Structural Study of a La(III) Complex of a 1,2,3-Triazole Ligand with Antioxidant Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Details
2.2. Materials
2.3. Methods
2.3.1. Deoxyribose Degradation Assay
2.3.2. Fenton Reaction MTT assay
2.3.3. DPPH Assay
2.3.4. ABTS Assay
2.3.5. LDCL in Presence of KO2
2.3.6. LDCL in Presence of NaClO
2.4. Computational Details
3. Results
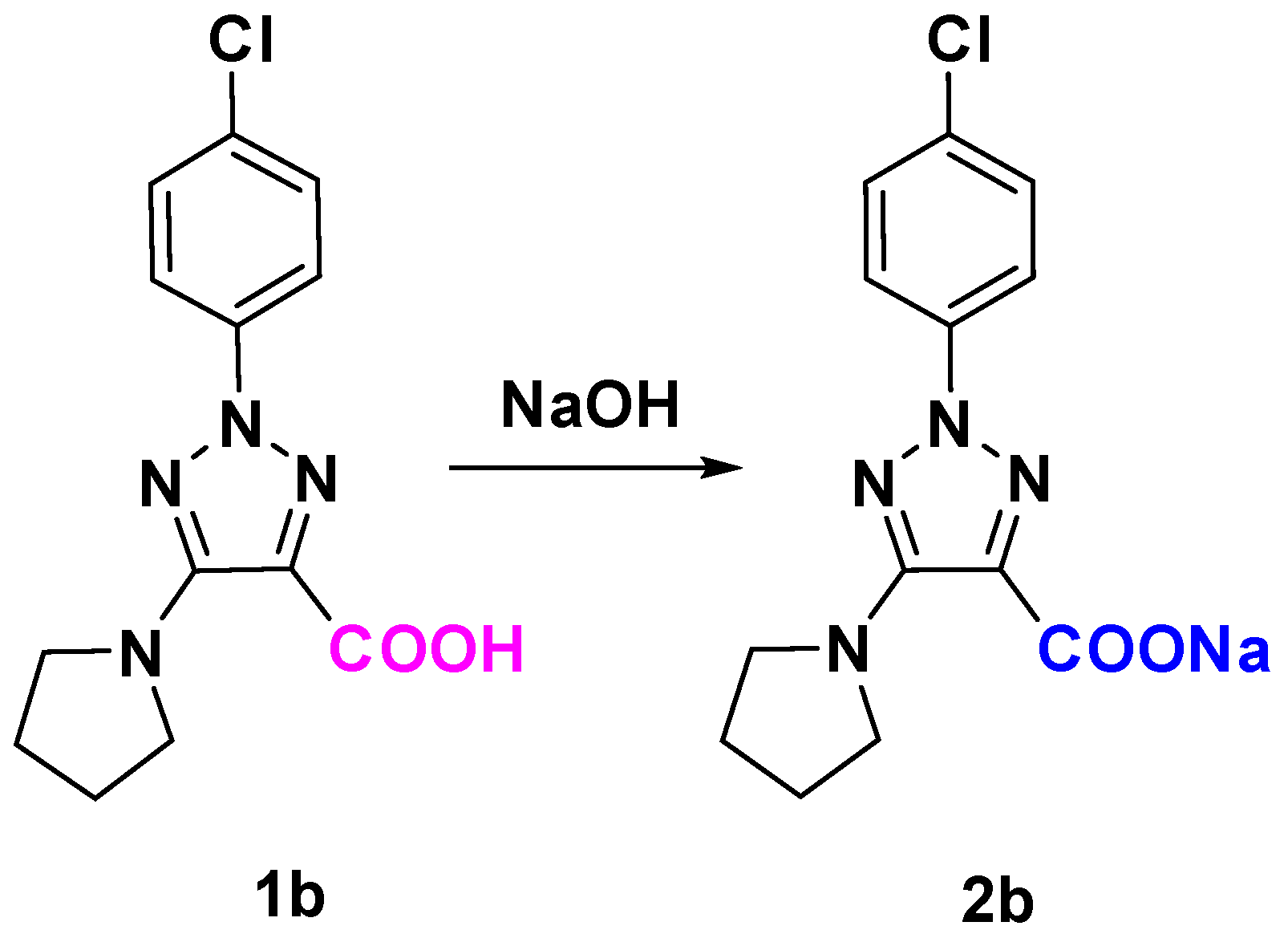
