Methionine Sulfoxide Reductases Suppress the Formation of the [PSI+] Prion and Protein Aggregation in Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Plasmids
2.2. Growth and Stress Conditions
2.3. Protein and Western Blot Analysis
2.4. Protein Identification and Statistical Analysis
2.5. Fluorescence Microscopy
2.6. Analysis of Prion Formation
3. Results
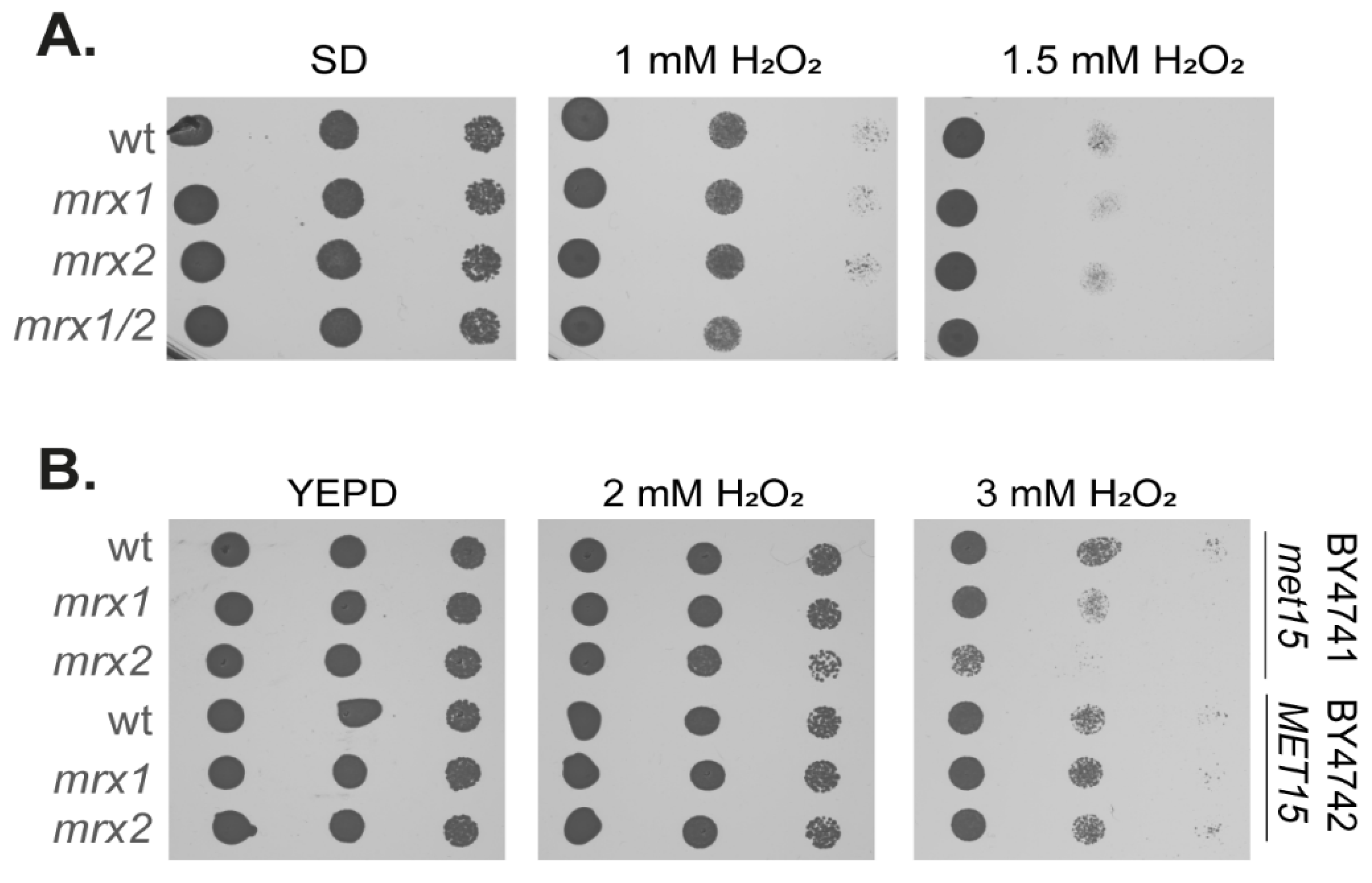
3.1. Strains Lacking Methionine Sulfoxide Reductases Are Unaffected in Hydrogen Peroxide Sensitivity
3.2. The Frequency of [PSI+] Prion Formation Is Increased in MXR Mutants
3.3. Mutation of Met124 in Sup35 Decreases the Frequency of [PSI+] Prion Formation
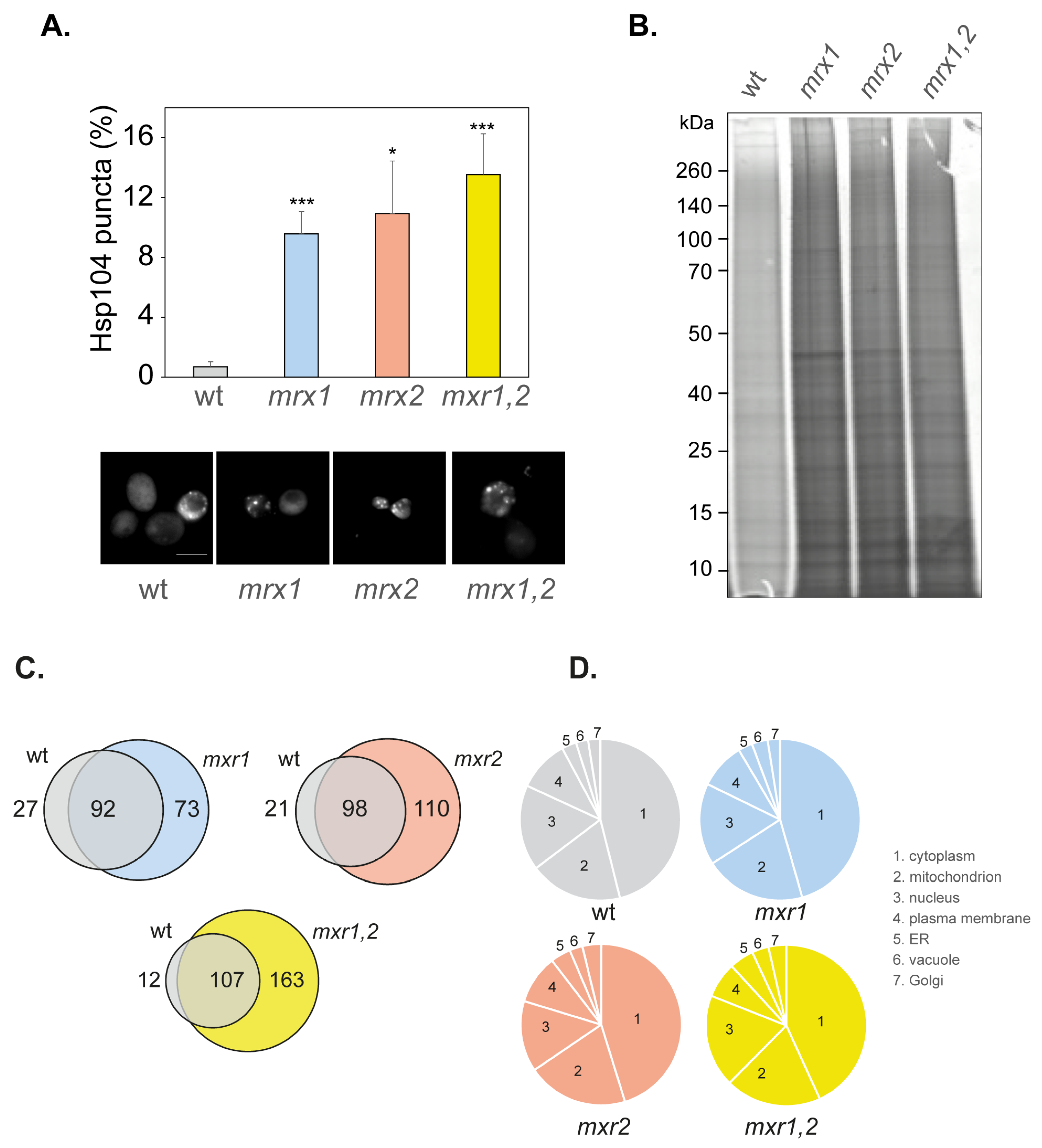
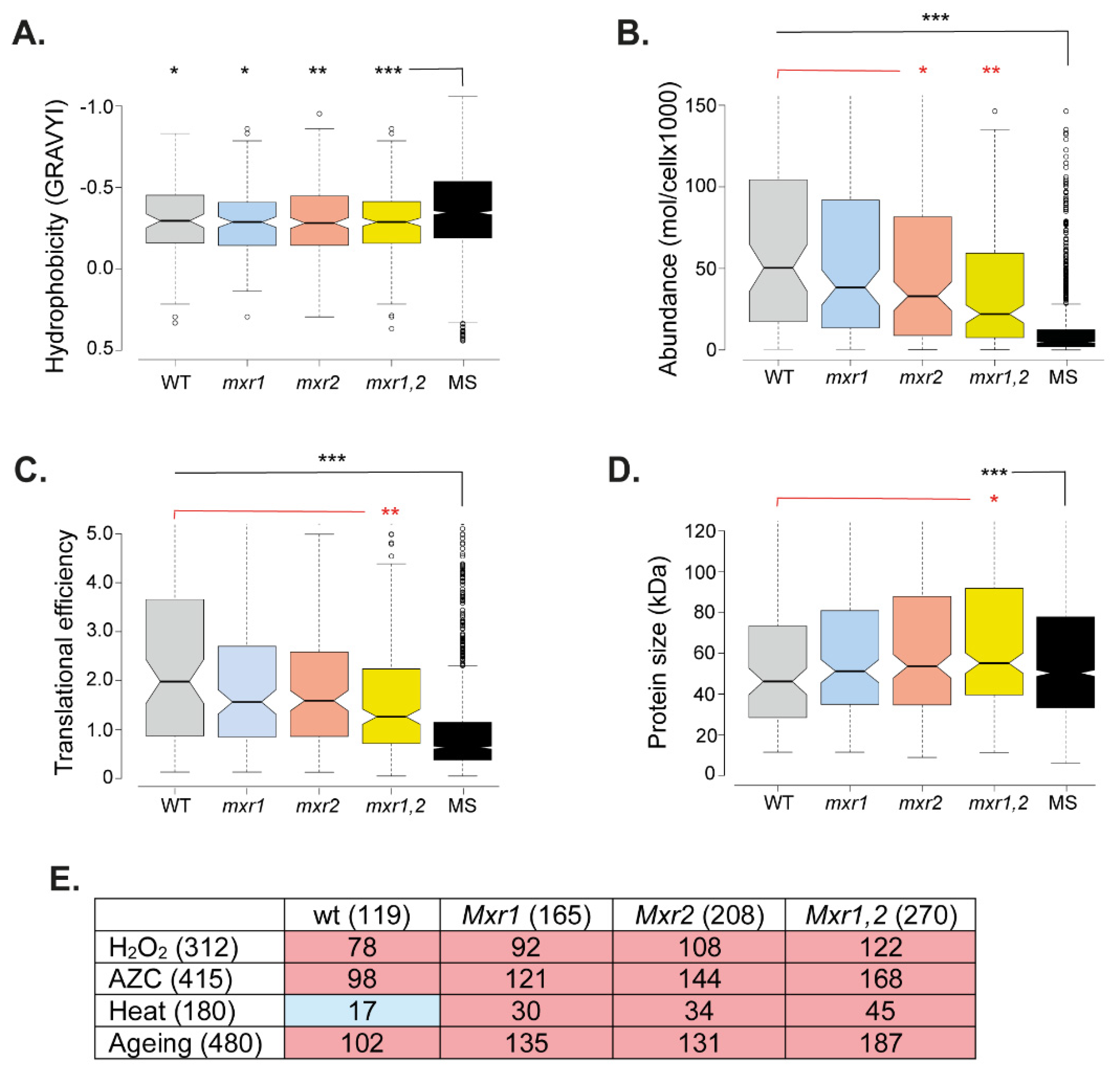
3.4. Amorphous Protein Aggregation Is Increased in MXR Mutant Strains
3.5. Sup35 Aggregation Is Increased in MXR Mutants following Sup35 Overexpression
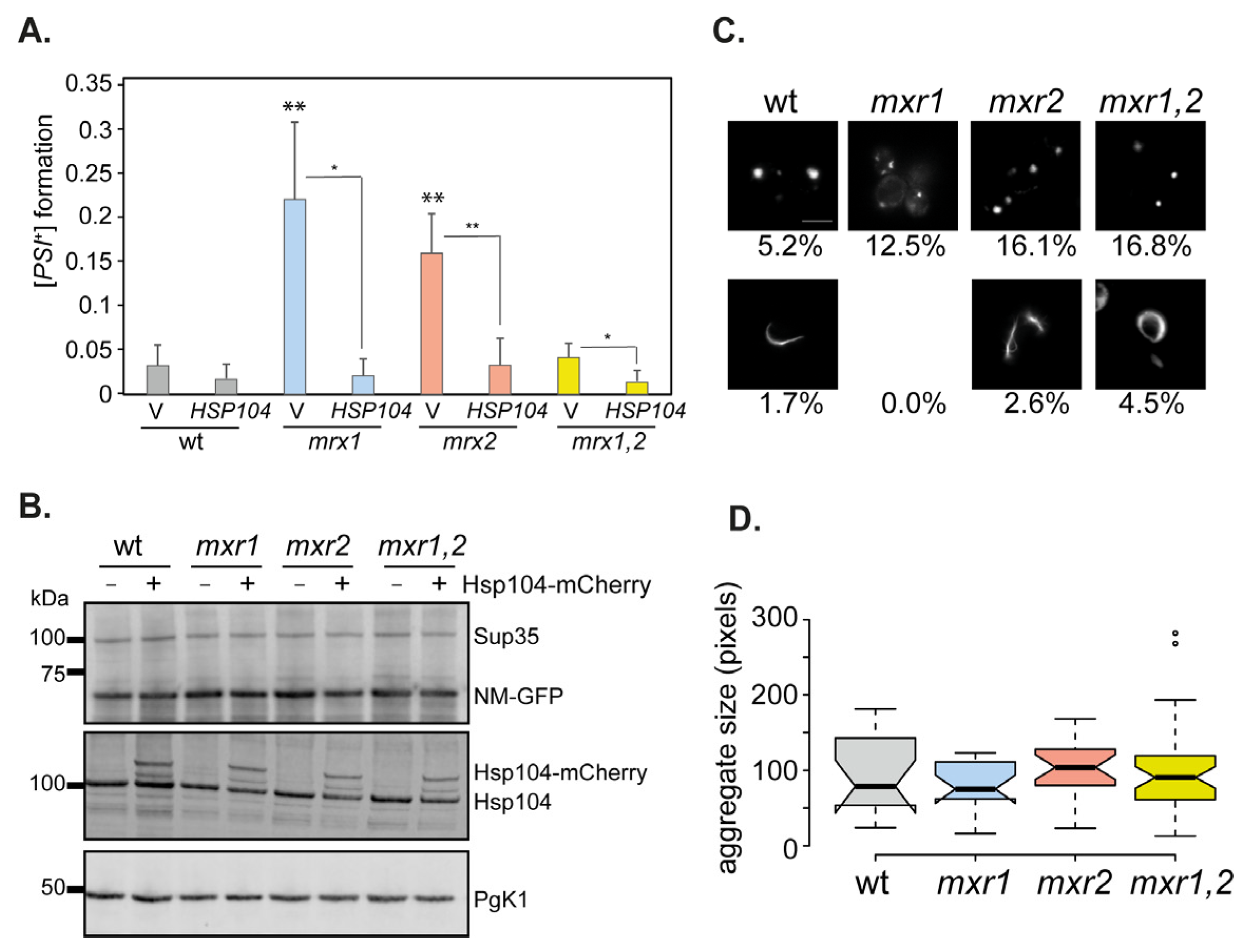
3.6. Overexpression-Induced [PSI+] Prion Formation Is Increased in Single but Not Double MXR Mutant Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stadtman, E.R.; Levine, R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Moskovitz, J.; Levine, R.L. Oxidation of Methionine Residues of Proteins: Biological Consequences. Antioxid. Redox Signal 2003, 5, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Weissbach, H.; Resnick, L.; Brot, N. Methionine sulfoxide reductases: History and cellular role in protecting against oxidative damage. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2005, 1703, 203–212. [Google Scholar] [CrossRef]
- Lim, J.M.; Kim, G.; Levine, R.L. Methionine in Proteins: It’s Not Just for Protein Initiation Anymore. Neurochem. Res. 2019, 44, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Weiss, S.J.; Levine, R.L. Methionine oxidation and reduction in proteins. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Moskovitz, J.; Stadtman, E.R. Oxidation of Methionine in Proteins: Roles in Antioxidant Defense and Cellular Regulation. IUBMB Life 2000, 50, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Smith, A. Methionine sulfoxide and the methionine sulfoxide reductase system as modulators of signal transduction pathways: A review. Amino Acids 2021, 53, 1011–1020. [Google Scholar] [CrossRef]
