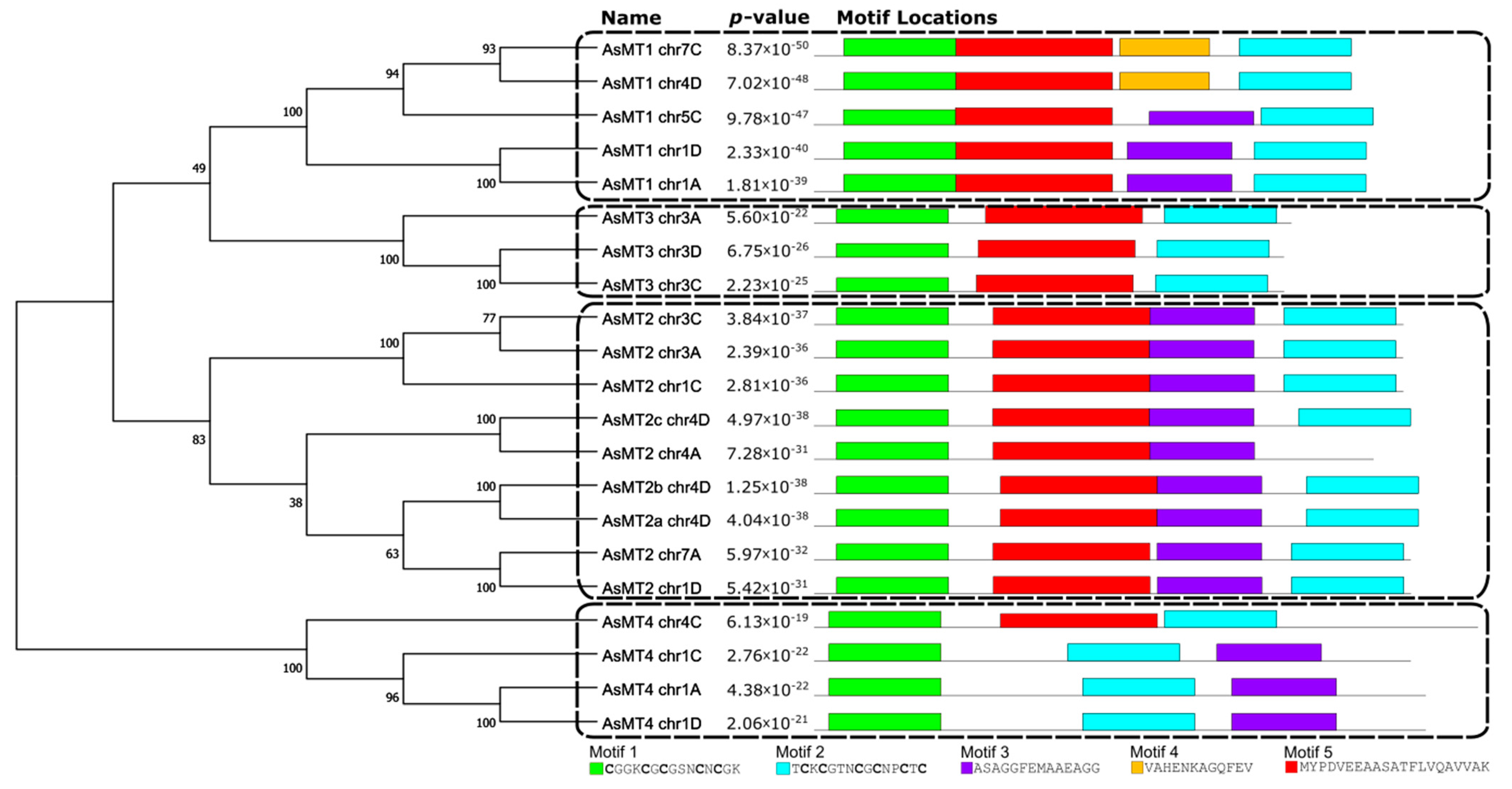
3.2. Analysis of Conserved Motifs in the Amino Acid Sequences of AsMTs
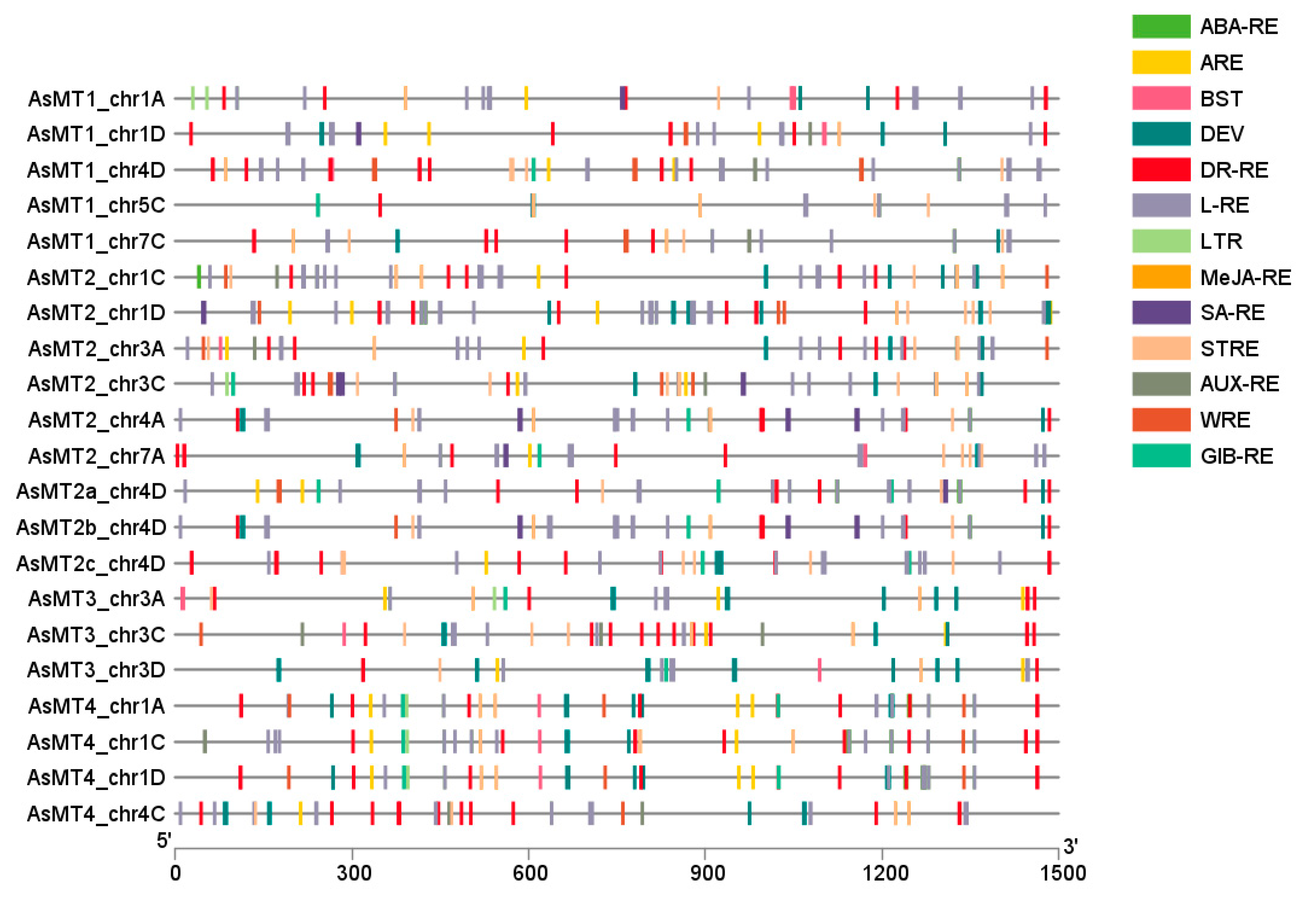
3.3. Prediction of Cis-Responsive Elements in AsMT Promoters
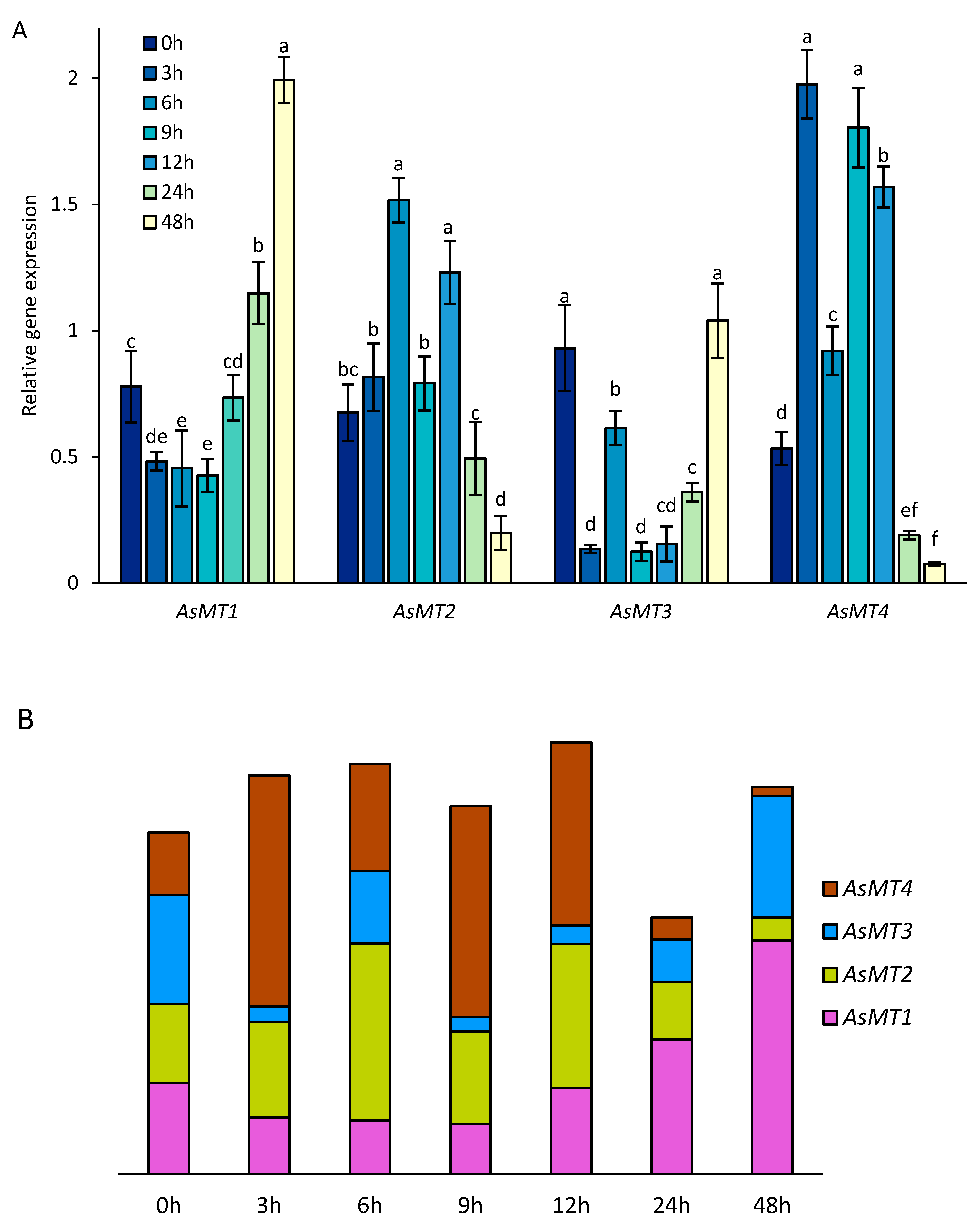
3.4. AsMT1-4 Expression during Seed Germination
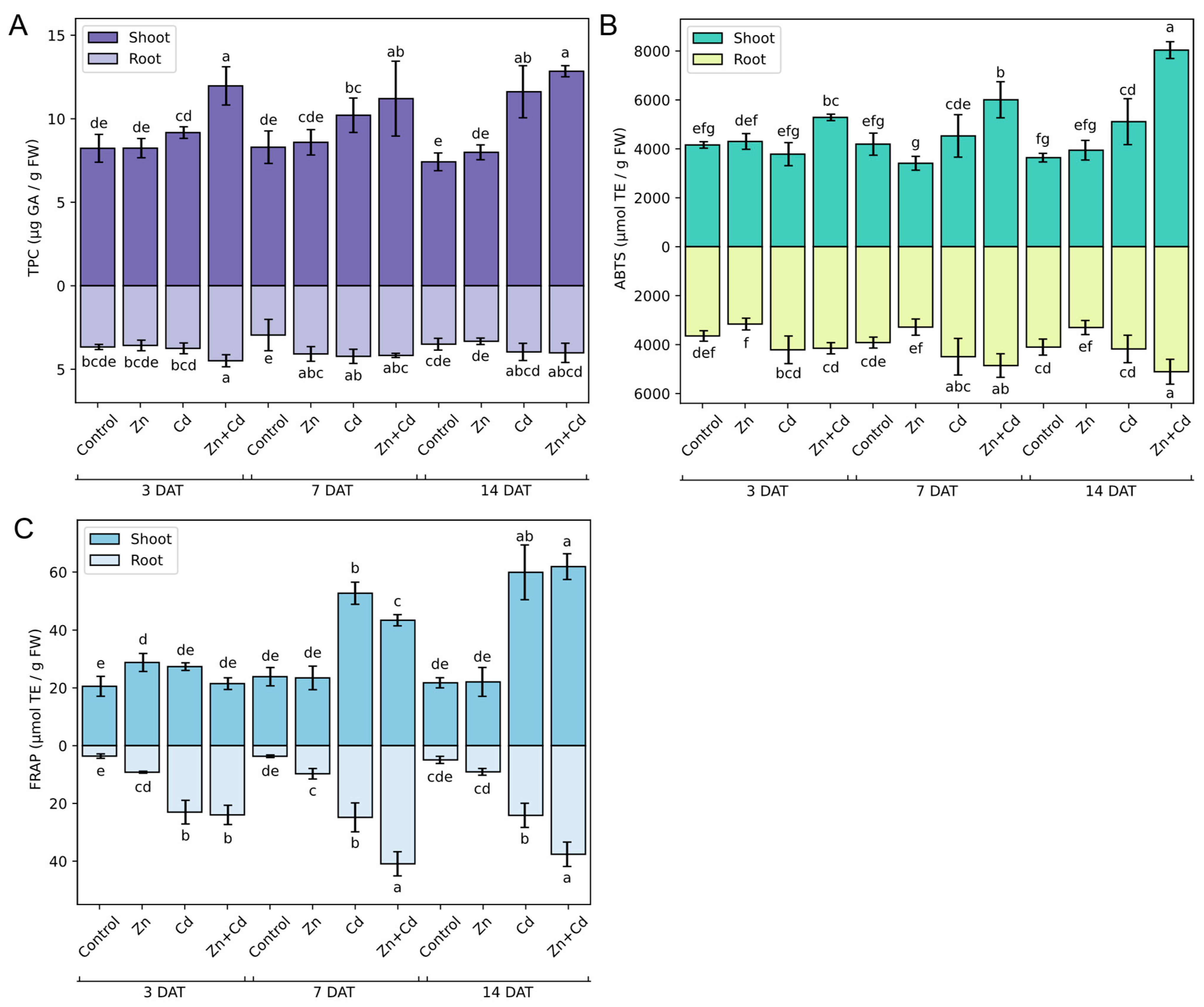
3.5. Effect of Heavy Metal on Oat Seedling Growth
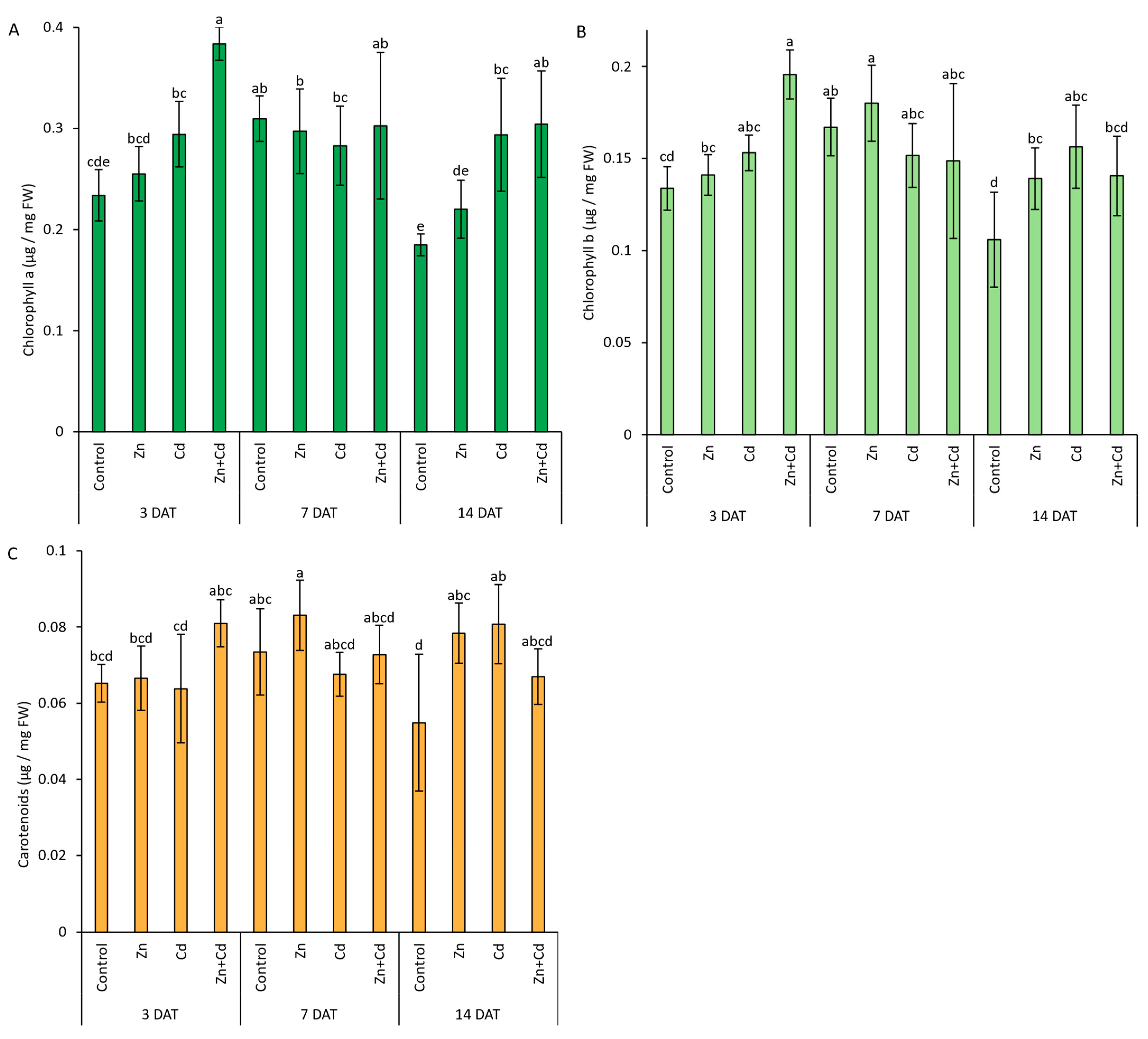
3.6. Chlorophyll a and b and Carotenoid Content in Response to Heavy Metal
3.7. Antioxidant Capacity of Oat Seedlings in Response to Heavy Metal
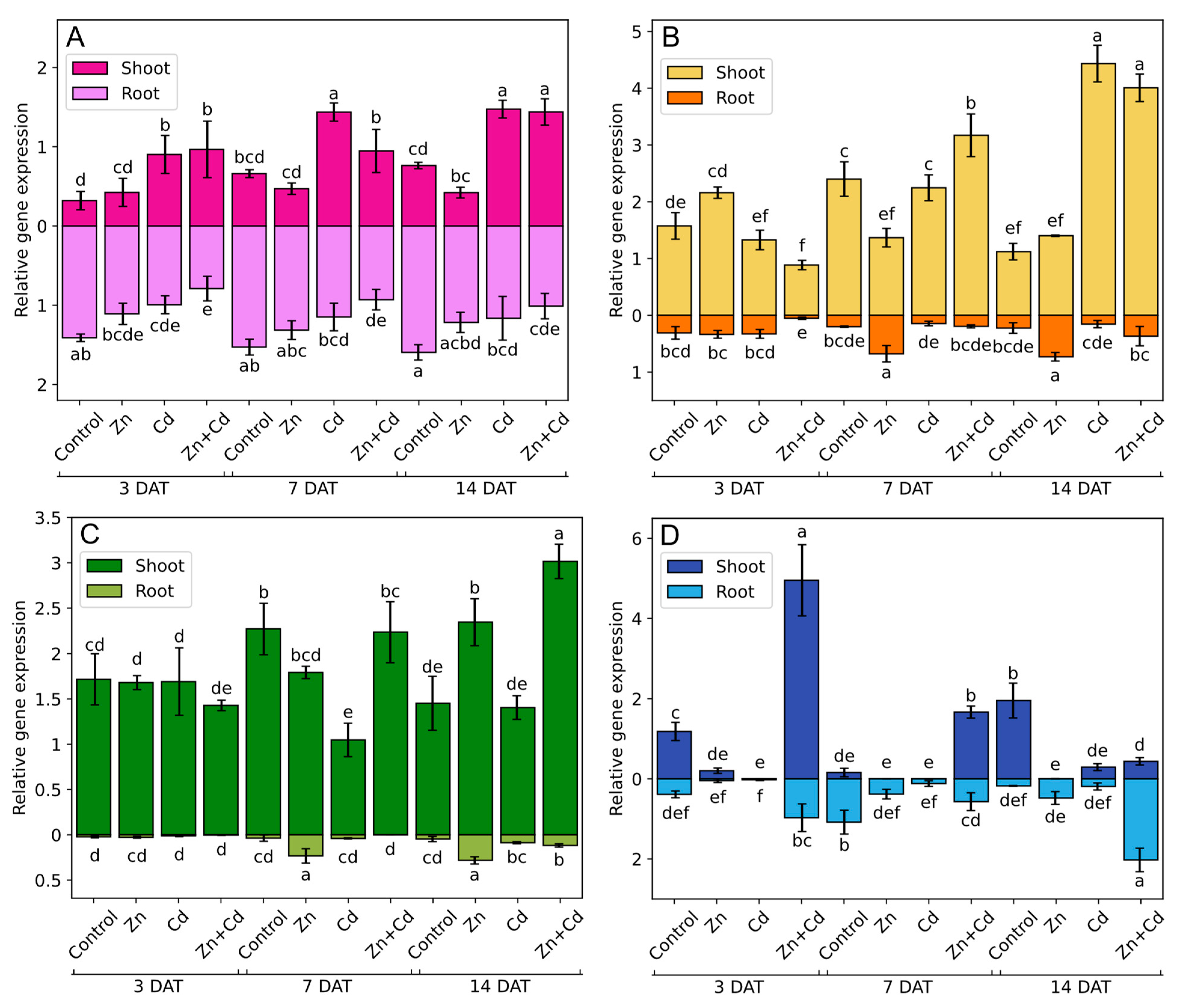
3.8. AsMT1-4 Expression during Heavy Metal Stress
3.9. Correlations among AsMT1-4 Gene Expression, the Content of Photosynthetic Pigments, and Antioxidant Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blindauer, C.A. Bacterial metallothioneins: Past, present, and questions for the future. J. Biol. Inorg. Chem. 2011, 16, 1011–1024. [Google Scholar] [CrossRef]
- Dąbrowska, G.; Mierek-Adamska, A.; Goc, A. Plant metallothioneins: Putative functions identified by promoter analysis in silico. Acta Biol. Crac. Ser. Bot. 2012, 54, 109–120. [Google Scholar] [CrossRef]
- Parameswari, E.; Ilakiya, T.; Davamani, V.; Kalaiselvi, P.; Sebastian, S.P. Metallothioneins: Diverse Protein Family to Bind Metallic Ions. In Heavy Metals-Their Environmental Impacts and Mitigation; IntechOpen: London, UK, 2021; ISBN 978-1-83968-122-6. [Google Scholar]
- Blindauer, C.A.; Leszczyszyn, O.I. Metallothioneins: Unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat. Prod. Rep. 2010, 27, 720–741. [Google Scholar] [CrossRef] [PubMed]
- Margoshes, M.; Valiee, B.L. A cadmium protein from equine kidney cortex. J. Am. Chem. Soc. 1957, 79, 4813–4814. [Google Scholar] [CrossRef]
- Lane, B.; Kajioka, R.; Kennedy, T. The wheat-germ Ec protein is a zinc-containing metallothionein. Biochem. Cell Biol. 1987, 65, 1001–1005. [Google Scholar] [CrossRef]
- Kawashima, I.; Kennedy, T.D.; Chino, M.; Lane, B.G. Wheat Ec metallothionein genes: Like mammalian Zn2+ metallothionein genes, wheat Zn2+ metallothionein genes are conspicuously expressed during embryogenesis. Eur. J. Biochem. 1992, 209, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, K.; Jakob, U. The role of thiols in antioxidant systems. Free Radic. Biol. Med. 2019, 140, 14. [Google Scholar] [CrossRef]