3.1. Synthesis
3.2. Molecular Structure of the Complex
3.3. Atomic Charges
3.4. Vibrational Analysis
3.5. Radical Scavenging Assays
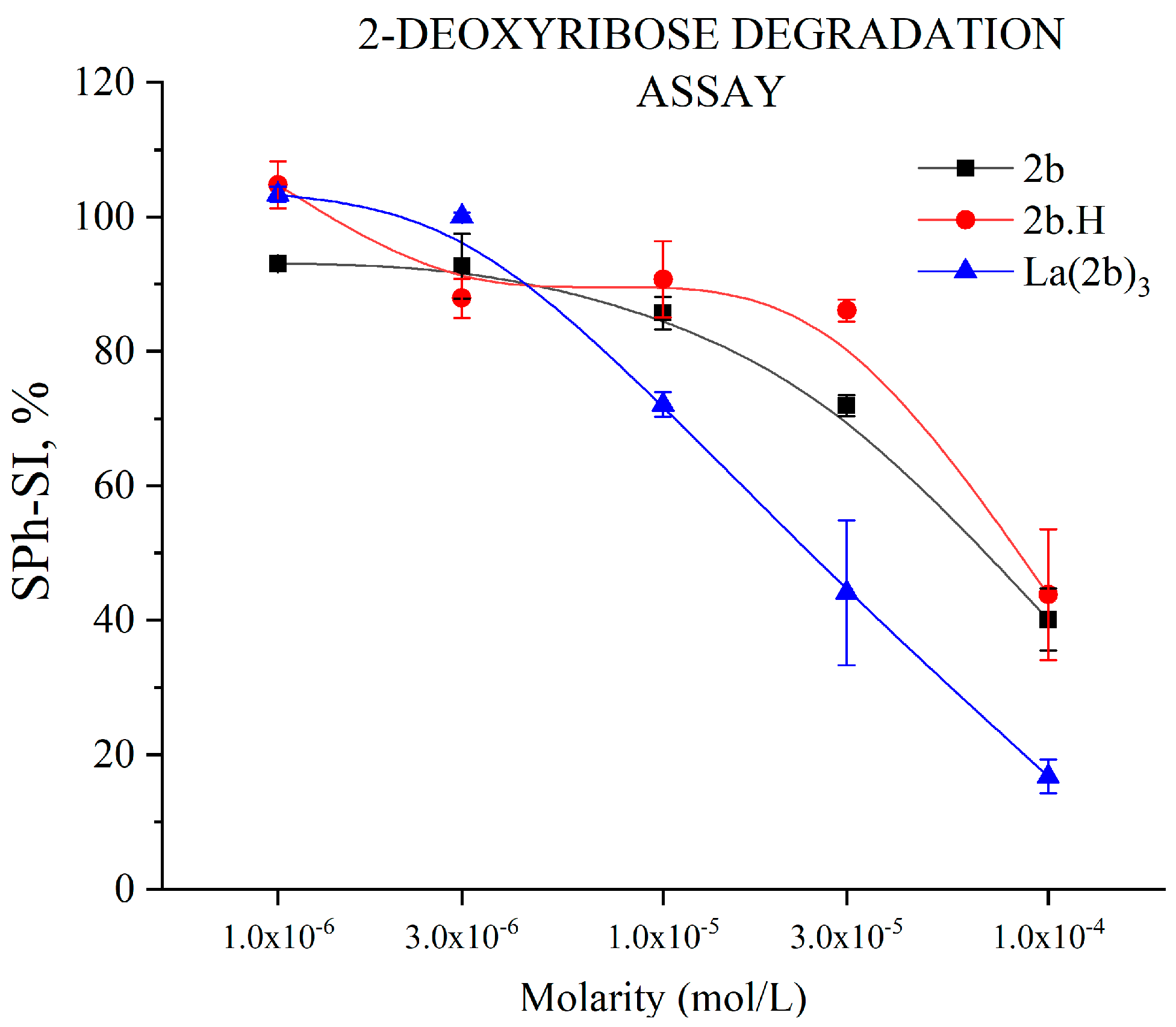
3.5.1. Impact of 1b, 2b, and La(2b’)3 on 2-Deoxyribose Degradation
3.5.2. Impact of 1b, 2b, and La(2b’)3 on a Model System Containing the Stable Radical DPPH●
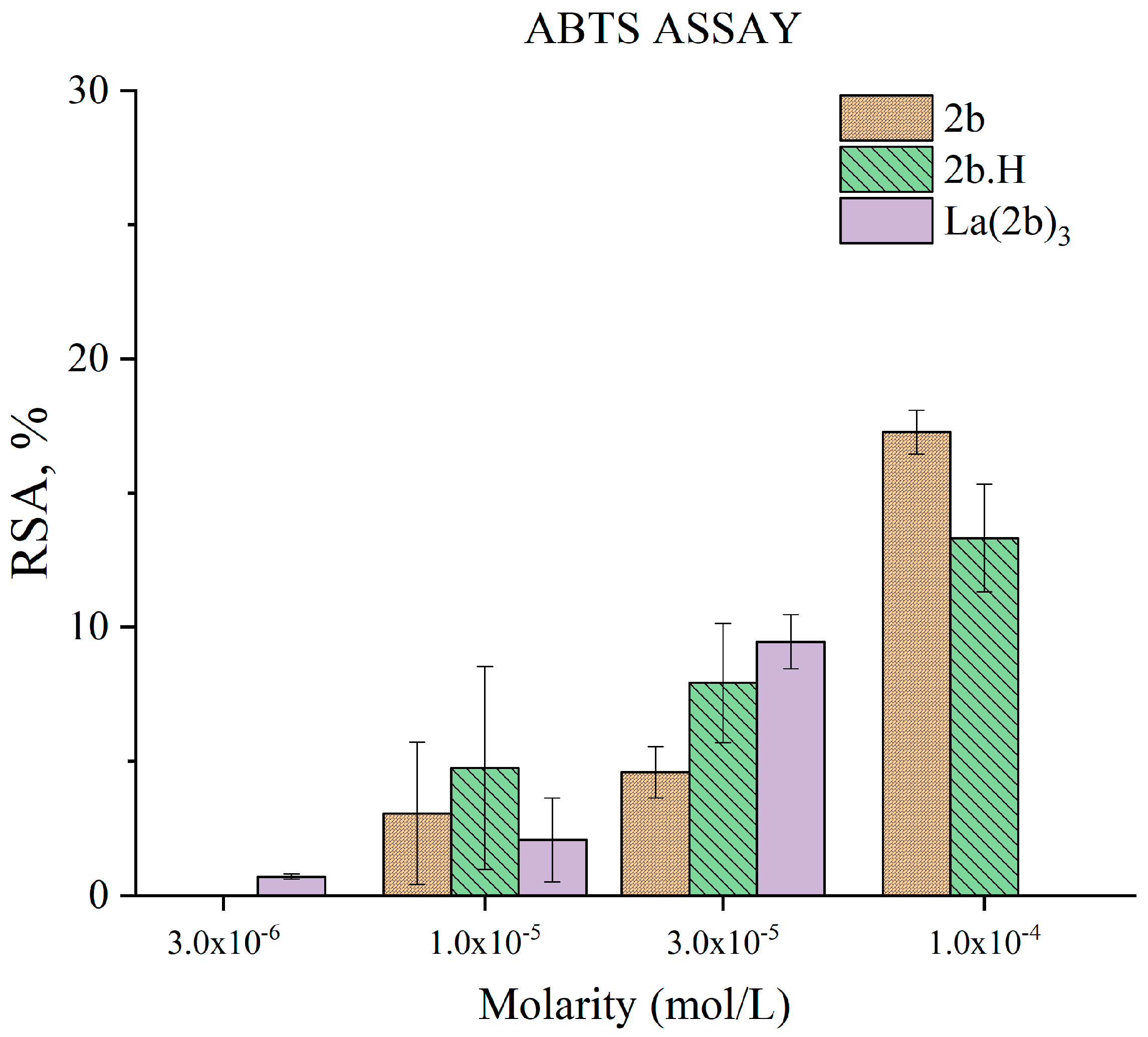
