Ascorbate Peroxidase 2 (APX2) of Chlamydomonas Binds Copper and Modulates the Copper Insertion into Plastocyanin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Chloroplast- and Mitochondrion-Enriched Fractions of Wild-Type Strain
2.2. Gel Electrophoresis and Blotting
2.3. Expression of Recombinant APX2 in Escherichia coli and Purification
2.4. Trp Fluorescence Assay
2.5. Circular Dichroism
2.6. APX2/Cu+ Stoichiometry Determination
2.7. AlphaFold2 Prediction for the Interaction between APX2 and Plastocyanin
2.8. Recombinant Plastocyanin
2.9. NMR Spectroscopy
2.10. Peroxidase Activity Assay
2.11. Structural Predictions
3. Results
3.1. APX2 Resides in Chloroplasts, Presents a TAT Motif for Translocation to the Thylakoid Lumen and a MxxM Motif for Metal Binding
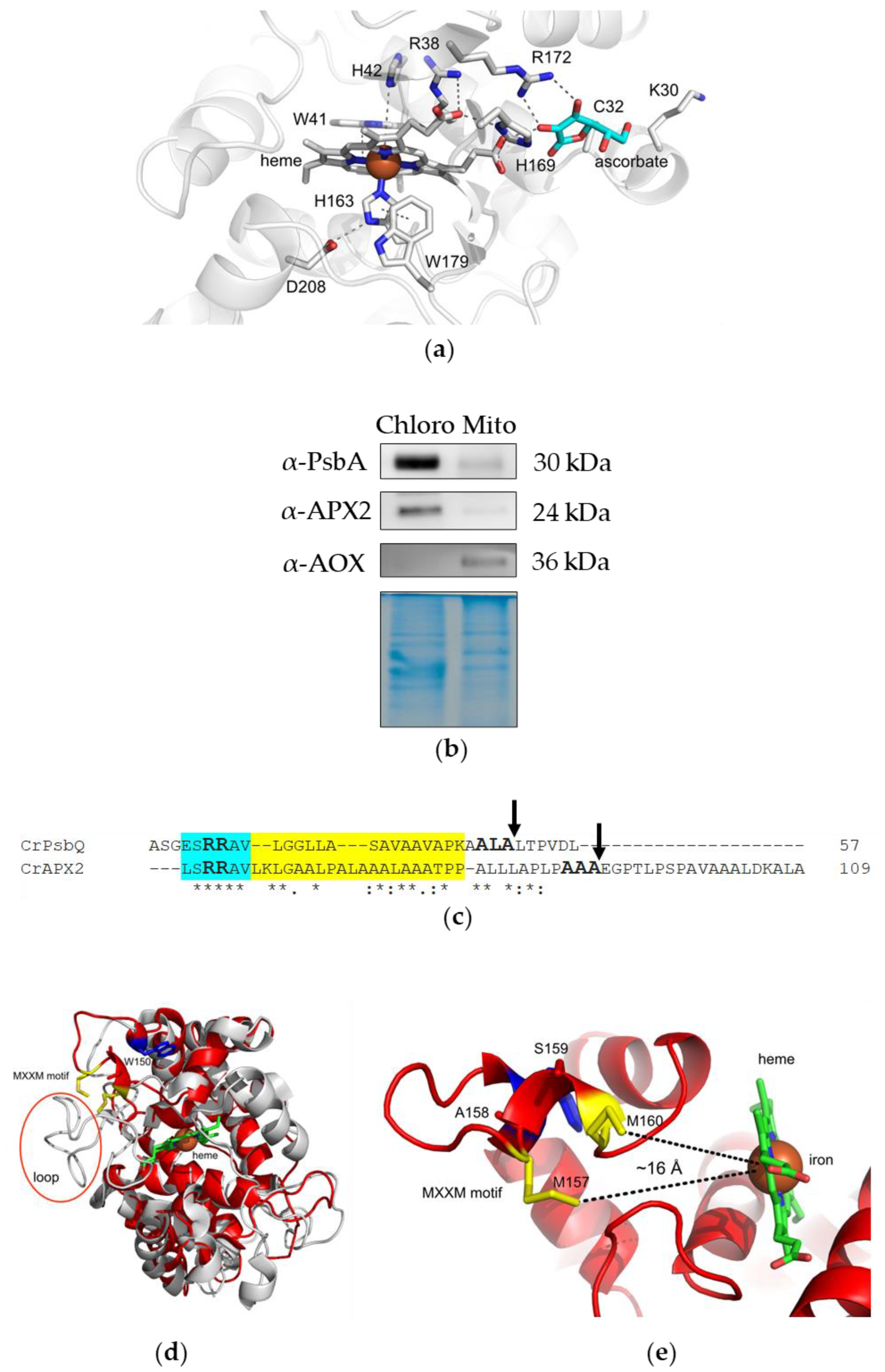
3.2. Recombinant APX2 Does Not Use Ascorbate as Electron Donor and Binds Copper
3.3. Recombinant APX2 Modulates the Binding of Copper to Plastocyanin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macdonald, I.K.; Badyal, S.K.; Ghamsari, L.; Moody, P.C.; Raven, E.L. Interaction of ascorbate peroxidase with substrates: A mechanistic and structural analysis. Biochemistry 2006, 45, 7808–7817. [Google Scholar] [CrossRef] [PubMed]
- Raven, E.L. Understanding functional diversity and substrate specificity in haem peroxidases: What can we learn from ascorbate peroxidase? Nat. Prod. Rep. 2003, 20, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Bursey, E.H.; Poulos, T.L. Two substrate binding sites in ascorbate peroxidase: The role of arginine 172. Biochemistry 2000, 39, 7374–7379. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Asada, K. THE WATER-WATER CYCLE IN CHLOROPLASTS: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Hossain, M.A.; Nakano, Y.; Asada, K. Monodehydroascorbate Reductase in Spinach Chloroplasts and Its Participation in Regeneration of Ascorbate for Scavenging Hydrogen Peroxide. Plant Cell Physiol. 1984, 25, 385–395. [Google Scholar] [CrossRef]
- Hossain, M.A.; Asada, K. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J. Biol. Chem. 1985, 260, 12920–12926. [Google Scholar] [CrossRef]
- Hossain, M.A.; Asada, K. Purification of Dehydroascorbate Reductase from Spinach and Its Characterization as a Thiol Enzyme. Plant Cell Physiol. 1984, 25, 85–92. [Google Scholar] [CrossRef]
- Lazzarotto, F.; Teixeira, F.K.; Rosa, S.B.; Dunand, C.; Fernandes, C.L.; de Vasconcelos Fontenele, A.; Silveira, J.A.G.; Verli, H.; Margis, R.; Margis-Pinheiro, M. Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 2011, 191, 234–250. [Google Scholar] [CrossRef]
- Lazzarotto, F.; Menguer, P.K.; Del-Bem, L.E.; Zamocky, M.; Margis-Pinheiro, M. Ascorbate Peroxidase Neofunctionalization at the Origin of APX-R and APX-L: Evidence from Basal Archaeplastida. Antioxidants 2021, 10, 597. [Google Scholar] [CrossRef]