- Ziller, A.; Fraissinet-Tachet, L. Metallothionein diversity and distribution in the tree of life: A multifunctional protein. Metallomics 2018, 10, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Freisinger, E. Plant MTs-Long neglected members of the metallothionein superfamily. Dalton Trans. 2008, 47, 6663–6675. [Google Scholar] [CrossRef] [PubMed]
- Saeed-Ur-Rahman;Khalid, M.; Hui, N.; Kayani, S.I.; Tang, K. Diversity and versatile functions of metallothioneins produced by plants: A review. Pedosphere 2020, 30, 577–588. [Google Scholar] [CrossRef]
- Freisinger, E. Structural features specific to plant metallothioneins. J. Biol. Inorg. Chem. 2011, 16, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, J.; Liu, S.; Jiang, L.; Hu, L. Identification of the metallothionein gene family from cucumber and functional characterization of CsMT4 in Escherichia coli under salinity and osmotic stress. 3 Biotech 2019, 9, 394. [Google Scholar] [CrossRef] [PubMed]
- Leszczyszyn, O.I.; Schmid, R.; Blindauer, C.A. Toward a property/function relationship for metallothioneins: Histidine coordination and unusual cluster composition in a zinc-metallothionein from plants. Proteins Struct. Funct. Genet. 2007, 68, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Blindauer, C.A.; Razi, M.T.; Campopiano, D.J.; Sadler, P.J. Histidine ligands in bacterial metallothionein enhance cluster stability. J. Biol. Inorg. Chem. 2007, 12, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Perinelli, M.; Tegoni, M.; Freisinger, E. Different behavior of the histidine residue toward cadmium and zinc in a cadmium-specific metallothionein from an aquatic fungus. Inorg. Chem. 2020, 59, 16988–16997. [Google Scholar] [CrossRef]
- Mekawy, A.M.M.; Assaha, D.V.M.; Ueda, A. Constitutive overexpression of rice metallothionein-like gene OsMT-3a enhances growth and tolerance of Arabidopsis plants to a combination of various abiotic stresses. J. Plant Res. 2020, 133, 429–440. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Islam, M.R.; Xue, X.; Mao, S.; Ren, C.; Eneji, A.E.; Hu, Y. Effects of water-saving superabsorbent polymer on antioxidant enzyme activities and lipid peroxidation in oat (Avena sativa L.) under drought stress. J. Sci. Food Agric. 2011, 91, 680–686. [Google Scholar] [CrossRef]
- Skrzypek, E.; Szechyńska-Hebda, M.; Dąbrowska, G.B.; Goc, A. The role of osmotic stress during in vitro regeneration of Triticum aestivum L. and Vicia faba ssp. minor. Problemowe Postępów Nauk Rolniczych 2008, 524, 221–230. [Google Scholar]