3.5.3. Impact of 2b, 1b, and La(2b’)3 on a Model System Containing the Stable Radical ABTS●+
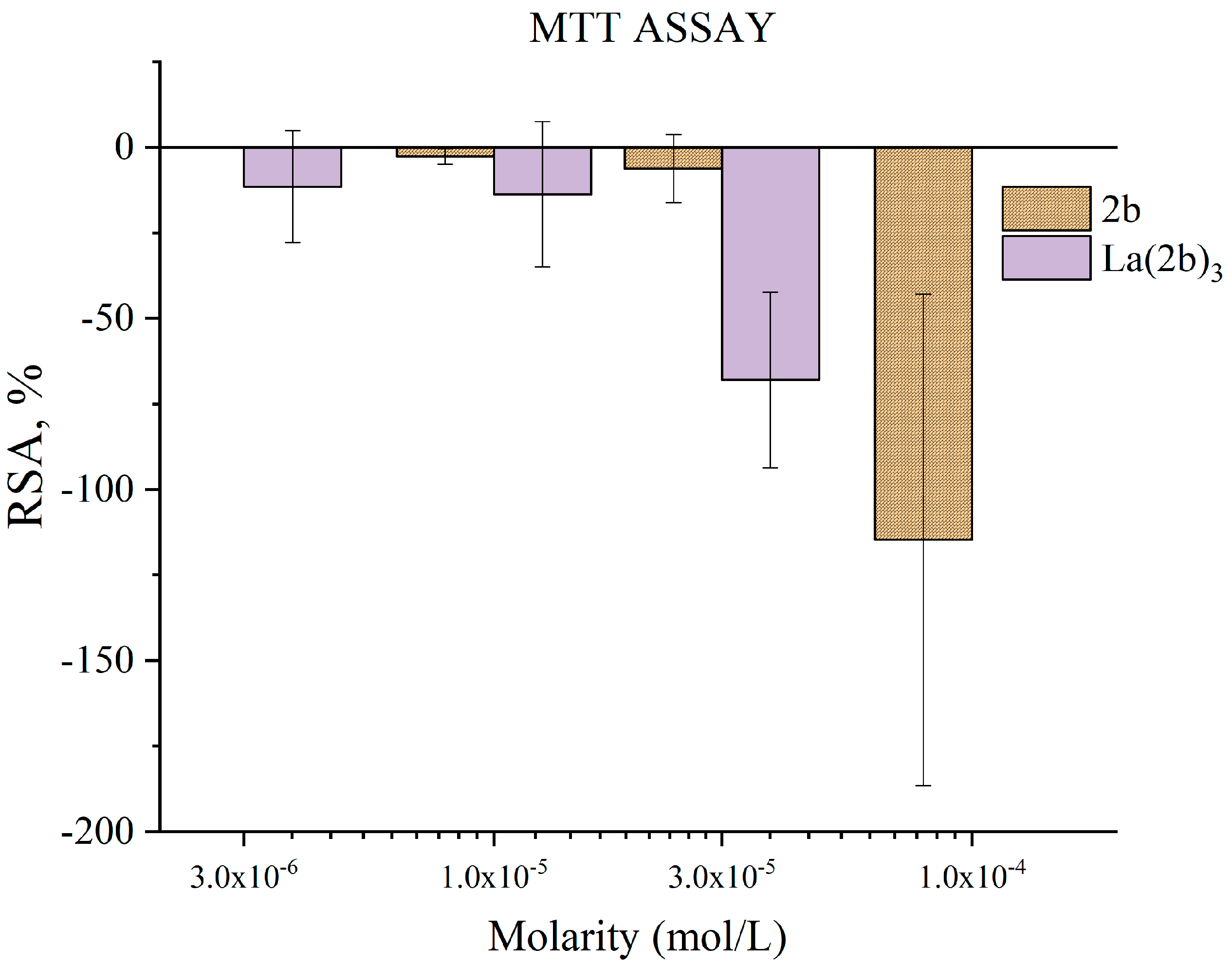
3.5.4. Impact of 2b and La(2b’)3 on MTT-Formazan Transformation via Fenton Reaction-Derived Hydroxyl Radicals
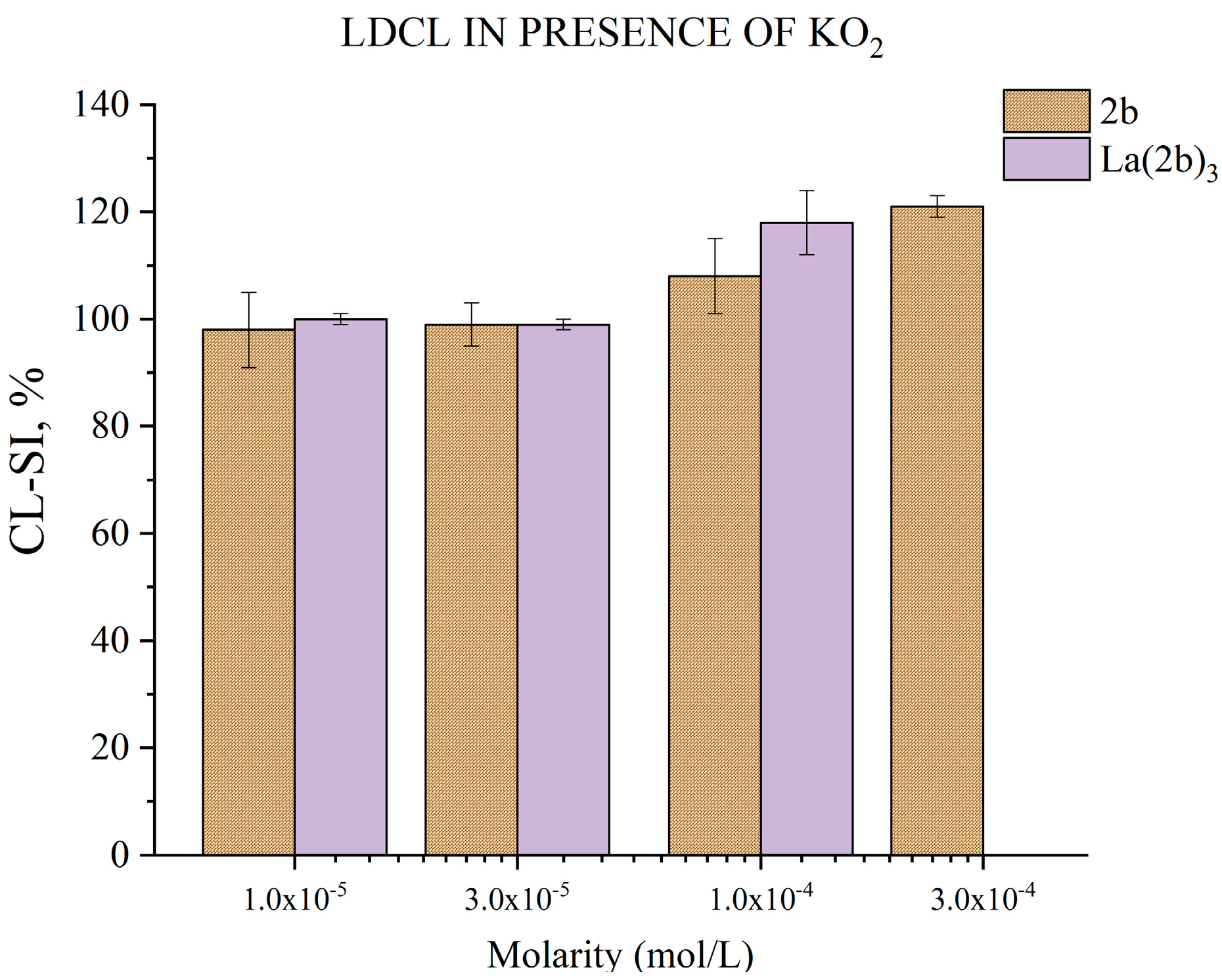
3.5.5. Impact of 2b and La(2b’)3 on LDCL in Presence of KO2
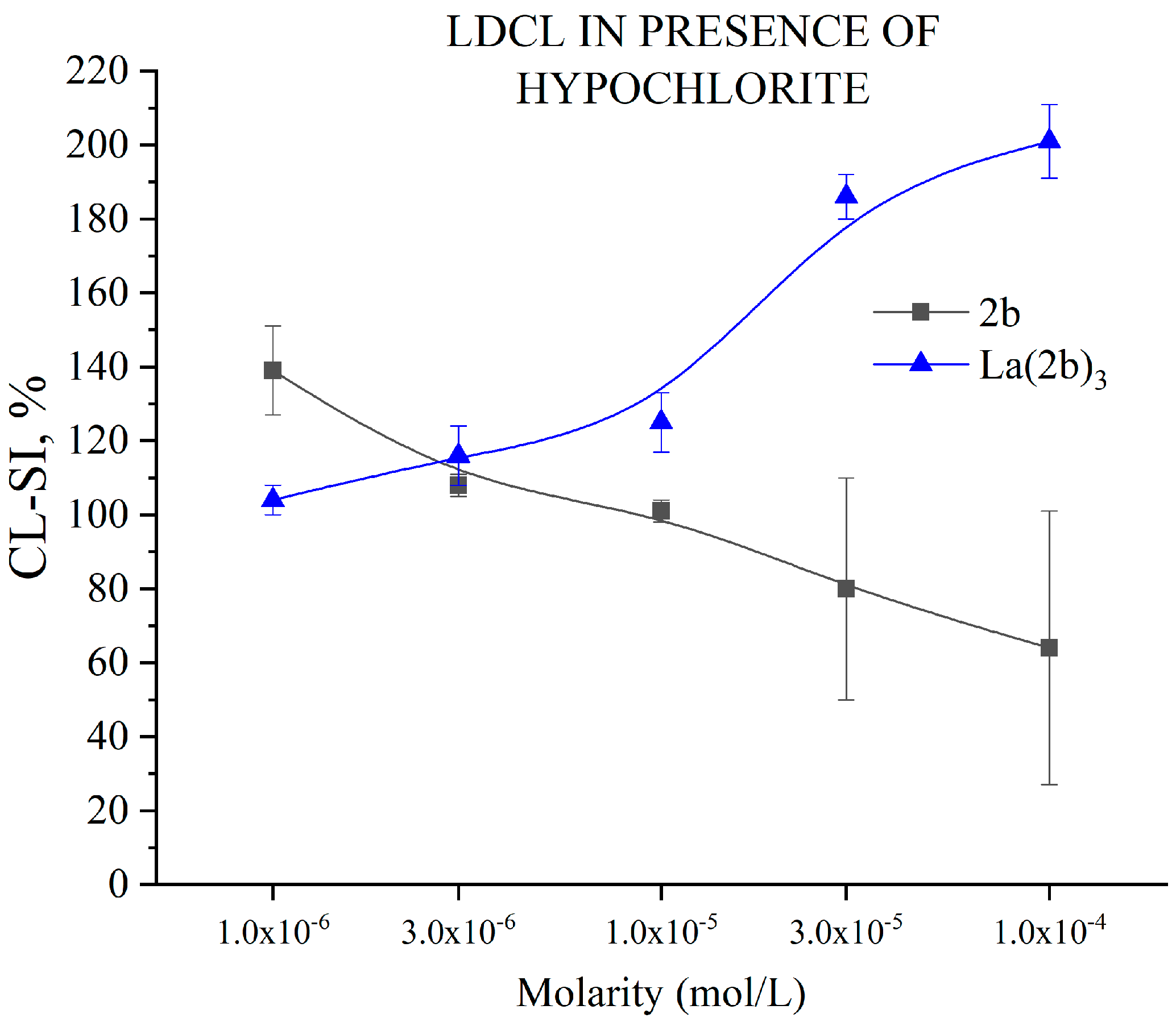
3.5.6. Impact of 2b and La(2b’)3 on LDCL in Presence of NaClO
4. Conclusions
- A structural study of the lanthanum(III) complex was carried out at three DFT levels. The starting optimized structure was that with the La(III) ion coordinated with three 2b ligands through the carboxylate group. The optimized structure at the M06-2X/lanl2dz level showed an almost symmetric arrangement. By rotation around the C9-C11 bond length, another conformer could be obtained, but it was less stable.