- Drazic, A.; Winter, J. The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2014, 1844, 1367–1382. [Google Scholar] [CrossRef]
- Kaya, A.; Lee, B.C.; Gladyshev, V.N. Regulation of Protein Function by Reversible Methionine Oxidation and the Role of Selenoprotein MsrB1. Antioxid. Redox Signal 2015, 23, 814–822. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Gladyshev, V.N. Methionine sulfoxide reductases: Selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem. J. 2007, 407, 321–329. [Google Scholar] [CrossRef]
- Minniti, A.N.; Arrazola, M.S.; Bravo-Zehnder, M.; Ramos, F.; Inestrosa, N.C.; Aldunate, R. The Protein Oxidation Repair Enzyme Methionine Sulfoxide Reductase A Modulates Aβ Aggregation and Toxicity In Vivo. Antioxid. Redox Signal 2015, 22, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hindupur, J.; Nguyen, J.L.; Ruf, K.J.; Zhu, J.; Schieler, J.L.; Bonham, C.C.; Wood, K.V.; Davisson, V.J.; Rochet, J.-C. Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson’s disease-related insults. Free Radic. Biol. Med. 2008, 45, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Bar-Noy, S.; Williams, W.M.; Requena, J.; Berlett, B.S.; Stadtman, E.R. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 12920–12925. [Google Scholar] [CrossRef]
- Moskovitz, J.; Du, F.; Bowman, C.F.; Yan, S.S. Methionine sulfoxide reductase A affects β-amyloid solubility and mitochondrial function in a mouse model of Alzheimer’s disease. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E388–E393. [Google Scholar] [CrossRef]
- Prusiner, S.B. Biology and Genetics of Prions Causing Neurodegeneration. Annu. Rev. Genet. 2013, 47, 601–623. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef]
- Colby, D.W.; Prusiner, S.B. De novo generation of prion strains. Nat. Rev. Genet. 2011, 9, 771–777. [Google Scholar] [CrossRef]
