Antioxidant Defense in Primary Murine Lung Cells following Short- and Long-Term Exposure to Plastic Particles
Abstract
:1. Introduction
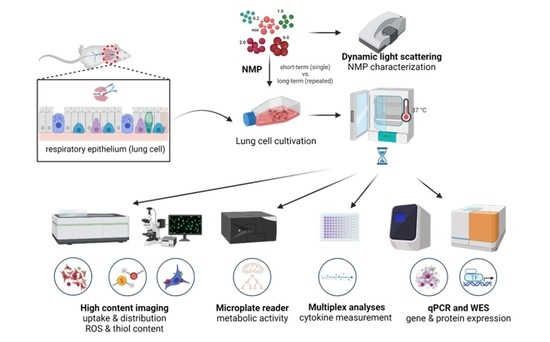
2. Materials and Methods
2.1. Lung Cell Culture
2.2. Polymer Particles and Lung Cell Exposure
2.3. Analysis of Intracellular ROS, Thiol Content, Cellular Metabolism, Viability, and Apoptosis
2.4. Quantification of Gene and Protein Expression
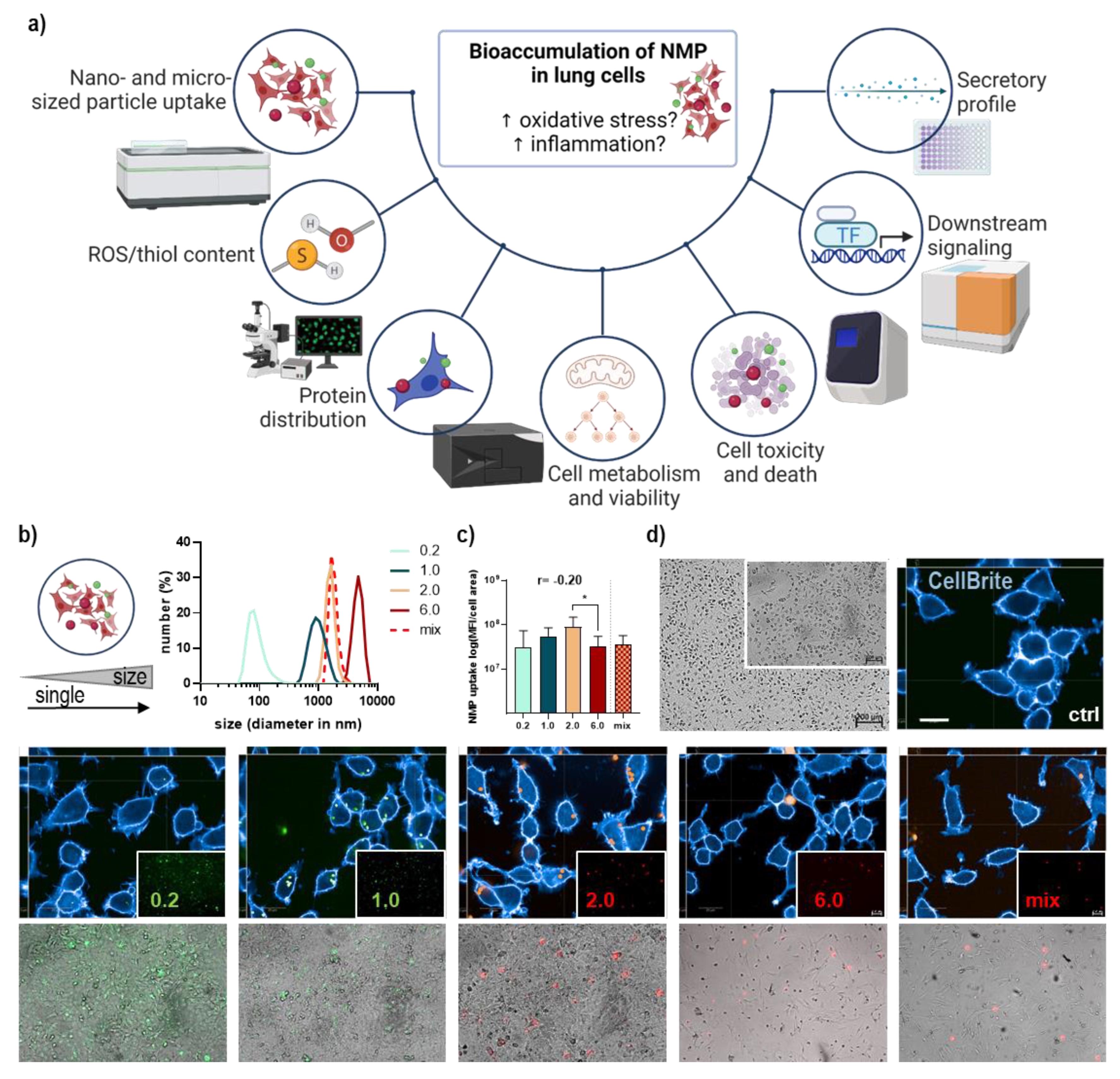
2.5. Imaging to Study the Bioaccumulation of NMP
2.6. Multiplex Chemokine and Cytokine Profiles
2.7. Statistical Analysis
3. Results
3.1. Lung Epithelial Cells’ NMP Uptake Affected Metabolism, ROS Formation, and Viability
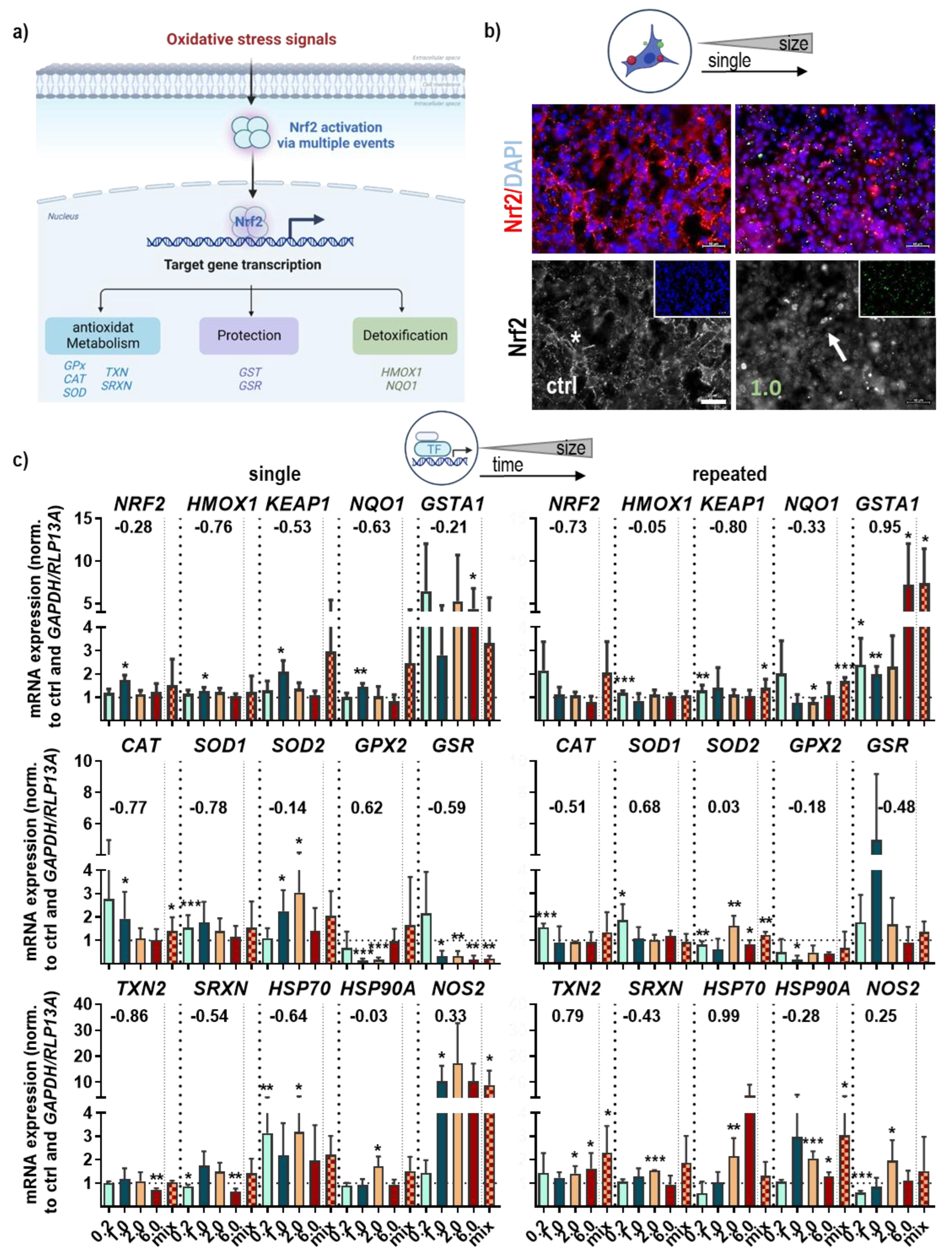
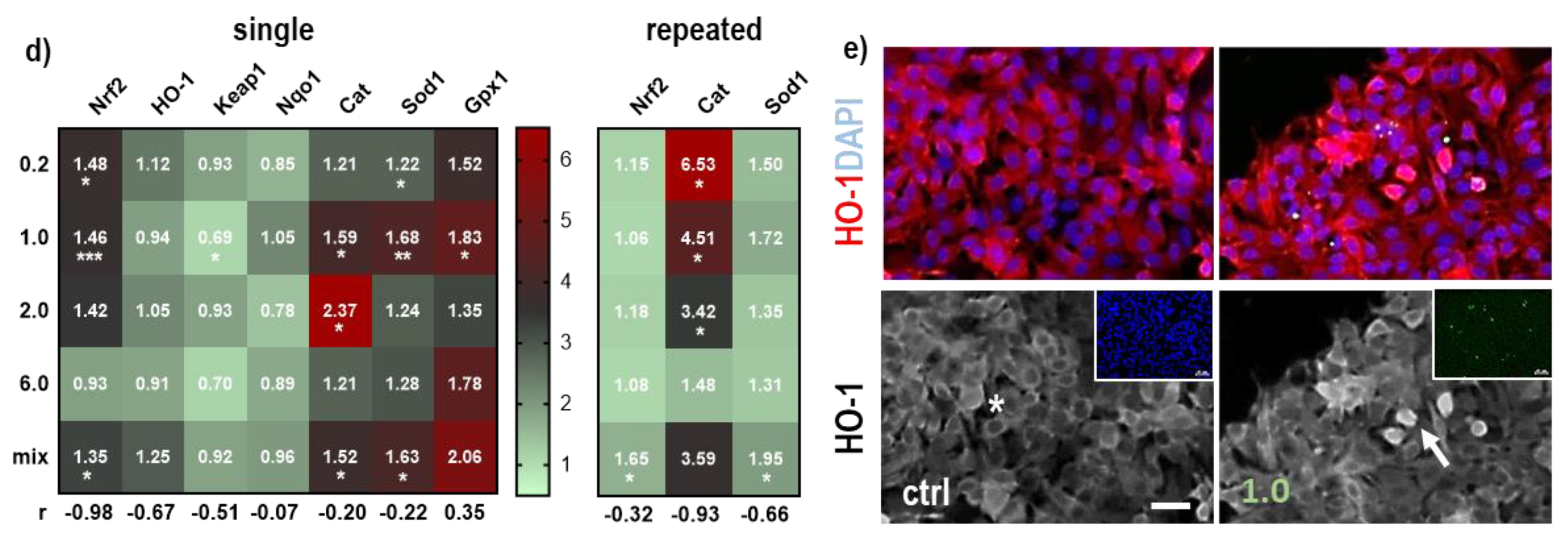
3.2. NMP-Affected Stress Signaling in Primary Lung Epithelial Cells
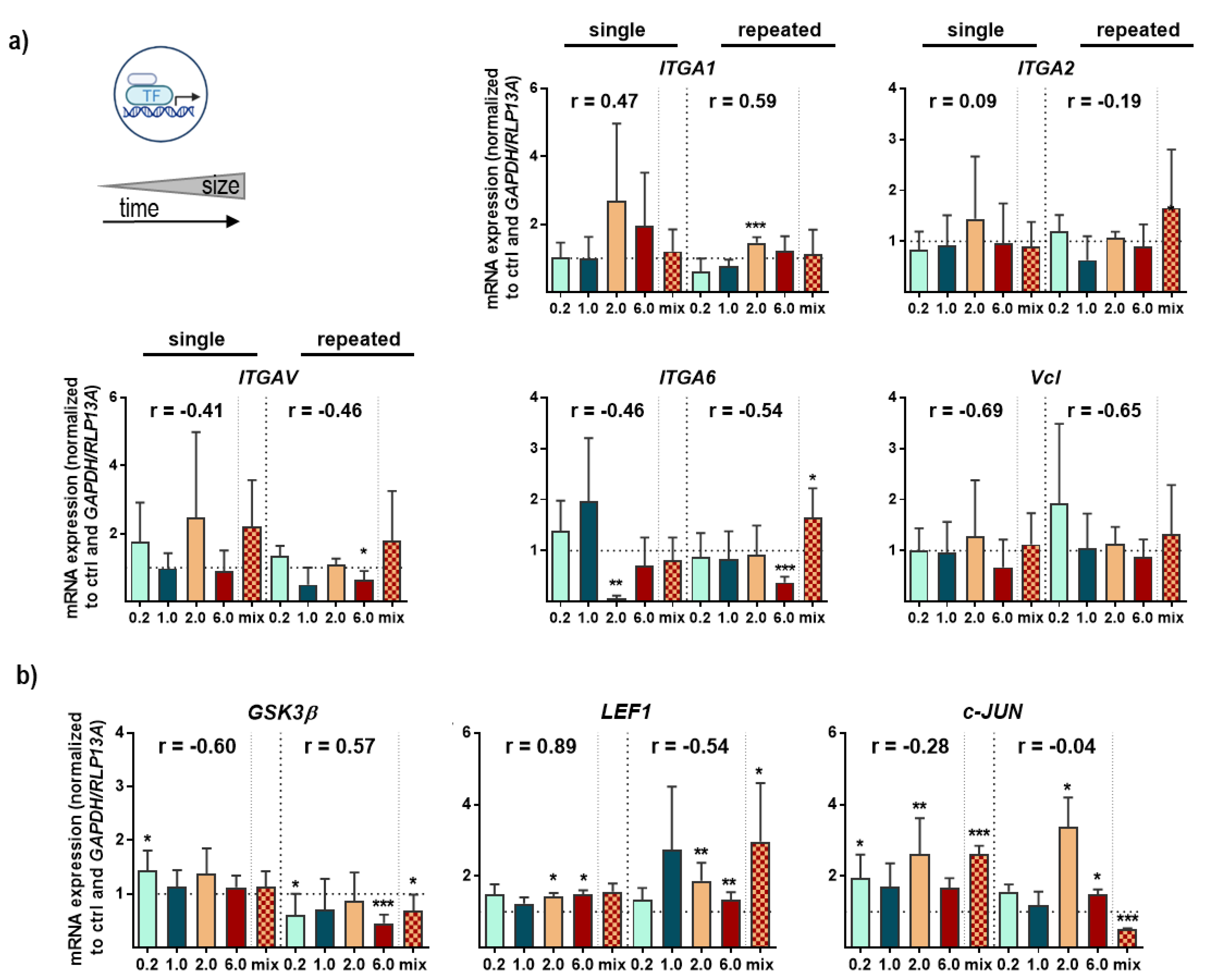
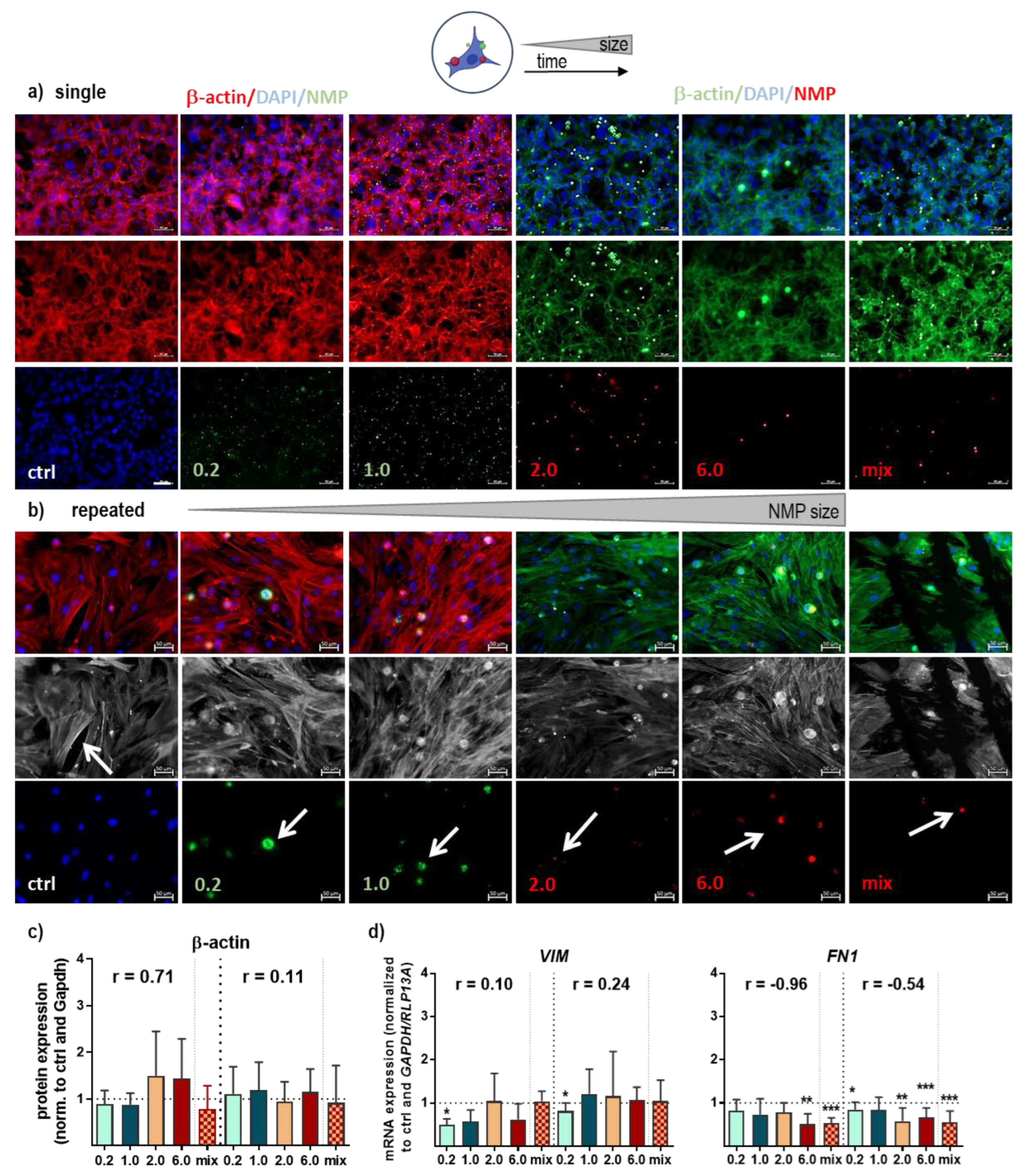
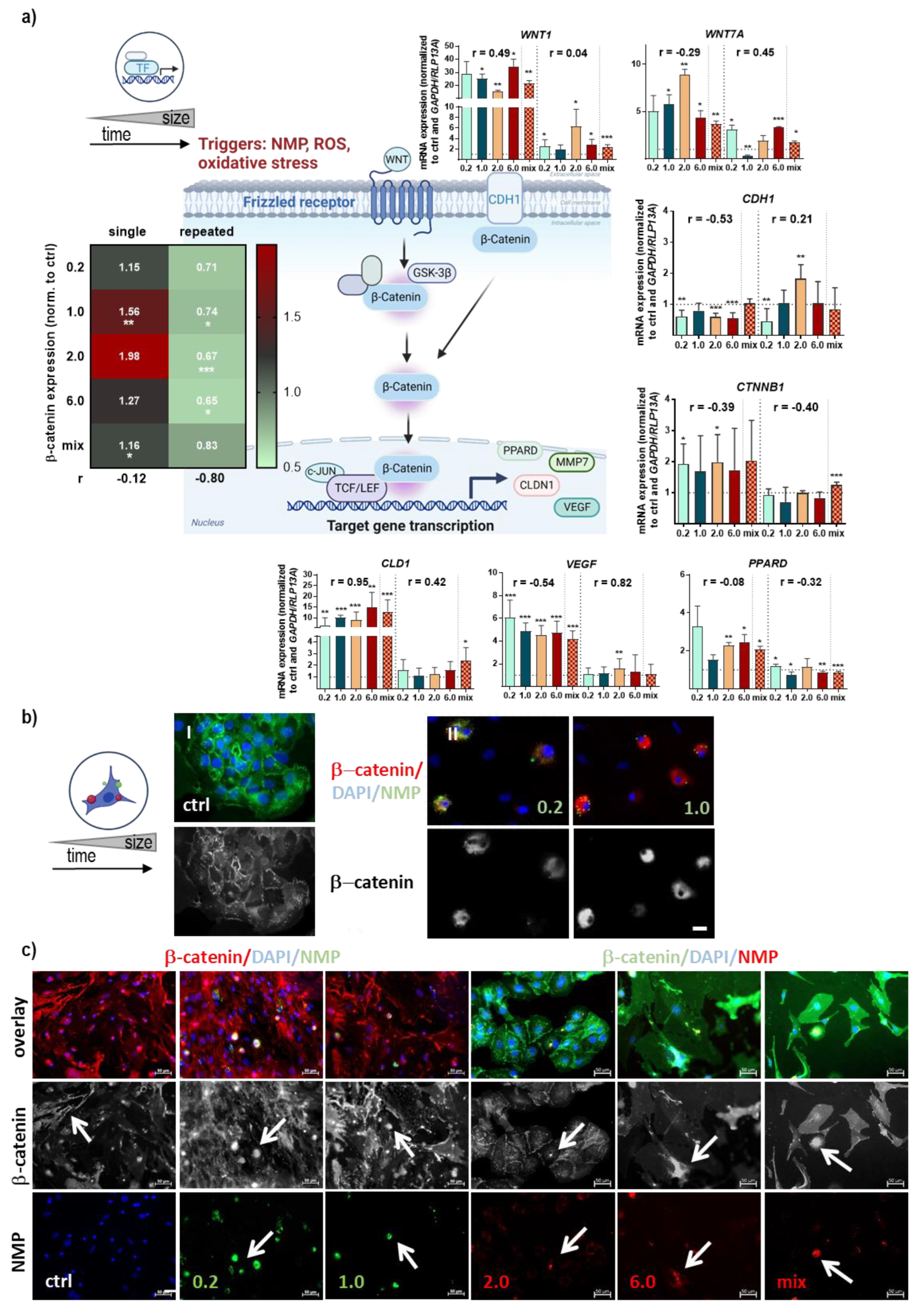
3.3. Structural and Functional Signaling Changes in NMP-Treated Lung Cells
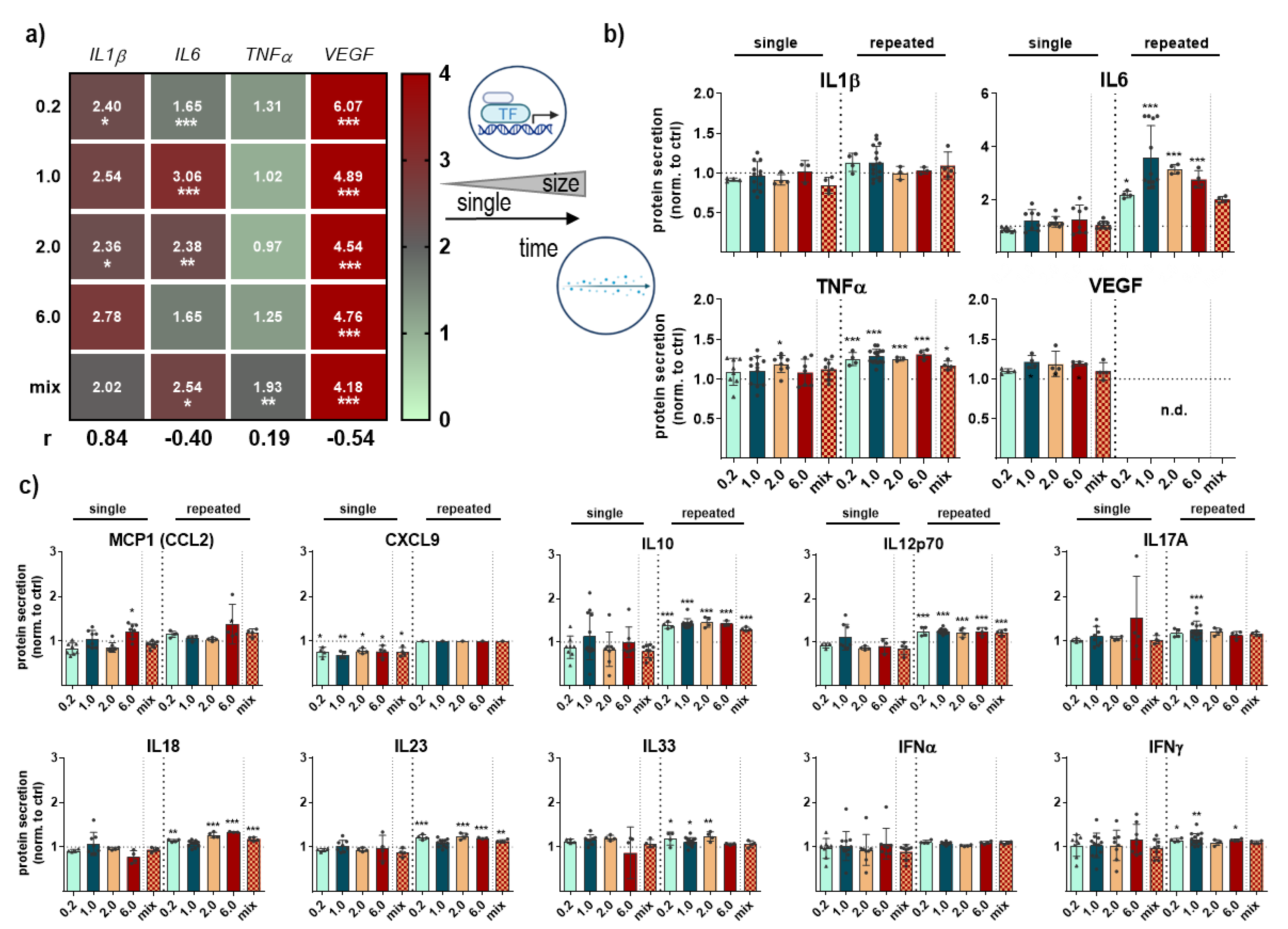
3.4. Cytokine Secretion Profile Changes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Gene Name | Alias with Gene ID | Primer Sequences (3′-5′) |
|---|---|---|
| nuclear factor erythroid 2-related factor 2 | NRF2_NM_010902 | GAG TCG CTT GCC CTG GAT ATC TCA TGG CTG CCT CCA GAG AA |
| heme oxygenase 1 | HMOX1_NM_010442 | TGA AGC AGG CAT CTG AGG G CGA AGG TGG AAG AGT GGG AG |
| NAD(P)H dehydrogenase [quinone] 1 | NQO1_NM_008706 | GGC ATC CAG TCC TCC ATC AA GTT AGT CCC TCG GCC ATT GTT |
| kelch-like ECH-associated protein 1 | KEAP1_NM_016679 | CGG GGA CGC AGT GAT GTA TG TGT GTA GCT GAA GGT TCG GTT A |
| catalase | CAT_NM_009804 | CAG AGA GCG GAT TCC TGA GAG A CTT TGC CTT GGA GTA TCT GGT GAT |
| superoxide dismutase [Cu-Zn] | SOD1_NM_011434 | GAA ACA AGA TGA CTT GGG CAA AG TTA CTG CGC AAT CCC AAT CA |
| glutathione peroxidase 1 | GPX2_NM_030677 | GTG GCG TCA CTC TGA GGA ACA CAG TTC TCC TGA TGT CCG AAC TG |
| glutathione-disulfide reductase | GSR_NM_010344 | TCG GAA TTC ATG CAC GAT CA GGC TCA CAT AGG CAT CCC TTT |
| tumor necrosis factor-alpha | TNFA_NM_013693 | TCT CAT GCA CCA CCA TCA AGG ACT ACC ACT CTC CCT TTG CAG AAC TCA |
| interleukin 1β | IL1β | GCA ACT GTT CCT GAA CTC AAC T ATC TTT TGG GGT CCG TCA ACT |
| interleukin 6 | IL6_NM_031168 | ATC CAG TTG CCT TCT TGG GAC TGA TAA GCC TCC GAC TTG TGA AGT GGT |
| β actin | ACTB_NM_007393 | TTG CTG ACA GGA TGC AGA AG ACA TCT GCT GGA AGG TGG AC |
| heat shock protein 70 | HSP70_NM_010479 | GGC TGA CAA GAA GAA GGT GC CTG GTA CAG CCC ACT GAT GA |
| heat shock protein 90α | HSP90A_NM_010480 | GAC GCT CTG GAT AAA ATC CGT T TGG GAA TGA GAT TGA TGT GCA G |
| vinculin | VCL_NM_009502 | GCT TCAGTC AGACCC ATA CTC G AGG TAA GCA GTA GGT CAG ATG T |
| focal adhesion kinase | FAK/PTK2_NM_007982 | GAG TAC GTC CCT ATG GTG AAG G CTC GAT CTC TCG ATG AGT GCT |
| integrin A1 | ITGA1_ NM_001033228 | GAC AGC CCT TGG AAT AGA CAC GTT GTC ATG CGA TTC TCC ATC A |
| integrin A2 | ITGA2_NM_008396 | TGT CTG GCG TAT AAT GTT GGC TGC TGT ACT GAA TAC CCA AAC TG |
| integrin A5 | ITGA5_NM_008402 | TGC AGT GGT TCG GAG CAA C TTT TCT GTG CGC CAG CTA TAC |
| integrin A6 | ITGA6_NM_008397 | GGG ATC GTC CGT GTA GAA CAA TCT CTC CAC CAA CTT CAT AGG G |
| fibronectin 1 | FN1_NM_010233 | GCT CAG CAA ATC GTG CAG C CTA GGT AGG TCC GTT CCC ACT |
| vimentin | VIM_NM_011701 | CGT CCA CAC GCA CCT ACA G GGG GGA TGA GGA ATA GAG GCT |
| wingless-type MMTV integration site 1 | WNT1_NM_021279 | CGACTGATCCGACAGAACCC CCATTTGCACTCTCGCACA |