- Global chemical reactivity descriptors were calculated in the La(2b’)3 complex. The low energy gap calculated indicated a large chemical reactivity and small excitation energies to the manifold of excited states.
- The coordination of the 2b ligand with the La(III) ion noticeably changed its IR and Raman spectra, which seemed different to those obtained with 2b alone.
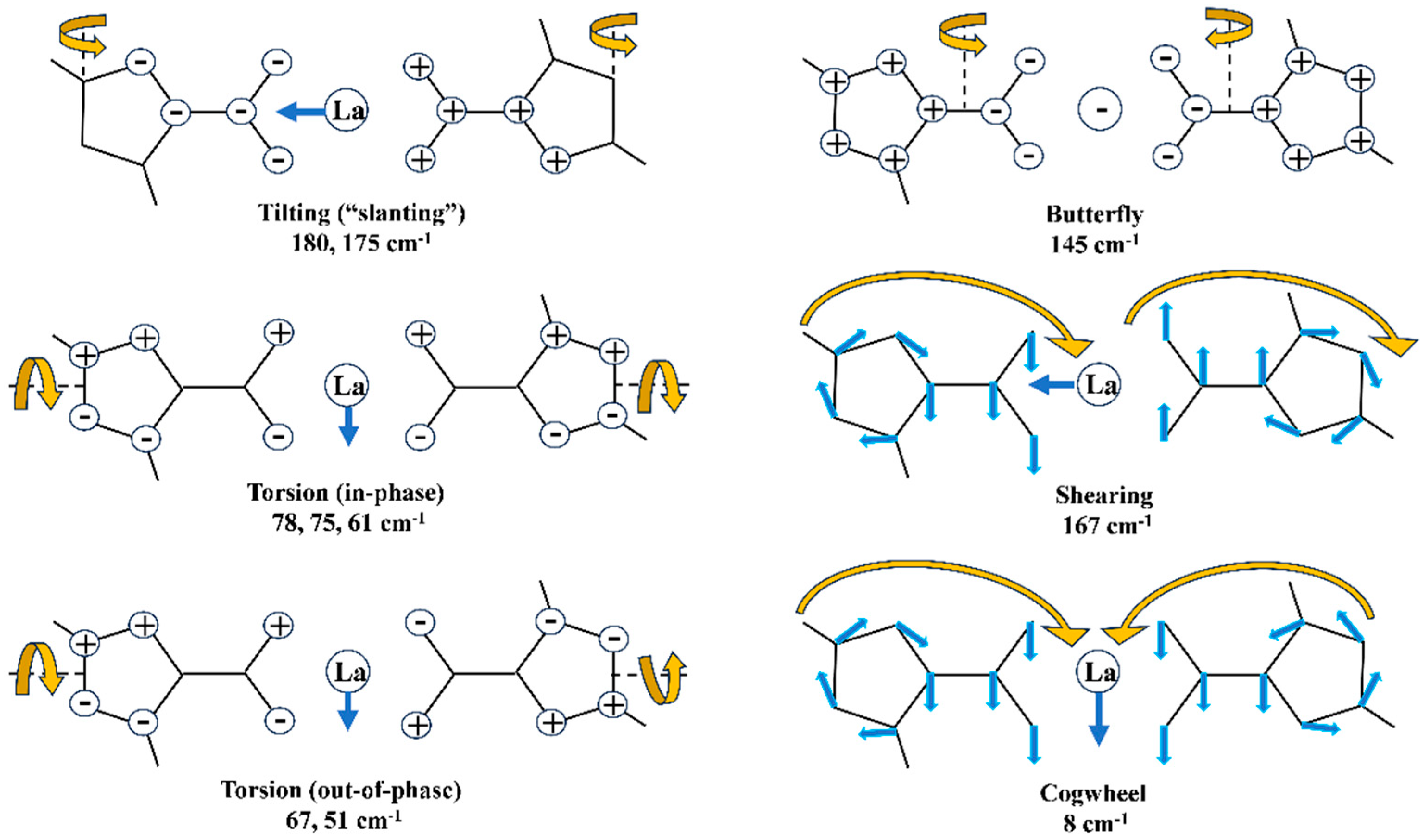
- The main low-lying molecular vibrations in the La(2b’)3 complex were characterized. Their number indicated a high flexibility in its molecular structure.
- The complex showed moderate HAT activity with the stable DPPH● radical, but the activity of ligand 2b in this model system was much lower, that is, almost zero. This lack of activity may be due to steric factors that hinder hydrogen transfer between the bulky DPPH● and the possible hydrogen-donating active sites in the 2b and La(2b’)3 molecules. When interacting with the small and mobile OH●, generated by UV-induced water radiolysis, both the ligand and complex behaved as scavengers (3 × 10−6 M or higher), with the latter being the more potent at equimolar concentrations.
- The ligand and the complex participated in SET with the ABTS●+ radical ion only moderately, in a concentration-dependent manner. Steric factors also seemed to be at play here, as the complex appeared to be less active than the ligand at three times the concentration.