- Wolschner, C.; Giese, A.; Kretzschmar, H.A.; Huber, R.; Moroder, L.; Budisa, N. Design of anti- and pro-aggregation variants to assess the effects of methionine oxidation in human prion protein. Proc. Natl. Acad. Sci. USA 2009, 106, 7756–7761. [Google Scholar] [CrossRef]
- Grant, C.M. Sup35 methionine oxidation is a trigger forde novo[PSI+] prion formation. Prion 2015, 9, 257–265. [Google Scholar] [CrossRef]
- Younan, N.D.; Nadal, R.C.; Davies, P.; Brown, D.R.; Viles, J.H. Methionine Oxidation Perturbs the Structural Core of the Prion Protein and Suggests a Generic Misfolding Pathway. J. Biol. Chem. 2012, 287, 28263–28275. [Google Scholar] [CrossRef]
- Requena, J.R.; Dimitrova, M.N.; Legname, G.; Teijeira, S.; Prusiner, S.B.; Levine, R.L. Oxidation of methionine residues in the prion protein by hydrogen peroxide. Arch. Biochem. Biophys. 2004, 432, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. [URE3] as an Altered URE2 Protein: Evidence for a Prion Analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381. [Google Scholar] [CrossRef]
- Tyedmers, J.; Madariaga, M.L.; Lindquist, S. Prion Switching in Response to Environmental Stress. PLoS Biol. 2008, 6, e294. [Google Scholar] [CrossRef] [PubMed]
- Sideri, T.C.; Stojanovski, K.; Tuite, M.F.; Grant, C.M. Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 6394–6399. [Google Scholar] [CrossRef]
- Sideri, T.C.; Koloteva-Levine, N.; Tuite, M.F.; Grant, C.M. Methionine Oxidation of Sup35 Protein Induces Formation of the [PSI+] Prion in a Yeast Peroxiredoxin Mutant. J. Biol. Chem. 2011, 286, 38924–38931. [Google Scholar] [CrossRef] [PubMed]
- Doronina, V.A.; Staniforth, G.L.; Speldewinde, S.H.; Tuite, M.F.; Grant, C.M. Oxidative stress conditions increase the frequency of de novo formation of the yeast [PSI+] prion. Mol. Microbiol. 2015, 96, 163–174. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef]
- Lee, R.E.C.; Brunette, S.; Puente, L.G.; Megeney, L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA 2010, 107, 13348–13353. [Google Scholar] [CrossRef] [PubMed]
- Osherovich, L.Z.; Cox, B.S.; Tuite, M.F.; Weissman, J.S. Dissection and Design of Yeast Prions. PLoS Biol. 2004, 2, e86. [Google Scholar] [CrossRef]