| wingless-type MMTV integration site 7a | WNT7A_NM_009527 | TCAGTTTCAGTTCCGAAATGGC CCCGACTCCCCACTTTGAG |
| β-catenin 1 | CTNNB1_NM_007614 | CCC AGT CCT TCA CGC AAG AG CAT CTA GCG TCT CAG GGA ACA |
| E-cadherin | CDH1_NM_009864 | CAC CTG GAG AGA GGC CAT GT TGG GAA ACA TGA GCA GCT CT |
| lymphoid enhancer-binding factor 1 | LEF1_NM_010703 | GCC ACC GAT GAG ATG ATC CC TTG ATG TCG GCT AAG TCG CC |
| Jnk for c-jun N-terminal kinase α | C-JUN_AJ315350 | GTCCTCCATAAATGCCTGTTCC GATGCAACCCACTGACCAGAT |
| peroxisome proliferator activator receptor delta | PPARD_NM_011145 | TCCATCGTCAACAAAGACGGG ACTTGGGCTCAATGATGTCAC |
| claudin 1 | CLD1_NM_016674 | GGG GAC AAC ATC GTG ACC G AGG AGT CGA AGA CTT TGC ACT |
| vascular endothelial growth factor | VEGF_NM_001025250 | AAC GAT GAA GCC CTG GAG TG GAC AAA CAA ATG CTT TCT CCG |
| glycogen synthase kinase 3 α | GSK3B_NM_019827 | AAG CTC TGC GAT TTT GGC AGT GAG TTC TGG AGC ACG GTA GTA |
| glyceraldehyde 3-phosphate dehydrogenase | GAPDH_NM_001289726 | CAT GGC CTC CAA GGA GTA AG TGT GAG GGA GAT GCT CAG TG |
| ribosomal protein 13A | RPL13A_NM_009438 | AGC CTA CCA GAA AGT TTG CTT AC GCT TCT TCT TCC GAT AGT GCA TC |

References
- Blair, R.M.; Waldron, S.; Phoenix, V.; Gauchotte-Lindsay, C. Micro- and Nanoplastic Pollution of Freshwater and Wastewater Treatment Systems. Springer Sci. Rev. 2017, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.; Reemtsma, T. Things We Know and Don’t Know About Nanoplastic in the Environment. Nat. Nanotechnol. 2019, 14, 300–301. [Google Scholar] [CrossRef] [PubMed]
- Karakolis, E.G.; Nguyen, B.; You, J.B.; Rochman, C.M.; Sinton, D. Fluorescent Dyes for Visualizing Microplastic Particles and Fibers in Laboratory-Based Studies. Environ. Sci. Technol. Lett. 2019, 6, 334. [Google Scholar] [CrossRef]
- Allen, S.; Allen, D.; Phoenix, V.R.; Le Roux, G.; Durántez Jiménez, P.; Simonneau, A.; Binet, S.; Galop, D. Atmospheric Transport and Deposition of Microplastics in a Remote Mountain Catchment. Nat. Geosci. 2019, 12, 339. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Halimu, G.; Zhang, Q.; Song, Y.; Fu, X.; Li, Y.; Li, Y.; Zhang, H. Internalization and Toxicity: A Preliminary Study of Effects of Nanoplastic Particles on Human Lung Epithelial Cell. Sci. Total Environ. 2019, 694, 133794. [Google Scholar] [CrossRef]
- Prata, J.C. Airborne Microplastics: Consequences to Human Health? Environ. Pollut. 2018, 234, 115–126. [Google Scholar] [CrossRef]
- Gasperi, J.; Wright, S.L.; Dris, R.; Collard, F.; Mandin, C.; Guerrouache, M.; Langlois, V.; Kelly, F.J.; Tassin, B. Microplastics in Air: Are We Breathing It In? Curr. Opin. Environ. Sci. Health 2018, 1, 1. [Google Scholar] [CrossRef] [Green Version]
- Manshoven, S.S.A.; Malarciuc, C.; Tenhunen, A. Microplastic Pollution from Textile Consumption in Europe. Eionet Rep. ETC/CE 2022, 1, 2022. [Google Scholar]
- Saavedra, J.; Stoll, S.; Slaveykova, V.I. Influence of Nanoplastic Surface Charge on Eco-Corona Formation, Aggregation and Toxicity to Freshwater Zooplankton. Environ. Pollut. 2019, 252, 715–722. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, P.M.; Saranya, V.; Vijayakumar, S.; Mythili Meera, M.; Ruprekha, S.; Kunal, R.; Pranay, A.; Thomas, J.; Mukherjee, A.; Chandrasekaran, N. Assessment on Interactive Prospectives of Nanoplastics with Plasma Proteins and the Toxicological Impacts of Virgin, Coronated and Environmentally Released-Nanoplastics. Sci. Rep. 2019, 9, 8860. [Google Scholar] [CrossRef] [Green Version]