- Both compounds appeared to have little impact on KO2-derived superoxide. Only at the highest tested concentrations, 3 × 10−4 M for 2b and 1 × 10−4 M for La(2b’)3, did they seem to show a slight significant prooxidant effect.
- Though 2b and La(2b’)3 scavenged OH●, a different model system (Fenton reaction), generating the same RS yielded completely opposite results. At low concentrations, both compounds seemed to be inactive. Increasing the molarity to 3 × 10−5 M and 1 × 10−4 M for La(2b’)3 and 2b, respectively, yielded results demonstrating that these compounds behave as prooxidants in this model system. As possible causes of this behavior, it is proposed that the ligand interacts with one or more components of the system rather than with the OH● generated by it.
- The La(III) ion dramatically changed the behavior of 2b toward hypochlorite once coordination took place. The higher the concentration, the better the hypochlorite scavenger 2b seemed to be. The behavior of its La(III) complex was the opposite, manifesting itself as a potent prooxidant at the highest concentration tested.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Franz, K.J.; Metzler-Nolte, N. Introduction: Metals in Medicine; ACS Publications: Washington, DC, USA, 2019; Volume 119, pp. 727–729. [Google Scholar]
- Gasser, G. Metal complexes and medicine: A successful combination. Chimia 2015, 69, 442. [Google Scholar] [CrossRef] [PubMed]
- Patyal, M.; Kaur, K.; Bala, N.; Gupta, N.; Malik, A.K. Innovative Lanthanide Complexes: Shaping the future of cancer/tumor Chemotherapy. J. Trace Elem. Med. Biol. 2023, 80, 127277. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, S. Applications of rare earth elements in cancer: Evidence mapping and scientometric analysis. Front. Med. 2022, 9, 946100. [Google Scholar] [CrossRef]
- Ascenzi, P.; Bettinelli, M.; Boffi, A.; Botta, M.; De Simone, G.; Luchinat, C.; Marengo, E.; Mei, H.; Aime, S. Rare earth elements (REE) in biology and medicine. Rend. Lincei. Sci. Fis. E Nat. 2020, 31, 821–833. [Google Scholar] [CrossRef]
- Bao, G. Lanthanide complexes for drug delivery and therapeutics. J. Lumin. 2020, 228, 117622. [Google Scholar] [CrossRef]
- Fouad, R. Synthesis and characterization of lanthanide complexes as potential therapeutic agents. J. Coord. Chem. 2020, 73, 2015–2028. [Google Scholar] [CrossRef]
- Paswan, S.; Anjum, A.; Yadav, N.; Jaiswal, N.; Singh, R.K.P. Synthesis, thermal, photo-physical, and biological properties of mononuclear Yb3+, Nd3+, and Dy3+ complexes derived from Schiff base ligands. J. Coord. Chem. 2020, 73, 686–701. [Google Scholar] [CrossRef]
- Taha, Z.A.; Hijazi, A.K.; Al Momani, W.M. Lanthanide complexes of the tridentate Schiff base ligand salicylaldehyde-2-picolinoylhydrazone: Synthesis, characterization, photophysical properties, biological activities and catalytic oxidation of aniline. J. Mol. Struct. 2020, 1220, 128712. [Google Scholar] [CrossRef]
- Wu, H.; Pan, G.; Bai, Y.; Zhang, Y.; Wang, H.; Shi, F.; Wang, X.; Kong, J. Study on synthesis, crystal structure, antioxidant and DNA-binding of mono-, di- and poly-nuclear lanthanides complexes with bis (N-salicylidene)-3-oxapentane-1,5-diamine. J. Photochem. Photobiol. B Biol. 2014, 135, 33–43. [Google Scholar] [CrossRef]
- Zou, H.-H.; Meng, T.; Chen, Q.; Zhang, Y.-Q.; Wang, H.-L.; Li, B.; Wang, K.; Chen, Z.-L.; Liang, F. Bifunctional mononuclear dysprosium complexes: Single-ion magnet behaviors and antitumor activities. Inorg. Chem. 2019, 58, 2286–2298. [Google Scholar] [CrossRef]
- Safronov, N.E.; Kostova, I.P.; Palafox, M.A.; Belskaya, N.P. Combined NMR Spectroscopy and Quantum-Chemical Calculations in Fluorescent 1,2,3-Triazole-4-carboxylic Acids Fine Structures Analysis. Int. J. Mol. Sci. 2023, 24, 8947. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M. 1,2,3-Triazole hybrids as anticancer agents: A review. Arch. Der Pharm. 2022, 355, 2100158. [Google Scholar] [CrossRef] [PubMed]
- Peica, N.; Kostova, I.; Kiefer, W. Theoretical and experimental studies on binding mode of 3,5-pyrazoledicarboxylic acid in its new La(III) complex. Chem. Phys. 2006, 325, 411–421. [Google Scholar] [CrossRef]
- Hrimla, M.; Oubella, A.; Laamari, M.R.; Bahsis, L.; Ghaleb, A.; Auhmani, A.; Morjani, H.; Julve Olcina, M.; Stiriba, S.E.; Itto, M.Y.A. Click synthesis, anticancer activity, and molecular docking investigation of some functional 1,2,3-triazole derivatives. Biointerface Res. Appl. Chem. 2022, 12, 7633–7667. [Google Scholar]
- Poonia, N.; Kumar, A.; Kumar, V.; Yadav, M.; Lal, K. Recent progress in 1H-1,2,3-triazoles as potential antifungal agents. Curr. Top. Med. Chem. 2021, 21, 2109–2133. [Google Scholar] [CrossRef]