- Parham, S.N.; Resende, C.G.; Tuite, M.F. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 2001, 20, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Ness, F.; Ferreira, P.; Cox, B.S.; Tuite, M.F. Guanidine Hydrochloride Inhibits the Generation of Prion “Seeds” but Not Prion Protein Aggregation in Yeast. Mol. Cell. Biol. 2002, 22, 5593–5605. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, N.; Kritsiligkou, P.; Grant, C.M. ER stress causes widespread protein aggregation and prion formation. J. Cell Biol. 2017, 216, 2295–2304. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; Lindquist, S. Biochemical, Cell Biological, and Genetic Assays to Analyze Amyloid and Prion Aggregation in Yeast. Methods Enzymol. 2010, 470, 709–734. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Speldewinde, S.H.; Doronina, V.A.; Tuite, M.F.; Grant, C.M. Disrupting the cortical actin cytoskeleton points to two distinct mechanisms of yeast [PSI+] prion formation. PLoS Genet. 2017, 13, e1006708. [Google Scholar] [CrossRef]
- Ferreira, P.C.; Ness, F.; Edwards, S.R.; Cox, B.S.; Tuite, M.F. The elimination of the yeast [PSI+] prion by guanidine hydro-chloride is the result of Hsp104 inactivation. Mol Microbiol 2001, 40, 1357–1369. [Google Scholar] [CrossRef]
- Jung, G.; Masison, D.C. Guanidine Hydrochloride Inhibits Hsp104 Activity In Vivo: A Possible Explanation for Its Effect in Curing Yeast Prions. Curr. Microbiol. 2001, 43, 7–10. [Google Scholar] [CrossRef] [PubMed]
- LE, D.T.; Lee, B.C.; Marino, S.M.; Zhang, Y.; Fomenko, D.E.; Kaya, A.; Hacioglu, E.; Kwak, G.-H.; Koc, A.; Kim, H.-Y.; et al. Functional Analysis of Free Methionine-R-sulfoxide Reductase from Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 4354–4364. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, A.K.; Bardill, J.P.; True, H.L.; Masel, J. The Spontaneous Appearance Rate of the Yeast Prion [PSI+] and Its Implications for the Evolution of the Evolvability Properties of the [PSI+] System. Genetics 2010, 184, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Lund, P.M.; Cox, B.S. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet. Res. 1981, 37, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Speldewinde, S.H.; Doronina, V.A.; Grant, C.M. Autophagy protects against de novo formation of the [PSI+] prion in yeast. Mol. Biol. Cell 2015, 26, 4541–4551. [Google Scholar] [CrossRef] [PubMed]
- Tessier, P.M.; Lindquist, S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+]. Nat. Struct. Mol. Biol. 2009, 16, 598–605. [Google Scholar] [CrossRef]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A Novel Chaperone System that Rescues Previously Aggregated Proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nyström, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef]
- Tomoyasu, T.; Mogk, A.; Langen, H.; Goloubinoff, P.; Bukau, B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 2001, 40, 397–413. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two Enzymes in One: Two Yeast Peroxiredoxins Display Oxidative Stress-Dependent Switching from a Peroxidase to a Molecular Chaperone Function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Rand, J.D.; Grant, C.M. The Thioredoxin System Protects Ribosomes against Stress-induced Aggregation. Mol. Biol. Cell 2006, 17, 387–401. [Google Scholar] [CrossRef]
- Koplin, A.; Preissler, S.; Ilina, Y.; Koch, M.; Scior, A.; Erhardt, M.; Deuerling, E. A dual function for chaperones SSB–RAC and the NAC nascent polypeptide–associated complex on ribosomes. J. Cell Biol. 2010, 189, 57–68. [Google Scholar] [CrossRef]
- Kaya, A.; Koc, A.; Lee, B.C.; Fomenko, D.E.; Rederstorff, M.; Krol, A.; Lescure, A.; Gladyshev, V.N. Compartmentalization and Regulation of Mitochondrial Function by Methionine Sulfoxide Reductases in Yeast. Biochemistry 2010, 49, 8618–8625. [Google Scholar] [CrossRef] [PubMed]