- Dąbrowska, G.; Kata, A.; Goc, A.; Szechyńska-Hebda, M.; Skrzypek, E. Characteristics of the plant ascorbate peroxidase family. Acta Biol. Crac. Ser. Bot. 2007, 49, 7–17. [Google Scholar]
- Konieczna, W.; Warchoł, M.; Mierek-Adamska, A.; Skrzypek, E.; Waligórski, P.; Piernik, A.; Dąbrowska, G.B. Changes in physio-biochemical parameters and expression of metallothioneins in Avena sativa L. in response to drought. Sci. Rep. 2023, 13, 2486. [Google Scholar] [CrossRef]
- Mierek-Adamska, A.; Kotowicz, K.; Goc, A.; Boniecka, J.; Berdychowska, J.; Dąbrowska, G.B. Potential involvement of rapeseed (Brassica napus L.) metallothioneins in the hydrogen peroxide-induced regulation of seed vigour. J. Agron. Crop Sci. 2019, 205, 598–607. [Google Scholar] [CrossRef]
- Konieczna, W.; Mierek-Adamska, A.; Warchoł, M.; Skrzypek, E.; Dąbrowska, G.B. The involvement of metallothioneins and stress markers in response to osmotic stress in Avena sativa L. J. Agron. Crop Sci. 2023, 209, 371–389. [Google Scholar] [CrossRef]
- Patankar, H.V.; Al-Harrasi, I.; Al Kharusi, L.; Jana, G.A.; Al-Yahyai, R.; Sunkar, R.; Yaish, M.W. Overexpression of a Metallothionein 2A Gene from Date Palm Confers Abiotic Stress Tolerance to Yeast and Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 2871. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Li, X.; Zhu, W.; Wu, C.; Yang, G.; Zheng, C. Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J. Exp. Bot. 2009, 60, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Anandhan, S.; Singh, S.; Patade, V.Y.; Ahmed, Z.; Pande, V. Metallothionein-like gene from Cicer microphyllum is regulated by multiple abiotic stresses. Protoplasma 2011, 248, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, G.; Baum, C.; Trejgell, A.; Hrynkiewicz, K. Impact of arbuscular mycorrhizal fungi on the growth and expression of gene encoding stress protein-metallothionein BnMT2 in the non-host crop Brassica napus L. J. Plant Nutr. Soil Sci. 2014, 177, 459–467. [Google Scholar] [CrossRef]
- Zhi, J.; Liu, X.; Yin, P.; Yang, R.; Liu, J.; Xu, J. Overexpression of the metallothionein gene PaMT3-1 from Phytolacca americana enhances plant tolerance to cadmium. Plant Cell Tissue Organ. Cult. 2020, 143, 211–218. [Google Scholar] [CrossRef]
- Gu, C.-S.; Liu, L.-Q.; Zhao, Y.-H.; Deng, Y.-M.; Zhu, X.-D.; Huang, S.-Z. Overexpression of Iris. lactea var. chinensis metallothionein llMT2a enhances cadmium tolerance in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2014, 105, 22–28. [Google Scholar] [CrossRef]