- Verla, A.W.; Enyoh, C.E.; Verla, E.N.; Nwarnorh, K.O. Microplastic–Toxic Chemical Interaction: A Review Study on Quantified Levels, Mechanism and Implication. SN Appl. Sci. 2019, 1, 1400. [Google Scholar] [CrossRef]
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovidou, E.; Purnell, P. An Overview of Chemical Additives Present in Plastics: Migration, Release, Fate and Environmental Impact During Their Use, Disposal and Recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, F.; Adeyemi, J.A.; Bocato, M.Z.; Comas, A.; Campiglia, A. A Critical Viewpoint on Current Issues, Limitations, and Future Research Needs on Micro-and Nanoplastic Studies: From the Detection to the Toxicological Assessment. Environ. Res. 2020, 182, 109089. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Guo, H.; Cui, M.; Huang, R.; Su, R.; Qi, W.; He, Z. Interaction of Particles with Mucosae and Cell Membranes. Colloids Surf. B Biointerfaces 2020, 186, 110657. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.F.; Chen, C.Y.; Lu, T.H.; Liao, C.M. Toxicity-Based Toxicokinetic/Toxicodynamic Assessment for Bioaccumulation of Polystyrene Microplastics in Mice. J. Hazard. Mater. 2019, 366, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Q.Y.; Valiyaveetill, S.; Tang, B.L. Toxicity of Microplastics and Nanoplastics in Mammalian Systems. Int. J. Environ. Res. Public Health 2020, 17, 1509. [Google Scholar] [CrossRef] [Green Version]
- Chang, X.; Xue, Y.; Li, J.; Zou, L.; Tang, M. Potential Health Impact of Environmental Micro- and Nanoplastics Pollution. J. Appl. Toxicol. 2020, 40, 4–15. [Google Scholar] [CrossRef]
- Elsaesser, A.; Howard, C.V. Toxicology of Nanoparticles. Adv. Drug Deliv. Rev. 2012, 64, 129–137. [Google Scholar] [CrossRef]
- Van Cauwenberghe, L.; Janssen, C.R. Microplastics in Bivalves Cultured for Human Consumption. Environ. Pollut. 2014, 193, 65–70. [Google Scholar] [CrossRef]
- Stock, V.; Laurisch, C.; Franke, J.; Donmez, M.H.; Voss, L.; Bohmert, L.; Braeuning, A.; Sieg, H. Uptake and Cellular Effects of Pe, Pp, Pet and Pvc Microplastic Particles. Toxicol. In Vitro 2021, 70, 105021. [Google Scholar] [CrossRef]
- Stock, V.; Bohmert, L.; Lisicki, E.; Block, R.; Cara-Carmona, J.; Pack, L.K.; Selb, R.; Lichtenstein, D.; Voss, L.; Henderson, C.J.; et al. Uptake and Effects of Orally Ingested Polystyrene Microplastic Particles in Vitro and in Vivo. Arch. Toxicol. 2019, 93, 1817–1833. [Google Scholar] [CrossRef] [PubMed]
- Salvi, S. Health Effects of Ambient Air Pollution in Children. Paediatr. Respir. Rev. 2007, 8, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.L.; Becker, L.G.; Huber, A.M.; Catalano, R.F. Review of Risk and Protective Factors of Substance Use and Problem Use in Emerging Adulthood. Addict. Behav. 2012, 37, 747–775. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.C.; Krewski, D.; Diver, W.R.; Pope, C.A., 3rd; Burnett, R.T.; Jerrett, M.; Marshall, J.D.; Gapstur, S.M. Ambient Air Pollution and Cancer Mortality in the Cancer Prevention Study Ii. Environ. Health Perspect. 2017, 125, 087013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato-Lourenco, L.F.; Dos Santos Galvao, L.; de Weger, L.A.; Hiemstra, P.S.; Vijver, M.G.; Mauad, T. An Emerging Class of Air Pollutants: Potential Effects of Microplastics to Respiratory Human Health? Sci. Total Environ. 2020, 749, 141676. [Google Scholar] [CrossRef]
- Lee, J.; Jang, J.; Park, S.M.; Yang, S.R. An Update on the Role of Nrf2 in Respiratory Disease: Molecular Mechanisms and Therapeutic Approaches. Int. J. Mol. Sci. 2021, 22, 8406. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, D.K.; Long, N.P.; Kwon, S.W.; Park, J.H. Uptake of Nanopolystyrene Particles Induces Distinct Metabolic Profiles and Toxic Effects in Caenorhabditis Elegans. Environ. Pollut. 2019, 246, 578–586. [Google Scholar] [CrossRef]