- Liang, T.; Sun, X.; Li, W.; Hou, G.; Gao, F. 1,2,3-Triazole-containing compounds as anti–lung cancer agents: Current developments, mechanisms of action, and structure–activity relationship. Front. Pharmacol. 2021, 12, 661173. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Jiang, Y.; Zhang, F.; Zhu, F.; Liu, J.; Xu, Z. Recent updates on 1,2,3-, 1,2,4-, and 1,3,5-triazine hybrids (2017–present): The anticancer activity, structure–activity relationships, and mechanisms of action. Arch. Der Pharm. 2023, 356, 2200479. [Google Scholar] [CrossRef]
- Wu, X.; Wang, J.; Xia, S.; Cheng, S.; Shi, Y. 1,2,3-Triazole Derivatives with Anti-breast Cancer Potential. Curr. Top. Med. Chem. 2022, 22, 1406–1425. [Google Scholar] [CrossRef]
- Matin, M.M.; Matin, P.; Rahman, M.R.; Ben Hadda, T.; Almalki, F.A.; Mahmud, S.; Ghoneim, M.M.; Alruwaily, M.; Alshehri, S. Triazoles and their derivatives: Chemistry, synthesis, and therapeutic applications. Front. Mol. Biosci. 2022, 9, 864286. [Google Scholar] [CrossRef]
- Kostova, I.; Valcheva-Traykova, M. New samarium(III) complex of 5-aminoorotic acid with antioxidant activity. Appl. Organomet. Chem. 2015, 29, 815–824. [Google Scholar] [CrossRef]
- Todorov, L.; Hristova, N.; Belskaya, N.; Kostova, I. Antioxidant properties of a novel triazole ligand. Maced. Pharm. Bull. 2022, 68, 2. [Google Scholar] [CrossRef]
- Kedare, S.B.; Singh, R. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 2011, 48, 412–422. [Google Scholar] [CrossRef]
- Molyneux, P. The use of the stable free radical diphenylpicrylhydrazyl (DPPH) for estimating antioxidant activity. Songklanakarin J. Sci. Technol. 2004, 26, 211–219. [Google Scholar]
- Erel, O. A novel automated direct measurement method for total antioxidant capacity using a new generation, more stable ABTS radical cation. Clin. Biochem. 2004, 37, 277–285. [Google Scholar] [CrossRef]
- Erel, O. A novel automated method to measure total antioxidant response against potent free radical reactions. Clin. Biochem. 2004, 37, 112–119. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.; Aruoma, O.I. The deoxyribose method: A simple “test-tube” assay for determination of rate constants for reactions of hydroxyl radicals. Anal. Biochem. 1987, 165, 215–219. [Google Scholar] [CrossRef]
- Burns, W.G.; Sims, H.E. Effect of radiation type in water radiolysis. J. Chem. Soc. Faraday Trans. 1 Phys. Chem. Condens. Phases 1981, 77, 2803–2813. [Google Scholar] [CrossRef]
- Seminario, J.M. Modern Density Functional Theory: A Tool for Chemistry; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Szafran, M.; Katrusiak, A.; Koput, J.; Dega-Szafran, Z. X-ray, MP2 and DFT studies of the structure, vibrational and NMR spectra of homarine. J. Molec. Struct. 2007, 846, 1–12. [Google Scholar] [CrossRef]
- Rasheed, T.; Ahmad, S. Computational studies of vibrational spectra and molecular properties of 6-methyluracil using HF, DFT and MP2 methods. Ind. J. Phys. 2011, 85, 239–260. [Google Scholar] [CrossRef]
- Riley, K.E.; Hobza, P. Noncovalent interactions in biochemistry. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 3–17. [Google Scholar] [CrossRef]
- Riley, K.E.; Pitonák, M.; Jurecka, P.; Hobza, P. Stabilization and structure calculations for noncovalent interactions in extended molecular systems based on wave function and density functional theories. Chem. Rev. 2010, 110, 5023–5063. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Truhlar, D.G. Applications and validations of the Minnesota density functionals. Chem. Phys. Lett. 2011, 502, 1–13. [Google Scholar] [CrossRef]
- Sert, Y.; Puttaraju, K.B.; Keskinoʇlu, S.; Shivashankar, K.; Ucun, F. FT-IR and Raman vibrational analysis, B3LYP and M06-2X simulations of 4-bromomethyl-6-tert-butyl-2H-chromen-2-one. J. Mol. Struct. 2015, 1079, 194–202. [Google Scholar] [CrossRef]
- Palafox, M.A. DFT computations on vibrational spectra: Scaling procedures to improve the wavenumbers. Phys. Sci. Rev. 2018, 3, 20170184. [Google Scholar]
- Frisch, M.e.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G.; Nakatsuji, H. Gaussian 16, Revision C. 01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Srivastav, G.; Yadav, B.; Yadav, R.K.; Yadav, R. DFT studies of molecular structures conformers and vibrational characteristics of sulfanilamide. Comput. Theor. Chem. 2019, 1167, 112588. [Google Scholar] [CrossRef]
- Alcolea Palafox, M.; Belskaya, N.P.; Kostova, I.P. Peculiarities of the Spatial and Electronic Structure of 2-Aryl-1,2,3-Triazol-5-Carboxylic Acids and Their Salts on the Basis of Spectral Studies and DFT Calculations. Int. J. Mol. Sci. 2023, 24, 14001. [Google Scholar] [CrossRef]
- Rogachev, A.; Kuzmina, N.; Nemukhin, A. Theoretical modeling of the heterobimetallic complex [La(pta)3Cu(salen)] and its precursors. J. Alloys Compd. 2004, 374, 335–338. [Google Scholar] [CrossRef]
- Mehring, M.; Mansfeld, D.; Schürmann, M. The Stereochemical Activity of the Lone Pair in [Bi(NO3)3{(iPrO)2(O)PCH2P(O)(OiPr)2}2]—Comparison of Bismuth, Lanthanum and Praseodymium Nitrate Complexes. Z. Für Anorg. Und Allg. Chem. 2004, 630, 452–461. [Google Scholar] [CrossRef]