- Kumar, S.; Yadav, A.; Verma, R.; Dubey, A.K.; Narayan, S.; Pandey, A.; Sahu, A.; Srivastava, S.; Sanyal, I. Metallothionein (MT1): A molecular stress marker in chickpea enhances drought and heavy metal stress adaptive efficacy in transgenic plants. Environ. Exp. Bot. 2022, 199, 104871. [Google Scholar] [CrossRef]
- Feng, M.; Yu, Q.; Chen, Y.; Fu, Z.; Xu, L.; Guo, J. ScMT10, a metallothionein-like gene from sugarcane, enhances freezing tolerance in Nicotiana tabacum transgenic plants. Environ. Exp. Bot. 2022, 194, 104750. [Google Scholar] [CrossRef]
- Yuan, J.; Chen, D.; Ren, Y.; Zhang, X.; Zhao, J. Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 2008, 146, 1637–1650. [Google Scholar] [CrossRef]
- Moyle, R.; Fairbairn, D.J.; Ripi, J.; Crowe, M.; Botella, J.R. Developing pineapple fruit has a small transcriptome dominated by metallothionein. J. Exp. Bot. 2005, 56, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chu, P.; Chen, H.; Li, Y.; Liu, J.; Ding, Y.; Tsang, E.W.T.; Jiang, L.; Wu, K.; Huang, S. Overexpression of Nelumbo nucifera metallothioneins 2a and 3 enhances seed germination vigor in Arabidopsis. Planta 2012, 235, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, G.; Mierek-Adamska, A.; Goc, A. Characterisation of Brassica napus L. metallothionein genes (BnMTs) expression in organs and during seed germination. Aust. J. Crop Sci. 2013, 7, 1324–1332. [Google Scholar]
- Bundithya, W.; Goldsbrough, P.B.; Guo, W.-J. Characterization of the Arabidopsis metallothionein gene family: Tissue-specific expression and induction during senescence and in response to copper. New Phytol. 2003, 159, 369–381. [Google Scholar] [CrossRef]
- Cheng, M.; Yuan, H.; Wang, R.; Zou, J.; Liang, T.; Yang, F.; Li, S. Genome-wide identification and analysis of the metallothionein genes in oryza genus. Int. J. Mol. Sci. 2021, 22, 9651. [Google Scholar] [CrossRef]
- Zhou, G.; Xu, Y.; Li, J.; Yang, L.; Liu, J.Y. Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). J. Biochem. Mol. Biol. 2006, 39, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; He, L.; Huo, C.; Jiang, X.; Chen, H.; Wang, R.; Tang, M.; Dong, L.; Chen, J.; Li, Y.; et al. Genome-Wide Identification and Expression Analysis of Heavy Metal Stress–Responsive Metallothionein Family Genes in Nicotiana tabacum. Plant Mol. Biol. Report. 2020, 39, 443–454. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, M.; Wang, S.; Ma, G.; Huang, X.; Qiao, C.; Wang, R.; Xu, X.; Liang, Y.; Lu, K.; et al. Genome-wide characterization and analysis of metallothionein family genes that function in metal stress tolerance in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2181. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.; Zhou, Z.; Kaur, R.; Bekele, W.; Tinker, N.A.; Singh, J. Toward the development of Ac/Ds transposon-mediated gene tagging system for functional genomics in oat (Avena sativa L.). Funct. Integr. Genom. 2022, 22, 669–681. [Google Scholar] [CrossRef]
- Zwer, P. Oats: Characteristics and Quality Requirements. In Cereal Grains: Assessing and Managing Quality; Woodhead Publishing: Sawston, UK, 2010; pp. 163–182. [Google Scholar]
- Alfieri, M.; Redaelli, R. Oat phenolic content and total antioxidant capacity during grain development. J. Cereal Sci. 2015, 65, 39–42. [Google Scholar] [CrossRef]
- Butt, M.S.; Tahir-Nadeem, M.; Khan, M.K.I.; Shabir, R.; Butt, M.S. Oat: Unique among the cereals. Eur. J. Nutr. 2008, 47, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Othman, R.A.; Moghadasian, M.H.; Jones, P.J.H. Cholesterol-lowering effects of oat β-glucan. Nutr. Rev. 2011, 69, 299–309. [Google Scholar] [CrossRef]
- Quiñones-Muñoz, T.A.; Villanueva-Rodríguez, S.J.; Torruco-Uco, J.G. Nutraceutical Properties of Medicago sativa L., Agave spp., Zea mays L. and Avena sativa L.: A Review of Metabolites and Mechanisms. Metabolites 2022, 12, 806. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jiang, C.; Yuan, W.; Zhang, M.; Fang, Z.; Li, Y.; Li, G.; Jia, J.; Yang, Z. A universal karyotypic system for hexaploid and diploid Avena species brings oat cytogenetics into the genomics era. BMC Plant Biol. 2021, 21, 213. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, A.S.; Huang, Y.; Smith, S.; Bekele, W.A.; Babiker, E.; Gnanesh, B.N.; Foresman, B.J.; Blanchard, S.G.; Jay, J.J.; Reid, R.W.; et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Lee, R.; Walstead, R.; Vickerstaff, R.J.; Fogarty, M.C.; Brouwer, C.R.; Reid, R.R.; Jay, J.J.; Bekele, W.A.; Jackson, E.W.; et al. Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 2019, 17, 92. [Google Scholar] [CrossRef]
- PepsiCo Avena Sativa-OT3098 v2, PepsiCo. Available online: https://wheat.pw.usda.gov/jb/?data=%2Fggds%2Foat-ot3098v2-pepsico&loc=chr7A%3A5130802..5132383&tracks=genes&highlight= (accessed on 1 May 2022).