- Nicklow, E.E.; Sevier, C.S. Activity of the yeast cytoplasmic Hsp70 nucleotide-exchange factor Fes1 is regulated by reversible methionine oxidation. J. Biol. Chem. 2020, 295, 552–569. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Schymkowitz, J.; Rousseau, F.; Diella, F.; Serrano, L. A Comparative Study of the Relationship Between Protein Structure and β-Aggregation in Globular and Intrinsically Disordered Proteins. J. Mol. Biol. 2004, 342, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Vabulas, R.M.; Raychaudhuri, S.; Hayer-Hartl, M.; Hartl, F.U. Protein Folding in the Cytoplasm and the Heat Shock Response. Cold Spring Harb. Perspect. Biol. 2010, 2, a004390. [Google Scholar] [CrossRef]
- Tamás, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy Metals and Metalloids as a Cause for Protein Misfolding and Aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Weids, A.J.; Ibstedt, S.; Tamás, M.J.; Grant, C.M. Distinct stress conditions result in aggregation of proteins with similar properties. Sci. Rep. 2016, 6, 24554. [Google Scholar] [CrossRef] [PubMed]
- Ibstedt, S.; Sideri, T.C.; Grant, C.M.; Tamás, M.J. Global analysis of protein aggregation in yeast during physiological conditions and arsenite stress. Biol. Open 2014, 3, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.W.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef]
- Peters, T.W.; Rardin, M.J.; Czerwieniec, G.; Evani, U.S.; Reis-Rodrigues, P.; Lithgow, G.J.; Mooney, S.D.; Gibson, B.W.; Hughes, R.E. Tor1 regulates protein solubility in Saccharomyces cerevisiae. Mol. Biol. Cell 2012, 23, 4679–4688. [Google Scholar] [CrossRef] [PubMed]
- Ganusova, E.E.; Ozolins, L.N.; Bhagat, S.; Newnam, G.P.; Wegrzyn, R.D.; Sherman, M.Y.; Chernoff, Y.O. Modulation of Prion Formation, Aggregation, and Toxicity by the Actin Cytoskeleton in Yeast. Mol. Cell. Biol. 2006, 26, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Derkatch, I.L.; Liebman, S.W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+]. Mol. Microbiol. 2001, 39, 37–46. [Google Scholar] [CrossRef]
- Mathur, V.; Taneja, V.; Sun, Y.; Liebman, S.W. Analyzing the Birth and Propagation of Two Distinct Prions, [PSI+] and [Het-s]y, in Yeast. Mol. Biol. Cell 2010, 21, 1449–1461. [Google Scholar] [CrossRef]