- Granlund, I.; Storm, P.; Schubert, M.; Garcia-Cerdan, J.G.; Funk, C.; Schroder, W.P. The TL29 protein is lumen located, associated with PSII and not an ascorbate peroxidase. Plant Cell Physiol. 2009, 50, 1898–1910. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, E.; Storm, P.; Schroder, W.P.; Funk, C. Crystal structure of the TL29 protein from Arabidopsis thaliana: An APX homolog without peroxidase activity. J. Struct. Biol. 2011, 176, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Lazzarotto, F.; Wahni, K.; Piovesana, M.; Maraschin, F.; Messens, J.; Margis-Pinheiro, M. Arabidopsis APx-R Is a Plastidial Ascorbate-Independent Peroxidase Regulated by Photomorphogenesis. Antioxidants 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Shumayla; Verma, P.C.; Singh, K.; Upadhyay, S.K. Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 2020, 112, 4208–4223. [Google Scholar] [CrossRef]
- Verma, D.; Upadhyay, S.K.; Singh, K. Characterization of APX and APX-R gene family in Brassica juncea and B. rapa for tolerance against abiotic stresses. Plant Cell Rep. 2022, 41, 571–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Letnik, I.; Hacham, Y.; Dobrev, P.; Ben-Daniel, B.H.; Vankova, R.; Amir, R.; Miller, G. ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 2014, 166, 370–383. [Google Scholar] [CrossRef]
- Chen, C.; Galon, Y.; Rahmati Ishka, M.; Malihi, S.; Shimanovsky, V.; Twito, S.; Rath, A.; Vatamaniuk, O.K.; Miller, G. ASCORBATE PEROXIDASE6 delays the onset of age-dependent leaf senescence. Plant Physiol. 2021, 185, 441–456. [Google Scholar] [CrossRef]
- Kuo, E.Y.; Cai, M.S.; Lee, T.M. Ascorbate peroxidase 4 plays a role in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Sci. Rep. 2020, 10, 13287. [Google Scholar] [CrossRef] [PubMed]
- Rubino, J.T.; Riggs-Gelasco, P.; Franz, K.J. Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg. Chem. 2010, 15, 1033–1049. [Google Scholar] [CrossRef] [PubMed]
- Cline, K. Mechanistic Aspects of Folded Protein Transport by the Twin Arginine Translocase (Tat). J. Biol. Chem. 2015, 290, 16530–16538. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Patena, W.; Fauser, F.; Jinkerson, R.E.; Saroussi, S.; Meyer, M.T.; Ivanova, N.; Robertson, J.M.; Yue, R.; Zhang, R.; et al. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 2019, 51, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Mason, C.B.; Bricker, T.M.; Moroney, J.V. A rapid method for chloroplast isolation from the green alga Chlamydomonas reinhardtii. Nat. Protoc. 2006, 1, 2227–2230. [Google Scholar] [CrossRef] [PubMed]
- Cardol, P.; Matagne, R.F.; Remacle, C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 2002, 319, 1211–1221. [Google Scholar] [CrossRef]
- Sylvestre-Gonon, E.; Schwartz, M.; Girardet, J.M.; Hecker, A.; Rouhier, N. Is there a role for tau glutathione transferases in tetrapyrrole metabolism and retrograde signalling in plants? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190404. [Google Scholar] [CrossRef]
- Blaby-Haas, C.E.; Padilla-Benavides, T.; Stube, R.; Arguello, J.M.; Merchant, S.S. Evolution of a plant-specific copper chaperone family for chloroplast copper homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, E5480–E5487. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Kuhlgert, S.; Drepper, F.; Fufezan, C.; Sommer, F.; Hippler, M. Residues PsaB Asp612 and PsaB Glu613 of photosystem I confer pH-dependent binding of plastocyanin and cytochrome c(6). Biochemistry 2012, 51, 7297–7303. [Google Scholar] [CrossRef]