- Tinker, N.A.; Wight, C.P.; Bekele, W.A.; Yan, W.; Jellen, E.N.; Renhuldt, N.T.; Sirijovski, N.; Lux, T.; Spannagl, M.; Mascher, M. Genome analysis in Avena sativa reveals hidden breeding barriers and opportunities for oat improvement. Commun. Biol. 2022, 5, 474. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H. bin Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.; Arnon, D. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1938, 347, 1–39. [Google Scholar]
- Wang, G.G.; Wang, G.G.; Zhang, X.; Wang, F.; Song, R. Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 2012, 23, 159–163. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, K.; Aziz, U.; Zhao, C.; Zhang, M. Evaluation of duplicated reference genes for quantitative real-time PCR analysis in genome unknown hexaploid oat (Avena sativa L.). Plant Methods 2020, 16, 138. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.H.H.K.; Wellburn, A.R.A.A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
- Morello, L.; Breviario, D. Plant spliceosomal introns: Not only cut and paste. Curr. Genom. 2008, 9, 227–238. [Google Scholar] [CrossRef]
- Ren, X.; Vorst, O.; Fiers, M.; Stiekema, W.; Nap, J. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Mierek-Adamska, A.; Dąbrowska, G.B.; Blindauer, C.A. The type 4 metallothionein from Brassica napus seeds folds in a metal-dependent fashion and favours zinc over other metals. Metallomics 2018, 10, 1430–1443. [Google Scholar] [CrossRef]
- Tomas, M.; Pagani, M.A.; Andreo, C.S.; Capdevila, M.; Bofill, R.; Atrian, S. His-containing plant metallothioneins: Comparative study of divalent metal-ion binding by plant MT3 and MT4 isoforms. J. Biol. Inorg. Chem. 2014, 19, 1149–1164. [Google Scholar] [CrossRef]
- Pagani, M.A.; Tomas, M.; Carrillo, J.; Bofill, R.; Capdevila, M.; Atrian, S.; Andreo, C.S. The response of the different soybean metallothionein isoforms to cadmium intoxication. J. Inorg. Biochem. 2012, 117, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Krężel, A.; Maret, W. The bioinorganic chemistry of mammalian metallothioneins. Chem. Rev. 2021, 121, 14594–14684. [Google Scholar] [CrossRef]
- Capdevila, M.; Bofill, R.; Palacios, O.; Atrian, S. State-of-the-art of metallothioneins at the beginning of the 21st century. Coord. Chem. Rev. 2012, 256, 46–62. [Google Scholar] [CrossRef]
- Salim, A.; Chesnov, S.; Freisinger, E. Metallation pathway of a plant metallothionein: Cicer arietinum MT2. J. Inorg. Biochem. 2020, 210, 111157. [Google Scholar] [CrossRef]
- Rono, J.K.; Le Wang, L.; Wu, X.C.; Cao, H.W.; Zhao, Y.N.; Khan, I.U.; Yang, Z.M. Identification of a new function of metallothionein-like gene OsMT1e for cadmium detoxification and potential phytoremediation. Chemosphere 2021, 265, 129136. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, F.; Wang, F.; Dong, Z.; Cao, Q.; Chen, M. Characterization of a type 1 metallothionein gene from the stresses-tolerant plant Ziziphus jujuba. Int. J. Mol. Sci. 2015, 16, 16750–16762. [Google Scholar] [CrossRef]
- Zhang, P.-H.; Zhang, X.-J.; Tang, T.-W.; Hu, H.-L.; Bai, N.-N.; Zhang, D.-W.; Meng, S.; Peng, J.-S. Isolation of three metallothionein genes and their roles in mediating cadmium resistance. Agronomy 2022, 12, 2971. [Google Scholar] [CrossRef]
- Woo, E.S.; Dellapiazza, D.; Wang, A.S.; Lazo, J.S. Energy-dependent nuclear binding dictates metallothionein localization. J. Cell Physiol. 2000, 182, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Maret, W.; Vallee, B.L. Zinc metallothionein imported into liver mitochondria modulates respiration. Proc. Natl. Acad. Sci. USA 2001, 98, 2317–2322. [Google Scholar] [CrossRef]
- Mariño-Ramírez, L.; Tharakaraman, K.; Spouge, J.L.; Landsman, D. Promoter analysis: Gene regulatory motif identification with A-GLAM. Methods Mol. Biol. 2009, 537, 263–276. [Google Scholar] [CrossRef]
- Lü, S.; Gu, H.; Yuan, X.; Wang, X.; Wu, A.M.; Qu, L.; Liu, J.Y. The GUS reporter-aided analysis of the promoter activities of a rice metallothionein gene reveals different regulatory regions responsible for tissue-specific and inducible expression in transgenic Arabidopsis. Transgenic Res. 2007, 16, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhao, J. Functional analysis of the rice metallothionein gene OsMT2b promoter in transgenic Arabidopsis plants and rice germinated embryos. Plant Sci. 2009, 176, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-J.; Wang, Y.; Yu, S.-S.; Liu, J.-Y. Characterization of a novel rice metallothionein gene promoter: Its tissue specificity and heavy metal responsiveness. J. Integr. Plant Biol. 2010, 52, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.K.; Xu, Y.F.; Liu, J.Y. Characterization of a rice class II metallothionein gene: Tissue expression patterns and induction in response to abiotic factors. J. Plant Physiol. 2005, 162, 686–696. [Google Scholar] [CrossRef]