- Leslie, H.A.; van Velzen, M.J.M.; Brandsma, S.H.; Vethaak, A.D.; Garcia-Vallejo, J.J.; Lamoree, M.H. Discovery and Quantification of Plastic Particle Pollution in Human Blood. Environ. Int. 2022, 163, 107199. [Google Scholar] [CrossRef]
- Leslie, H.A.; Depledge, M.H. Where Is the Evidence That Human Exposure to Microplastics Is Safe? Environ. Int. 2020, 142, 105807. [Google Scholar] [CrossRef]
- Da Silva Brito, W.A.; Singer, D.; Miebach, L.; Saadati, F.; Wende, K.; Schmidt, A.; Bekeschus, S. Comprehensive in Vitro Polymer Type, Concentration, and Size Correlation Analysis to Microplastic Toxicity and Inflammation. Sci. Total Environ. 2022, 854, 158731. [Google Scholar] [CrossRef]
- Cheon, S.; Poon, R.; Yu, C.; Khoury, M.; Shenker, R.; Fish, J.; Alman, B.A. Prolonged Beta-Catenin Stabilization and Tcf-Dependent Transcriptional Activation in Hyperplastic Cutaneous Wounds. Lab. Investig. 2005, 85, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Schwabl, P.; Koppel, S.; Konigshofer, P.; Bucsics, T.; Trauner, M.; Reiberger, T.; Liebmann, B. Detection of Various Microplastics in Human Stool a Prospective Case Series. Ann. Intern. Med. 2019, 171, 453–457. [Google Scholar] [CrossRef]
- Horvatits, T.; Tamminga, M.; Liu, B.; Sebode, M.; Carambia, A.; Fischer, L.; Puschel, K.; Huber, S.; Fischer, E.K. Microplastics Detected in Cirrhotic Liver Tissue. EBioMedicine 2022, 82, 104147. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, A.; Svelato, A.; Santacroce, C.; Catalano, P.; Notarstefano, V.; Carnevali, O.; Papa, F.; Rongioletti, M.C.A.; Baiocco, F.; Draghi, S.; et al. Plasticenta: First Evidence of Microplastics in Human Placenta. Environ. Int. 2021, 146, 106274. [Google Scholar] [CrossRef]
- Lee, T.R.; Choi, M.; Kopacz, A.M.; Yun, S.H.; Liu, W.K.; Decuzzi, P. On the near-Wall Accumulation of Injectable Particles in the Microcirculation: Smaller Is Not Better. Sci. Rep. 2013, 3, srep02079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, S.B.; D’Errico, J.N.; Adler, D.S.; Kollontzi, S.; Goedken, M.J.; Fabris, L.; Yurkow, E.J.; Stapleton, P.A. Nanopolystyrene Translocation and Fetal Deposition after Acute Lung Exposure During Late-Stage Pregnancy. Part. Fibre Toxicol. 2020, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Ríos, A.L.; Tejeda-Benítez, L.P.; Bustillo-Lecompte, C.F. Sources, Characteristics, Toxicity, and Control of Ultrafine Particles: An Overview. Geosci. Front. 2022, 13, 101147. [Google Scholar] [CrossRef]
- Deville, S.; Penjweini, R.; Smisdom, N.; Notelaers, K.; Nelissen, I.; Hooyberghs, J.; Ameloot, M. Intracellular Dynamics and Fate of Polystyrene Nanoparticles in A549 Lung Epithelial Cells Monitored by Image (Cross-) Correlation Spectroscopy and Single Particle Tracking. Biochim. Biophys. Acta 2015, 1853. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.L.; Ng, C.T.; Zou, L.; Lu, Y.; Chen, J.; Bay, B.H.; Shen, H.M.; Ong, C.N. Targeted Metabolomics Reveals Differential Biological Effects of Nanoplastics and Nanozno in Human Lung Cells. Nanotoxicology 2019, 13, 1117–1132. [Google Scholar] [CrossRef]
- Chiu, H.W.; Xia, T.; Lee, Y.H.; Chen, C.W.; Tsai, J.C.; Wang, Y.J. Cationic Polystyrene Nanospheres Induce Autophagic Cell Death through the Induction of Endoplasmic Reticulum Stress. Nanoscale 2015, 7, 736–746. [Google Scholar] [CrossRef]