- Arslan, F.; Hong, J.Y.; Kanneganti, V.; Park, S.-K.; Liebman, S.W. Heterologous Aggregates Promote De Novo Prion Appearance via More than One Mechanism. PLoS Genet. 2015, 11, e1004814. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast [PSI+] Prion Aggregates Are Formed by Small Sup35 Polymers Fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef]
- Satpute-Krishnan, P.; Langseth, S.X.; Serio, T.R. Hsp104-Dependent Remodeling of Prion Complexes Mediates Protein-Only Inheritance. PLoS Biol. 2007, 5, e24. [Google Scholar] [CrossRef]
- Hung, G.-C.; Masison, D.C. N-Terminal Domain of Yeast Hsp104 Chaperone Is Dispensable for Thermotolerance and Prion Propagation but Necessary for Curing Prions by Hsp104 Overexpression. Genetics 2006, 173, 611–620. [Google Scholar] [CrossRef]
- Labbadia, J.; Morimoto, R.I. The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef]
- Hipp, M.S.; Park, S.-H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Nyström, T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005, 24, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Mulinacci, F.; Capelle, M.A.; Gurny, R.; Drake, A.F.; Arvinte, T. Stability of Human Growth Hormone: Influence of Methionine Oxidation on Thermal Folding. J. Pharm. Sci. 2011, 100, 451–463. [Google Scholar] [CrossRef]
- Smith, C.D.; Carney, J.M.; Starke-Reed, P.E.; Oliver, C.N.; Stadtman, E.R.; Floyd, R.A.; Markesbery, W.R. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1991, 88, 10540–10543. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Gagliano, N.; Di Simplicio, P.; Colombo, R.; Milzani, A.D.G. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free Radic. Biol. Med. 2002, 32, 927–937. [Google Scholar] [CrossRef]
- Xu, K.; Uversky, V.N.; Xue, B. Local flexibility facilitates oxidization of buried methionine residues. Protein Pept. Lett. 2012, 19, 688–697. [Google Scholar] [CrossRef]
- Walker, E.J.; Bettinger, J.Q.; Welle, K.A.; Hryhorenko, J.R.; Vargas, A.M.M.; O’Connell, M.R.; Ghaemmaghami, S. Protein folding stabilities are a major determinant of oxidation rates for buried methionine residues. J. Biol. Chem. 2022, 298, 101872. [Google Scholar] [CrossRef]