- Ahn, Y.O.; Kim, S.H.; Lee, J.; Ran Kim, H.; Lee, H.-S.S.; Kwak, S.-S.S.; Kim, H.; Lee, H.-S.S.; Kwak, S.-S.S.; Ran Kim, H.; et al. Three Brassica rapa metallothionein genes are differentially regulated under various stress conditions. Mol. Biol. Rep. 2012, 39, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Usha, B.; Venkataraman, G.; Parida, A. Heavy metal and abiotic stress inducible metallothionein isoforms from Prosopis juliflora (SW) D.C. show differences in binding to heavy metals in vitro. Mol. Genet. Genom. 2009, 281, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-O.; Kang, H. Comparative expression analysis of genes encoding metallothioneins in response to heavy metals and abiotic stresses in rice (Oryza sativa) and Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2018, 82, 1656–1665. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Leszczyszyn, O.I.; Imam, H.T.; Blindauer, C.A. Diversity and distribution of plant metallothioneins: A review of structure, properties and functions. Metallomics 2013, 5, 1146–1169. [Google Scholar] [CrossRef]
- Matsusaki, M.; Okuda, A.; Matsuo, K.; Gekko, K.; Masuda, T.; Naruo, Y.; Hirose, A.; Kono, K.; Tsuchi, Y.; Urade, R. Regulation of plant ER oxidoreductin 1 (ERO1) activity for efficient oxidative protein folding. J. Biol. Chem. 2019, 294, 18820–18835. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef]
- Anand, A.; Kumari, A.; Thakur, M.; Koul, A. Hydrogen peroxide signaling integrates with phytohormones during the germination of magnetoprimed tomato seeds. Sci. Rep. 2019, 9, 8814. [Google Scholar] [CrossRef]
- King, J.C. Zinc: An essential but elusive nutrient. Am. J. Clin. Nutr. 2011, 94, 679S. [Google Scholar] [CrossRef]
- Clemens, S.; Aarts, M.G.M.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Palmgren, M.G.; Clemens, S.; Williams, L.E.; Krämer, U.; Borg, S.; Schjørring, J.K.; Sanders, D. Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 2008, 13, 464–473. [Google Scholar] [CrossRef]
- Shi, G.; Cai, Q. Zinc tolerance and accumulation in eight oil crops. J. Plant Nutr. 2010, 33, 982–997. [Google Scholar] [CrossRef]
- Cherif, J.; Mediouni, C.; Ben Ammar, W.; Jemal, F. Interactions of zinc and cadmium toxicity in their effects on growth and in antioxidative systems in tomato plants (Solarium lycopersicum). J. Environ. Sci. 2011, 23, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Dawar, K.; Ullah, I.; Irfan, M.; Zhang, Z.; Mian, I.A.; Khan, B.; Gul, N.; Fahad, S.; Jalal, A.; et al. Zinc foliar application mitigates cadmium-induced growth inhibition and enhances wheat growth, chlorophyll contents, and yield. ACS Omega 2023, 8, 32372–32381. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Rehman, M.Z.U.; Maqbool, A. A critical review on the effects of zinc at toxic levels of cadmium in plants. Environ. Sci. Pollut. Res. 2019, 26, 6279–6289. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Gupta, P.; Seth, C.S. Foliar application of α-lipoic acid attenuates cadmium toxicity on photosynthetic pigments and nitrogen metabolism in Solanum lycopersicum L. Acta Physiol. Plant 2022, 44, 112. [Google Scholar] [CrossRef]
- Manzoor, H.; Mehwish; Bukhat, S.; Rasul, S.; Rehmani, M.I.A.; Noreen, S.; Athar, H.-R.; Zafar, Z.U.; Skalicky, M.; Soufan, W.; et al. Methyl jasmonate alleviated the adverse effects of cadmium stress in pea (Pisum sativum L.): A nexus of photosystem II activity and dynamics of redox balance. Front. Plant Sci. 2022, 13, 860664. [Google Scholar] [CrossRef]
- Adamakis, I.-D.S.; Sperdouli, I.; Hanć, A.; Dobrikova, A.; Apostolova, E.; Moustakas, M. Rapid hormetic responses of photosystem II photochemistry of clary sage to cadmium exposure. Int. J. Mol. Sci. 2020, 22, 41. [Google Scholar] [CrossRef]
- Jain, R.; Srivastava, S.; Solomon, S.; Shrivastava, A.K.; Chandra, A. Impact of excess zinc on growth parameters, cell division, nutrient accumulation, photosynthetic pigments and oxidative stress of sugarcane (Saccharum spp.). Acta Physiol. Plant 2010, 32, 979–986. [Google Scholar] [CrossRef]
- Fatima, N.; Ahmad, N.; Anis, M. Enhanced in vitro regeneration and change in photosynthetic pigments, biomass and proline content in Withania somnifera L. (Dunal) induced by copper and zinc ions. Plant Physiol. Biochem. 2011, 49, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Drążkiewicz, M.; Baszyński, T. Growth parameters and photosynthetic pigments in leaf segments of Zea mays exposed to cadmium, as related to protection mechanisms. J. Plant Physiol. 2005, 162, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Xiang, P.; Liu, X.; Ma, L.Q. Closely-related species of hyperaccumulating plants and their ability in accumulation of As, Cd, Cu, Mn, Ni, Pb and Zn. Chemosphere 2020, 251, 126334. [Google Scholar] [CrossRef]
- Apak, R.; Özyürek, M.; Güçlü, K.; Çapanoğlu, E. Antioxidant activity/capacity measurement. 2. Hydrogen atom transfer (HAT)-based, mixed-mode (electron transfer (ET)/HAT), and lipid peroxidation assays. J. Agric. Food Chem. 2016, 64, 1028–1045. [Google Scholar] [CrossRef]
- Apak, R.; Özyürek, M.; Güçlü, K.; Çapanoğlu, E. Antioxidant activity/capacity measurement. 1. Classification, physicochemical principles, mechanisms, and electron transfer (ET)-based assays. J. Agric. Food Chem. 2016, 64, 997–1027. [Google Scholar] [CrossRef] [PubMed]
- Naikoo, M.I.; Dar, M.I.; Raghib, F.; Jaleel, H.; Ahmad, B.; Raina, A.; Khan, F.A.; Naushin, F. Role and Regulation of Plants Phenolics in Abiotic Stress Tolerance. In Plant Signaling Molecules; Khan, M.I.R., Reddy, P.S., Ferrante, A., Khan, N.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 157–168. [Google Scholar]