- Xia, T.; Kovochich, M.; Liong, M.; Zink, J.I.; Nel, A.E. Cationic Polystyrene Nanosphere Toxicity Depends on Cell-Specific Endocytic and Mitochondrial Injury Pathways. ACS Nano 2008, 2, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Paget, V.; Dekali, S.; Kortulewski, T.; Grall, R.; Gamez, C.; Blazy, K.; Aguerre-Chariol, O.; Chevillard, S.; Braun, A.; Rat, P.; et al. Specific Uptake and Genotoxicity Induced by Polystyrene Nanobeads with Distinct Surface Chemistry on Human Lung Epithelial Cells and Macrophages. PLoS ONE 2015, 10, e0123297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, R.F.; Dobbs, L.G. Isolation and Culture of Alveolar Epithelial Type I and Type Ii Cells from Rat Lungs. Methods Mol. Biol. 2013, 945, 45–159. [Google Scholar] [CrossRef] [Green Version]
- Garbuzenko, O.M.G.; Taratula, O.; Minko, T. Inhalation Treatment of Lung Cancer: The Influence of Composition, Size and Shape of Nanocarriers on Their Lung Accumulation and Retention. Cancer Biol. Med. 2014, 11, 756–765. [Google Scholar]
- Mohammad, A.K.; Amayreh, L.K.; Mazzara, J.M.; Reineke, J.J. Rapid Lymph Accumulation of Polystyrene Nanoparticles Following Pulmonary Administration. Pharm. Res. 2013, 30, 424–434. [Google Scholar] [CrossRef]
- Alipour, S.; Montaseri, H.; Tafaghodi, M. Preparation and Characterization of Biodegradable Paclitaxel Loaded Alginate Microparticles for Pulmonary Delivery. Colloids Surf. B Biointerfaces 2010, 81, 521–529. [Google Scholar] [CrossRef]
- Kuzmov, A.; Minko, T. Nanotechnology Approaches for Inhalation Treatment of Lung Diseases. J. Control. Release 2015, 219, 500–518. [Google Scholar] [CrossRef] [Green Version]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano Based Drug Delivery Systems: Recent Developments and Future Prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Facciola, A.; Visalli, G.; Pruiti Ciarello, M.; Di Pietro, A. Newly Emerging Airborne Pollutants: Current Knowledge of Health Impact of Micro and Nanoplastics. Int. J. Environ. Res. Public Health 2021, 18, 2997. [Google Scholar] [CrossRef]
- Krug, H.F. Nanosafety Research--Are We on the Right Track? Angew Chem. Int. Ed. Engl. 2014, 53, 12304–12319. [Google Scholar] [CrossRef] [Green Version]
- Schirinzi, G.F.; Perez-Pomeda, I.; Sanchis, J.; Rossini, C.; Farre, M.; Barcelo, D. Cytotoxic Effects of Commonly Used Nanomaterials and Microplastics on Cerebral and Epithelial Human Cells. Environ. Res. 2017, 159, 579–587. [Google Scholar] [CrossRef]
- Hesler, M.; Aengenheister, L.; Ellinger, B.; Drexel, R.; Straskraba, S.; Jost, C.; Wagner, S.; Meier, F.; von Briesen, H.; Buchel, C.; et al. Multi-Endpoint Toxicological Assessment of Polystyrene Nano- and Microparticles in Different Biological Models in Vitro. Toxicol. In Vitro 2019, 61, 104610. [Google Scholar] [CrossRef]
- Domenech, J.; de Britto, M.; Velázquez, A.; Pastor, S.; Hernández, A.; Marcos, R.; Cortés, C. Long-Term Effects of Polystyrene Nanoplastics in Human Intestinal Caco-2 Cells. Biomolecules 2021, 11, 1442. [Google Scholar] [CrossRef]
- Brun, N.R.; Koch, B.E.V.; Varela, M.; Peijnenburg, W.J.G.M.; Spaink, H.P.; Vijver, M.G. Nanoparticles Induce Dermal and Intestinal Innate Immune System Responses in Zebrafish Embryos. Environ. Sci. Nano 2018, 5, 904–916. [Google Scholar] [CrossRef]
- Kang, T.; Park, C.; Lee, B.J. Investigation of Biomimetic Shear Stress on Cellular Uptake and Mechanism of Polystyrene Nanoparticles in Various Cancer Cell Lines. Arch. Pharmacal Res. 2016, 39, 1663–1670. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Meindl, C.; Roblegg, E.; Frohlich, E. Reaction of Monocytes to Polystyrene and Silica Nanoparticles in Short-Term and Long-Term Exposures. Toxicol. Res. (Camb) 2014, 3, 86–97. [Google Scholar] [CrossRef] [Green Version]