- Shishkin, O.V.; Pelmenschikov, A.; Hovorun, D.M.; Leszczynski, J. Theoretical analysis of low-lying vibrational modes of free canonical 2-deoxyribonucleosides. Chem. Phys. 2000, 260, 317–325. [Google Scholar] [CrossRef]
- Hovorun, D.; Mishchuk, Y.R.; Yurenko, Y.P. Low-frequency Raman spectra of polycrystalline ribonucleosides. Biopolym. Cell 2002, 18, 219–226. [Google Scholar] [CrossRef]
- Martel, P.; Hennion, B.; Durand, D.; Calmettes, P. Low-frequency vibrations of a nucleoside analog. J. Biomol. Struct. Dyn. 1994, 12, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Pelmenschikov, A.; Hovorun, D.M.; Shishkin, O.V.; Leszczynski, J. A density functional theory study of vibrational coupling between ribose and base rings of nucleic acids with ribosyl guanosine as a model system. J. Chem. Phys. 2000, 113, 5986–5990. [Google Scholar] [CrossRef]
- Palafox, M.A.; Nunez, J.; Gil, M. Theoretical quantum chemical study of benzoic acid: Geometrical parameters and vibrational wavenumbers. Int. J. Quantum Chem. 2002, 89, 1–24. [Google Scholar] [CrossRef]
- Zelsmann, H.; Mielke, Z. Far-infrared spectra of benzoic acid. Chem. Phys. Lett. 1991, 186, 501–508. [Google Scholar] [CrossRef]
- Galano, A. Free radicals induced oxidative stress at a molecular level: The current status, challenges and perspectives of computational chemistry based protocols. J. Mex. Chem. Soc. 2015, 59, 231–262. [Google Scholar] [CrossRef]
- Kell, D.B. Iron behaving badly: Inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med. Genom. 2009, 2, 2. [Google Scholar] [CrossRef]
- Che, M.; Wang, R.; Li, X.; Wang, H.-Y.; Zheng, X.S. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov. Today 2016, 21, 143–149. [Google Scholar] [CrossRef]
- Trist, B.G.; Hilton, J.B.; Hare, D.J.; Crouch, P.J.; Double, K.L. Superoxide dismutase 1 in health and disease: How a frontline antioxidant becomes neurotoxic. Angew. Chem. Int. Ed. 2021, 60, 9215–9246. [Google Scholar] [CrossRef]
- Pattison, D.; Davies, M. Reactions of myeloperoxidase-derived oxidants with biological substrates: Gaining chemical insight into human inflammatory diseases. Curr. Med. Chem. 2006, 13, 3271–3290. [Google Scholar] [CrossRef]
- Strzepa, A.; Pritchard, K.A.; Dittel, B.N. Myeloperoxidase: A new player in autoimmunity. Cell. Immunol. 2017, 317, 1–8. [Google Scholar] [CrossRef]
- Todorov, L.T.; Traykova, M.L.; Kostova, I.P. In Vitro Interaction of 5-aminoorotic Acid and Its Lanthanum (III) Complex with Superoxide and Hypochlorite Radicals. Der Pharma Chem. 2020, 12, 10. [Google Scholar]
- D’Haese, P.C.; Douglas, G.; Verhulst, A.; Neven, E.; Behets, G.J.; Vervaet, B.A.; Finsterle, K.; Lürling, M.; Spears, B. Human health risk associated with the management of phosphorus in freshwaters using lanthanum and aluminium. Chemosphere 2019, 220, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, C.; Zhang, W.; Liu, H.; Wang, M.; Li, F.; Li, Q.; Cao, Y. Effect and mechanism of PINK1/Parkin-mediated mitochondrial autophagy in rat lung injury induced by nano lanthanum oxide. Nanomaterials 2022, 12, 2594. [Google Scholar] [CrossRef]
- Abe, C.; Miyazawa, T.; Miyazawa, T. Current use of Fenton reaction in drugs and food. Molecules 2022, 27, 5451. [Google Scholar] [CrossRef] [PubMed]




| Control | Sample | Blank | |
|---|---|---|---|
| Tested compound | no | 200 μL | 200 μL |
| MTT | 200 μL | 200 μL | 200 μL |
| Fe2+/H2O2/Na2-EDTA | 100 μL | 100 μL | no |
| Ascorbic acid | 100 μL | 100 μL | no |
| Bi-distilled water | up to 2.0 mL | дo 2.0 mL | дo 2.0 mL |
| Blank | Control | Sample | |
|---|---|---|---|
| Tested compound | 200 μL | no | 200 μL |
| DPPH | no | 1800 μL | 1800 μL |
| Ethanol | 1800 μL | no | no |
| Bi-distilled water | no | 200 μL | no |
| Blank | Control | Sample | |
|---|---|---|---|
| Tested compound | 100 μL | no | 100 μL |
| R1 | 860 μL | 860 μL | 860 μL |
| R2 | no | 40 μL | 40 μL |
| Bi-distilled water | 40 μL | 100 μL | no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcolea Palafox, M.; Belskaya, N.P.; Todorov, L.T.; Kostova, I.P. Structural Study of a La(III) Complex of a 1,2,3-Triazole Ligand with Antioxidant Activity. Antioxidants 2023, 12, 1872. https://doi.org/10.3390/antiox12101872
Alcolea Palafox M, Belskaya NP, Todorov LT, Kostova IP. Structural Study of a La(III) Complex of a 1,2,3-Triazole Ligand with Antioxidant Activity. Antioxidants. 2023; 12(10):1872. https://doi.org/10.3390/antiox12101872
Chicago/Turabian StyleAlcolea Palafox, Mauricio, Nataliya P. Belskaya, Lozan T. Todorov, and Irena P. Kostova. 2023. "Structural Study of a La(III) Complex of a 1,2,3-Triazole Ligand with Antioxidant Activity" Antioxidants 12, no. 10: 1872. https://doi.org/10.3390/antiox12101872
APA StyleAlcolea Palafox, M., Belskaya, N. P., Todorov, L. T., & Kostova, I. P. (2023). Structural Study of a La(III) Complex of a 1,2,3-Triazole Ligand with Antioxidant Activity. Antioxidants, 12(10), 1872. https://doi.org/10.3390/antiox12101872