- Ubbink, M.; Lian, L.Y.; Modi, S.; Evans, P.A.; Bendall, D.S. Analysis of the 1H-NMR chemical shifts of Cu(I)-, Cu(II)- and Cd-substituted pea plastocyanin. Metal-dependent differences in the hydrogen-bond network around the copper site. Eur. J. Biochem. 1996, 242, 132–147. [Google Scholar] [CrossRef]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Ogola, H.J.; Kamiike, T.; Hashimoto, N.; Ashida, H.; Ishikawa, T.; Shibata, H.; Sawa, Y. Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Env. Microbiol. 2009, 75, 7509–7518. [Google Scholar] [CrossRef]
- Kumar, P. Stress amelioration response of glycine betaine and Arbuscular mycorrhizal fungi in sorghum under Cr toxicity. PLoS ONE 2021, 16, e0253878. [Google Scholar] [CrossRef]
- Janson, G.; Zhang, C.; Prado, M.G.; Paiardini, A. PyMod 2.0: Improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 2017, 33, 444–446. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef]
- Mayfield, S.P.; Schirmer-Rahire, M.; Frank, G.; Zuber, H.; Rochaix, J.D. Analysis of the genes of the OEE1 and OEE3 proteins of the photosystem II complex from Chlamydomonas reinhardtii. Plant Mol. Biol. 1989, 12, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Nadas, I.A.; Kim, M.A.; Franz, K.J. A Mets motif peptide found in copper transport proteins selectively binds Cu(I) with methionine-only coordination. Inorg. Chem. 2005, 44, 9787–9794. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Doerge, D.R.; Divi, R.L.; Churchwell, M.I. Identification of the colored guaiacol oxidation product produced by peroxidases. Anal. Biochem. 1997, 250, 10–17. [Google Scholar] [CrossRef]
- Aguirre, G.; Pilon, M. Copper Delivery to Chloroplast Proteins and its Regulation. Front. Plant Sci. 2015, 6, 1250. [Google Scholar] [CrossRef]
- Kropat, J.; Gallaher, S.D.; Urzica, E.I.; Nakamoto, S.S.; Strenkert, D.; Tottey, S.; Mason, A.Z.; Merchant, S.S. Copper economy in Chlamydomonas: Prioritized allocation and reallocation of copper to respiration vs. photosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 2644–2651. [Google Scholar] [CrossRef]
- Strenkert, D.; Schmollinger, S.; Gallaher, S.D.; Salome, P.A.; Purvine, S.O.; Nicora, C.D.; Mettler-Altmann, T.; Soubeyrand, E.; Weber, A.P.M.; Lipton, M.S.; et al. Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 2019, 116, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Schmollinger, S.; Strenkert, D.; Moseley, J.L.; Blaby-Haas, C.E. From economy to luxury: Copper homeostasis in Chlamydomonas and other algae. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118822. [Google Scholar] [CrossRef]
- Lazzarotto, F.; Turchetto-Zolet, A.C.; Margis-Pinheiro, M. Revisiting the Non-Animal Peroxidase Superfamily. Trends Plant Sci. 2015, 20, 807–813. [Google Scholar] [CrossRef]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef]