- Mrakovcic, M.; Absenger, M.; Riedl, R.; Smole, C.; Roblegg, E.; Frohlich, L.F.; Frohlich, E. Assessment of Long-Term Effects of Nanoparticles in a Microcarrier Cell Culture System. PLoS ONE 2013, 8, e56791. [Google Scholar] [CrossRef] [Green Version]
- Domenech, J.; Marcos, R. Pathways of Human Exposure to Microplastics, and Estimation of the Total Burden. Curr. Opin. Food Sci. 2021, 39, 144–151. [Google Scholar] [CrossRef]
- Vales, G.; Rubio, L.; Marcos, R. Long-Term Exposures to Low Doses of Titanium Dioxide Nanoparticles Induce Cell Transformation, but Not Genotoxic Damage in Beas-2b Cells. Nanotoxicology 2015, 9, 568–578. [Google Scholar] [CrossRef]
- Annangi, B.; Bach, J.; Vales, G.; Rubio, L.; Marcos, R.; Hernandez, A. Long-Term Exposures to Low Doses of Cobalt Nanoparticles Induce Cell Transformation Enhanced by Oxidative Damage. Nanotoxicology 2015, 9, 138–147. [Google Scholar] [CrossRef]
- Schroter, L.; Ventura, N. Nanoplastic Toxicity: Insights and Challenges from Experimental Model Systems. Small 2022, 18, 2201680. [Google Scholar] [CrossRef] [PubMed]
- Heddagaard, F.E.; Moller, P. Hazard Assessment of Small-Size Plastic Particles: Is the Conceptual Framework of Particle Toxicology Useful? Food Chem. Toxicol. 2020, 136, 111106. [Google Scholar] [CrossRef] [PubMed]
- Green, T.R.; Fisher, J.; Stone, M.; Wroblewski, B.M.; Ingham, E. Polyethylene Particles of a ‘Critical Size’ Are Necessary for the Induction of Cytokines by Macrophages in Vitro. Biomaterials 1998, 19, 2297–2302. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, A.S.; Jacobs, J.J.; Black, J.; Galante, J.O.; Glant, T.T. Macrophage/Particle Interactions: Effect of Size, Composition and Surface Area. J. Biomed. Mater. Res. 1994, 28, 81–90. [Google Scholar] [CrossRef]
- Zhang, J.M.; An, J. Cytokines, Inflammation, and Pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Wall, I.B.; Moseley, R.; Baird, D.M.; Kipling, D.; Giles, P.; Laffafian, I.; Price, P.E.; Thomas, D.W.; Stephens, P. Fibroblast Dysfunction Is a Key Factor in the Non-Healing of Chronic Venous Leg Ulcers. J. Investig. Dermatol. 2008, 128, 2526–2540. [Google Scholar] [CrossRef] [Green Version]
- Amsen, D.; Antov, A.; Flavell, R.A. The Different Faces of Notch in T-Helper-Cell Differentiation. Nat. Rev. Immunol. 2009, 9, 116–124. [Google Scholar] [CrossRef]
- Hardy, C.L.; Lemasurier, J.S.; Mohamud, R.; Yao, J.; Xiang, S.D.; Rolland, J.M.; O’Hehir, R.E.; Plebanski, M. Differential Uptake of Nanoparticles and Microparticles by Pulmonary Apc Subsets Induces Discrete Immunological Imprints. J. Immunol. 2013, 191, 5278–5290. [Google Scholar] [CrossRef] [Green Version]
- Claudia, M.; Kristin, O.; Jennifer, O.; Eva, R.; Eleonore, F. Comparison of Fluorescence-Based Methods to Determine Nanoparticle Uptake by Phagocytes and Non-Phagocytic Cells in Vitro. Toxicology 2017, 378, 25–36. [Google Scholar] [CrossRef]
- Poma, A.; Vecchiotti, G.; Colafarina, S.; Zarivi, O.; Aloisi, M.; Arrizza, L.; Chichiricco, G.; Di Carlo, P. In Vitro Genotoxicity of Polystyrene Nanoparticles on the Human Fibroblast Hs27 Cell Line. Nanomaterials 2019, 9, 1299. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Palic, D. Micro- and Nano-Plastics Activation of Oxidative and Inflammatory Adverse Outcome Pathways. Redox Biol. 2020, 37, 101620. [Google Scholar] [CrossRef]