- Veredas, F.J.; Cantón, F.R.; Aledo, J.C. Methionine residues around phosphorylation sites are preferentially oxidized in vivo under stress conditions. Sci. Rep. 2017, 7, 40403. [Google Scholar] [CrossRef]
- Aledo, J.C.; Cantón, F.R.; Veredas, F.J. A machine learning approach for predicting methionine oxidation sites. BMC Bioinform. 2017, 18, 430. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, S.L.; Petropoulos, I.; Friguet, B. The Oxidized Protein Repair Enzymes Methionine Sulfoxide Reductases and Their Roles in Protecting against Oxidative Stress, in Ageing and in Regulating Protein Function. Antioxidants 2018, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J. Methionine sulfoxide reductases: Ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2005, 1703, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Wu, S.; Niznikiewicz, M. Innate immunity to prions: Anti-prion systems turn a tsunami of prions into a slow drip. Curr. Genet. 2021, 67, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. Yeast and Fungal Prions. Cold Spring Harb. Perspect. Biol. 2016, 8, a023531. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schepers, J.; Carter, Z.; Kritsiligkou, P.; Grant, C.M. Methionine Sulfoxide Reductases Suppress the Formation of the [PSI+] Prion and Protein Aggregation in Yeast. Antioxidants 2023, 12, 401. https://doi.org/10.3390/antiox12020401
Schepers J, Carter Z, Kritsiligkou P, Grant CM. Methionine Sulfoxide Reductases Suppress the Formation of the [PSI+] Prion and Protein Aggregation in Yeast. Antioxidants. 2023; 12(2):401. https://doi.org/10.3390/antiox12020401
Chicago/Turabian StyleSchepers, Jana, Zorana Carter, Paraskevi Kritsiligkou, and Chris M. Grant. 2023. "Methionine Sulfoxide Reductases Suppress the Formation of the [PSI+] Prion and Protein Aggregation in Yeast" Antioxidants 12, no. 2: 401. https://doi.org/10.3390/antiox12020401
APA StyleSchepers, J., Carter, Z., Kritsiligkou, P., & Grant, C. M. (2023). Methionine Sulfoxide Reductases Suppress the Formation of the [PSI+] Prion and Protein Aggregation in Yeast. Antioxidants, 12(2), 401. https://doi.org/10.3390/antiox12020401