- Hill, K.L.; Hassett, R.; Kosman, D.; Merchant, S. Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 1996, 112, 697–704. [Google Scholar] [CrossRef]
- Pham, K.L.J.; Schmollinger, S.; Merchant, S.S.; Strenkert, D. Chlamydomonas ATX1 is essential for Cu distribution to multiple cupro-enzymes and maintenance of biomass in conditions demanding cupro-enzyme-dependent metabolic pathways. Plant Direct 2022, 6, e383. [Google Scholar] [CrossRef] [PubMed]
- Castruita, M.; Casero, D.; Karpowicz, S.J.; Kropat, J.; Vieler, A.; Hsieh, S.I.; Yan, W.; Cokus, S.; Loo, J.A.; Benning, C.; et al. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 2011, 23, 1273–1292. [Google Scholar] [CrossRef]
- Smeekens, S.; Bauerle, C.; Hageman, J.; Keegstra, K.; Weisbeek, P. The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell 1986, 46, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Castell, C.; Rodriguez-Lumbreras, L.A.; Hervas, M.; Fernandez-Recio, J.; Navarro, J.A. New Insights into the Evolution of the Electron Transfer from Cytochrome f to Photosystem I in the Green and Red Branches of Photosynthetic Eukaryotes. Plant Cell Physiol. 2021, 62, 1082–1093. [Google Scholar] [CrossRef]
- Miyake, C.; Michihata, F.; Asada, K. Scavenging of Hydrogen Peroxide in Prokaryotic and Eukaryotic Algae: Acquisition of Ascorbate Peroxidase during the Evolution of Cyanobacteria. Plant Cell Physiol. 1991, 32, 33–43. [Google Scholar] [CrossRef]
- Dunand, C.; Mathe, C.; Lazzarotto, F.; Margis, R.; Margis-Pinheiro, M. Ascorbate peroxidase-related (APx-R) is not a duplicable gene. Plant Signal Behav. 2011, 6, 1908–1913. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caccamo, A.; Vega de Luna, F.; Wahni, K.; Volkov, A.N.; Przybyla-Toscano, J.; Amelii, A.; Kriznik, A.; Rouhier, N.; Messens, J.; Remacle, C. Ascorbate Peroxidase 2 (APX2) of Chlamydomonas Binds Copper and Modulates the Copper Insertion into Plastocyanin. Antioxidants 2023, 12, 1946. https://doi.org/10.3390/antiox12111946
Caccamo A, Vega de Luna F, Wahni K, Volkov AN, Przybyla-Toscano J, Amelii A, Kriznik A, Rouhier N, Messens J, Remacle C. Ascorbate Peroxidase 2 (APX2) of Chlamydomonas Binds Copper and Modulates the Copper Insertion into Plastocyanin. Antioxidants. 2023; 12(11):1946. https://doi.org/10.3390/antiox12111946
Chicago/Turabian StyleCaccamo, Anna, Félix Vega de Luna, Khadija Wahni, Alexander N. Volkov, Jonathan Przybyla-Toscano, Antonello Amelii, Alexandre Kriznik, Nicolas Rouhier, Joris Messens, and Claire Remacle. 2023. "Ascorbate Peroxidase 2 (APX2) of Chlamydomonas Binds Copper and Modulates the Copper Insertion into Plastocyanin" Antioxidants 12, no. 11: 1946. https://doi.org/10.3390/antiox12111946
APA StyleCaccamo, A., Vega de Luna, F., Wahni, K., Volkov, A. N., Przybyla-Toscano, J., Amelii, A., Kriznik, A., Rouhier, N., Messens, J., & Remacle, C. (2023). Ascorbate Peroxidase 2 (APX2) of Chlamydomonas Binds Copper and Modulates the Copper Insertion into Plastocyanin. Antioxidants, 12(11), 1946. https://doi.org/10.3390/antiox12111946