- Liang, B.; Zhong, Y.; Huang, Y.; Lin, X.; Liu, J.; Lin, L.; Hu, M.; Jiang, J.; Dai, M.; Wang, B.; et al. Underestimated Health Risks: Polystyrene Micro- and Nanoplastics Jointly Induce Intestinal Barrier Dysfunction by Ros-Mediated Epithelial Cell Apoptosis. Part. Fibre Toxicol. 2021, 18, 20. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative Stress and Antioxidant Defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Aquilano, K.; Baldelli, S.; Ciriolo, M.R. Glutathione: New Roles in Redox Signaling for an Old Antioxidant. Front. Pharmacol. 2014, 5, 196. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Li, J.; Chen, J.; Miao, X.; Li, G.; He, Q.; Xu, H.; Li, H.; Wei, Y. Cytotoxic Effects of Polystyrene Nanoplastics with Different Surface Functionalization on Human Hepg2 Cells. Sci. Total Environ. 2020, 723, 138180. [Google Scholar] [CrossRef]
- Jeong, C.B.; Kang, H.M.; Lee, M.C.; Kim, D.H.; Han, J.; Hwang, D.S.; Souissi, S.; Lee, S.J.; Shin, K.H.; Park, H.G.; et al. Adverse Effects of Microplastics and Oxidative Stress-Induced Mapk/Nrf2 Pathway-Mediated Defense Mechanisms in the Marine Copepod Paracyclopina Nana. Sci. Rep. 2017, 7, srep41323. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.Y.; Reddy, S.P.; Kleeberger, S.R. Nrf2 Defends the Lung from Oxidative Stress. Antioxid. Redox Signal. 2006, 8, 76–87. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, W. The Nuclear Translocation of Heme Oxygenase-1 in Human Diseases. Front Cell Dev. Biol. 2022, 10, 890186. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, S.; Ge, Y.; Wan, X.; Zhu, Y.; Li, J.; Yin, L.; Pu, Y.; Liang, G. Polystyrene Nanoplastics Induce Lung Injury Via Activating Oxidative Stress: Molecular Insights from Bioinformatics Analysis. Nanomaterials 2022, 12, 3507. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, S.; Liu, Q.; Wei, J.; Jin, Y.; Wang, X.; Zhang, L. Polystyrene Microplastics Cause Cardiac Fibrosis by Activating Wnt/Beta-Catenin Signaling Pathway and Promoting Cardiomyocyte Apoptosis in Rats. Environ. Pollut. 2020, 265, 115025. [Google Scholar] [CrossRef]
- Kawano, Y.; Kypta, R. Secreted Antagonists of the Wnt Signalling Pathway. J. Cell Sci. 2003, 116, 2627–2634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Liu, J.; Jin, R.; Shen, J.; Liang, Y.; Ma, R.; Lin, H.; Liang, X.; Yu, H.; Cai, X. Cytoplasmic and/or Nuclear Expression of Beta-Catenin Correlate with Poor Prognosis and Unfavorable Clinicopathological Factors in Hepatocellular Carcinoma: A Meta-Analysis. PLoS ONE 2014, 9, e111885. [Google Scholar] [CrossRef]
- Gao, Z.H.; Lu, C.; Wang, M.X.; Han, Y.; Guo, L.J. Differential Beta-Catenin Expression Levels Are Associated with Morphological Features and Prognosis of Colorectal Cancer. Oncol. Lett. 2014, 8, 2069–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.Q.; Yang, X.L.; Zhang, G.; Wu, S.P.; Deng, X.B.; Xiao, S.J.; Liu, Q.Z.; Yao, K.T.; Xiao, G.H. Nuclear Beta-Catenin Accumulation Is Associated with Increased Expression of Nanog Protein and Predicts Poor Prognosis of Non-Small Cell Lung Cancer. J. Transl. Med. 2013, 11, 114. [Google Scholar] [CrossRef]

| Single (Gene Expression) | Repeated (Gene Expression) | ||||||
|---|---|---|---|---|---|---|---|
| Target | PCC (r) | Significance | Target | PCC (r) | Significance | ||
| FN1 | −0.96 | * | KEPA1 | −0.80 | ns | ||
| TXN2 | −0.86 | ns | NRF2 | −0.73 | ns | ||
| P53 | −0.82 | ns | VCL | −0.65 | ns | ||
| SOD1 | −0.78 | ns | FN1 | −0.54 | ns | ||
| CAT | −0.77 | ns | ITGA6 | −0.54 | ns | ||
| HMOX1 | −0.76 | ns | LEF1 | −0.54 | ns | ||
| VCL | −0.69 | ns | CAT | −0.51 | ns | ||