- Szydłowska-Czerniak, A.; Bartkowiak-Broda, I.; Karlović, I.; Karlovits, G.; Szłyk, E. Antioxidant capacity, total phenolics, glucosinolates and colour parameters of rapeseed cultivars. Food Chem. 2011, 127, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Vašák, M.; Meloni, G. Mammalian metallothionein-3: New functional and structural insights. Int. J. Mol. Sci. 2017, 18, 117. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jeong, J.C.; Ahn, Y.O.; Lee, H.S.; Kwak, S.S. Differential responses of three sweetpotato metallothionein genes to abiotic stress and heavy metals. Mol. Biol. Rep. 2014, 41, 6957–6966. [Google Scholar] [CrossRef]
- Ren, Y.; Liu, Y.; Chen, H.; Li, G.; Zhang, X.; Zhao, J. Type 4 metallothionein genes are involved in regulating Zn ion accumulation in late embryo and in controlling early seedling growth in Arabidopsis. Plant Cell Environ. 2012, 35, 770–789. [Google Scholar] [CrossRef]
- Collett, H.; Shen, A.; Gardner, M.; Farrant, J.M.; Denby, K.J.; Illing, N. Towards transcript profiling of desiccation tolerance in Xerophyta humilis: Construction of a normalized 11 k X. humilis cDNA set and microarray expression analysis of 424 cDNAs in response to dehydration. Physiol. Plant 2004, 122, 39–53. [Google Scholar] [CrossRef]






| Primer Name | Sequence 5′→ 3′ | Product Size [bp] | Target | Reference |
|---|---|---|---|---|
| AsMT1_qPCR_f AsMT1_qPCR_r | CAAACTGCAAGTGCGGGAAG TTGTTCTCATGAGCCACGCC | 103 | AsMT1 chr7C | [25] |
| AsMT2_qPCR_f AsMT2_qPCR_r | CTGCGGAGGGTGCAAGATG AACGATGGCTTGGAAGAGGG | 96 | AsMT2 chr1C | |
| AsMT3_qPCR_f AsMT3_qPCR_r | TCCACCATGTCGAACACCTG TGGCTCTTCTCGGTGTCAAC | 107 | AsMT3 chr3A | |
| AsMT4_qPCR_f | CACGTGCGGAGAGCACTG | 121 | AsMT4 chr1D | This study |
| AsMT4_qPCR_r | ACAGGAGGCGCAGTCACAG | |||
| EIF4A_f EIF4A_r | TCTCGCAGGATACGGATGTCG TCCATCGCATTGGTCGCTCT | 88 | Eukaryotic Initiation Factor 4A-3 | [60] |
| AsACT_f AsACT_r | CTTCCCCAGTATCGTCGGAC AGGGCAATGTAGGACAGCTT | 573 | Actin (MH260250.1) | This study |
| AsMT Type | Gene Name | Gene ID/Position | Strand | gDNA Size (bp) | Protein Length (aa) | MW (kDa) | pI | Cys Number | Cys Sequence Pattern | Length of Spacer between Cys Domains | Predicated Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MT1 | AsMT1_chr1A | AVESA.00001b.r3.1Ag0000996 | − | 474 | 74 | 7.45 | 5.11 | 11 (6 + 5) | CxC | 43 | Cyto/Nucl |
| AsMT1_chr1D | AVESA.00001b.r3.1Dg0000988 | + | 321 | 74 | 7.32 | 6.50 | 12 (6 + 6) | 43 | Cyto/Nucl | ||
| AsMT1_chr4D | AVESA.00001b.r3.4Dg0000092 | − | 489 | 72 | 7.26 | 5.05 | 12 (6 + 6) | 41 | Cell mem/Cyto/Nucl | ||
| AsMT1_chr5C | AVESA.00001b.r3.5Cg0000058 | + | 673 | 75 | 7.62 | 4.75 | 12 (6 + 6) | 44 | Cyto/Nucl | ||
| AsMT1_chr7C | AVESA.00001b.r3.7Cg0001922 | − | 487 | 72 | 7.29 | 5.00 | 13 (6 + 7) | 41 | Cell mem/Cyto/Nucl | ||
| MT2 | AsMT2_chr1C | AVESA.00001b.r3.1Cg0000164 | + | 529 | 79 | 7.59 | 5.10 | 14 (8 + 6) | CC, CxC, CxxC, C | 41 | Cell mem/Cyto/Nucl |
| AsMT2_chr3A | AVESA.00001b.r3.3Ag0000375 | − | 565 | 79 | 7.56 | 5.05 | 14 (8 + 6) | 41 | Cell mem/Chlo/Cyto/Nucl | ||
| AsMT2_chr3C | AVESA.00001b.r3.3Cg0000320 | + | 531 | 79 | 7.61 | 4.71 | 14 (8 + 6) | 41 | Cell mem/Chlo/Cyto/Nucl | ||
| AsMT2_chr1D | AVESA.00001b.r3.1Dg0002311 | − | 390 | 80 | 7.59 | 6.47 | 17 (8 + 9) | 37 | Cyto | ||
| AsMT2_chr4A | AVESA.00001b.r3.4Ag0003869 | − | 323 | 75 | 7.19 | 5.57 | 15 (8 + 7) | 38 | Cyto | ||
| AsMT2a_chr4D | AVESA.00001b.r3.4Dg0003379 | − | 339 | 81 | 7.72 | 4.96 | 17 (8 + 9) | 39 | Cyto | ||
| AsMT2b_chr4D | AVESA.00001b.r3.4Dg0003380 | − | 341 | 81 | 7.70 | 4.96 | 17 (8 + 9) | 39 | Cyto | ||
| AsMT2c_chr4D | AVESA.00001b.r3.4Dg0003934 | − | 334 | 80 | 7.63 | 4.96 | 17 (8 + 9) | 38 | Cyto | ||
| AsMT2_chr7A | AVESA.00001b.r3.7Ag0000076 | + | 388 | 80 | 7.53 | 6.47 | 17 (8 + 9) | 37 | Cyto | ||
| MT3 | AsMT3_chr3A | AVESA.00001b.r3.3Ag0000802 | − | 530 | 64 | 6.81 | 4.85 | 10 (4 + 6) | C, CxC | 32 | Nucl |
| AsMT3_chr3C | AVESA.00001b.r3.3Cg0000779 | − | 910 | 63 | 6.66 | 5.07 | 10 (4 + 6) | 31 | Nucl | ||
| AsMT3_chr3D | AVESA.00001b.r3.3Dg0000256 | − | 887 | 63 | 6.68 | 5.07 | 10 (4 + 6) | 31 | Nucl | ||
| MT4 | AsMT4_chr1A | chr1A:252750400..252750656 | + | 249 | 82 | 7.91 | 7.36 | 17 (6 + 6 + 5) | 16, 15 | Cyto | |
| AsMT4_chr1C | AVESA.00001b.r3.1Cg0001463 | + | 243 | 80 | 7.83 | 7.36 | 17 (6 + 6 + 5) | 14, 15 | Cyto | ||
| AsMT4_chr1D | chr1D:243956192..243956442 | + | 249 | 82 | 7.94 | 7.36 | 17 (6 + 6 + 5) | 16, 15 | Cell mem/Cyto | ||
| AsMT4_chr4C | chr4C:76127001..76127600 | + | 360 | 89 | 8.62 | 5.75 | 17 (6 + 6 + 5) | 26, 13 | Cell mem/Cyto |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konieczna, W.; Mierek-Adamska, A.; Chojnacka, N.; Antoszewski, M.; Szydłowska-Czerniak, A.; Dąbrowska, G.B. Characterization of the Metallothionein Gene Family in Avena sativa L. and the Gene Expression during Seed Germination and Heavy Metal Stress. Antioxidants 2023, 12, 1865. https://doi.org/10.3390/antiox12101865
Konieczna W, Mierek-Adamska A, Chojnacka N, Antoszewski M, Szydłowska-Czerniak A, Dąbrowska GB. Characterization of the Metallothionein Gene Family in Avena sativa L. and the Gene Expression during Seed Germination and Heavy Metal Stress. Antioxidants. 2023; 12(10):1865. https://doi.org/10.3390/antiox12101865
Chicago/Turabian StyleKonieczna, Wiktoria, Agnieszka Mierek-Adamska, Natalia Chojnacka, Marcel Antoszewski, Aleksandra Szydłowska-Czerniak, and Grażyna B. Dąbrowska. 2023. "Characterization of the Metallothionein Gene Family in Avena sativa L. and the Gene Expression during Seed Germination and Heavy Metal Stress" Antioxidants 12, no. 10: 1865. https://doi.org/10.3390/antiox12101865