| HSP70 | −0.64 | ns | GSR | −0.48 | ns | ||
| NQO1 | −0.63 | ns | HIF1A | −0.47 | ns | ||
| GSK3B | −0.60 | ns | ITGA5 | −0.46 | ns | ||
| GSR | −0.59 | ns | SXRN | −0.43 | ns | ||
| SRXN | −0.54 | ns | CTNNB1 | −0.40 | ns | ||
| VEGF | −0.54 | ns | NQO1 | −0.33 | ns | ||
| CDH1 | −0.53 | ns | PPARD | −0.32 | ns | ||
| KEAP1 | −0.53 | ns | HSP90A | −0.28 | ns | ||
| HIF1A | −0.50 | ns | ITGA2 | −0.19 | ns | ||
| ITGA6 | −0.46 | ns | GPx2 | −0.18 | ns | ||
| ITGAV | −0.41 | ns | HMOX1 | −0.05 | ns | ||
| IL6 | −0.41 | ns | C-JUN | −0.04 | ns | ||
| CTNNB1 | −0.39 | ns | SOD2 | 0.03 | ns | ||
| WNT7A | −0.29 | ns | WNT1 | 0.04 | ns | ||
| C-JUN | −0.28 | ns | P53 | 0.18 | ns | ||
| NRF2 | −0.28 | ns | CDH1 | 0.21 | ns | ||
| GSTA1 | −0.21 | ns | VIM | 0.24 | ns | ||
| P21 | −0.16 | ns | NOS2 (INOS) | 0.25 | ns | ||
| SOD2 | −0.14 | ns | CLD1 | 0.42 | ns | ||
| PPARD | −0.08 | ns | BAX | 0.44 | ns | ||
| HSP90A | −0.03 | ns | WNT7A | 0.45 | ns | ||
| BAX | 0.06 | ns | GSK3B | 0.57 | ns | ||
| ITGA2 | 0.09 | ns | P21 | 0.57 | ns | ||
| VIM | 0.10 | ns | ITGA1 | 0.59 | ns | ||
| TNFα | 0.19 | ns | GSTA1 | 0.59 | ns | ||
| NOS2 (INOS) | 0.33 | ns | SOD1 | 0.68 | ns | ||
| ITGA1 | 0.47 | ns | TXN2 | 0.79 | ns | ||
| WNT1 | 0.49 | ns | HSP70A | 0.99 | ** | ||
| GPX2 | 0.62 | ns | |||||
| IL1β | 0.84 | ns | r > 0.9 or r < −0.9 | strong | |||
| LEF1 | 0.89 | ns | r > 0.7 or r < −0.7 | moderate | |||
| CLD1 | 0.95 | ** | 0.7 > r < −0.7 | weak | |||
| Single (Protein Expression) | Repeated (Protein Expression) | ||||||
| Target | PCC (r) | p-Value | Significance | Target | PCC (r) | p-Value | Significance |
| Nrf2 | −0.98 | 0.02 | * | Cat | −0.93 | 0.05 | * |
| HO-1 | −0.67 | 0.33 | ns | β-catenin | −0.80 | 0.2 | ns |
| Cldn1 | −0.54 | 0.46 | ns | Sod1 | −0.66 | 0.34 | ns |
| Keap1 | −0.51 | 0.49 | ns | Nrf2 | −0.32 | 0.67 | ns |
| Sod1 | −0.22 | 0.78 | ns | β-actin | 0.11 | 0.89 | ns |
| integrin | −0.21 | 0.79 | ns | ||||
| Cat | −0.20 | 0.80 | ns | ||||
| β-catenin | −0.12 | 0.23 | ns | ||||
| Nqo1 | −0.07 | 0.93 | ns | ||||
| Vcl | 0.32 | 0.68 | ns | ||||
| Gpx1 | 0.35 | 0.65 | ns | ||||
| Wnt | 0.59 | 0.41 | ns | ||||
| β-actin | 0.71 | 0.29 | ns | ||||
| Cellular Process | |||||||
| Target | PCC (r) | p-Value | Significance | ||||
| NMP uptake | −0.53 | 0.8 | ns | ||||
| Metabolic activity (s) | −0.34 | 0.66 | ns | ||||
| Metabolic activity (r) | −0.24 | 0.76 | ns | ||||
| ROS | −0.17 | 0.83 | ns | ||||
| Thiol content | −0.39 | 0.61 | ns | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, A.; Mühl, M.; Brito, W.A.d.S.; Singer, D.; Bekeschus, S. Antioxidant Defense in Primary Murine Lung Cells following Short- and Long-Term Exposure to Plastic Particles. Antioxidants 2023, 12, 227. https://doi.org/10.3390/antiox12020227
Schmidt A, Mühl M, Brito WAdS, Singer D, Bekeschus S. Antioxidant Defense in Primary Murine Lung Cells following Short- and Long-Term Exposure to Plastic Particles. Antioxidants. 2023; 12(2):227. https://doi.org/10.3390/antiox12020227
Chicago/Turabian StyleSchmidt, Anke, Melissa Mühl, Walison Augusto da Silva Brito, Debora Singer, and Sander Bekeschus. 2023. "Antioxidant Defense in Primary Murine Lung Cells following Short- and Long-Term Exposure to Plastic Particles" Antioxidants 12, no. 2: 227. https://doi.org/10.3390/antiox12020227
APA StyleSchmidt, A., Mühl, M., Brito, W. A. d. S., Singer, D., & Bekeschus, S. (2023). Antioxidant Defense in Primary Murine Lung Cells following Short- and Long-Term Exposure to Plastic Particles. Antioxidants, 12(2), 227. https://doi.org/10.3390/antiox12020227