The Transcriptional Repressor PerR Senses Sulfane Sulfur by Cysteine Persulfidation at the Structural Zn2+ Site in Synechococcus sp. PCC7002
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Construction of PCC7002 Mutants
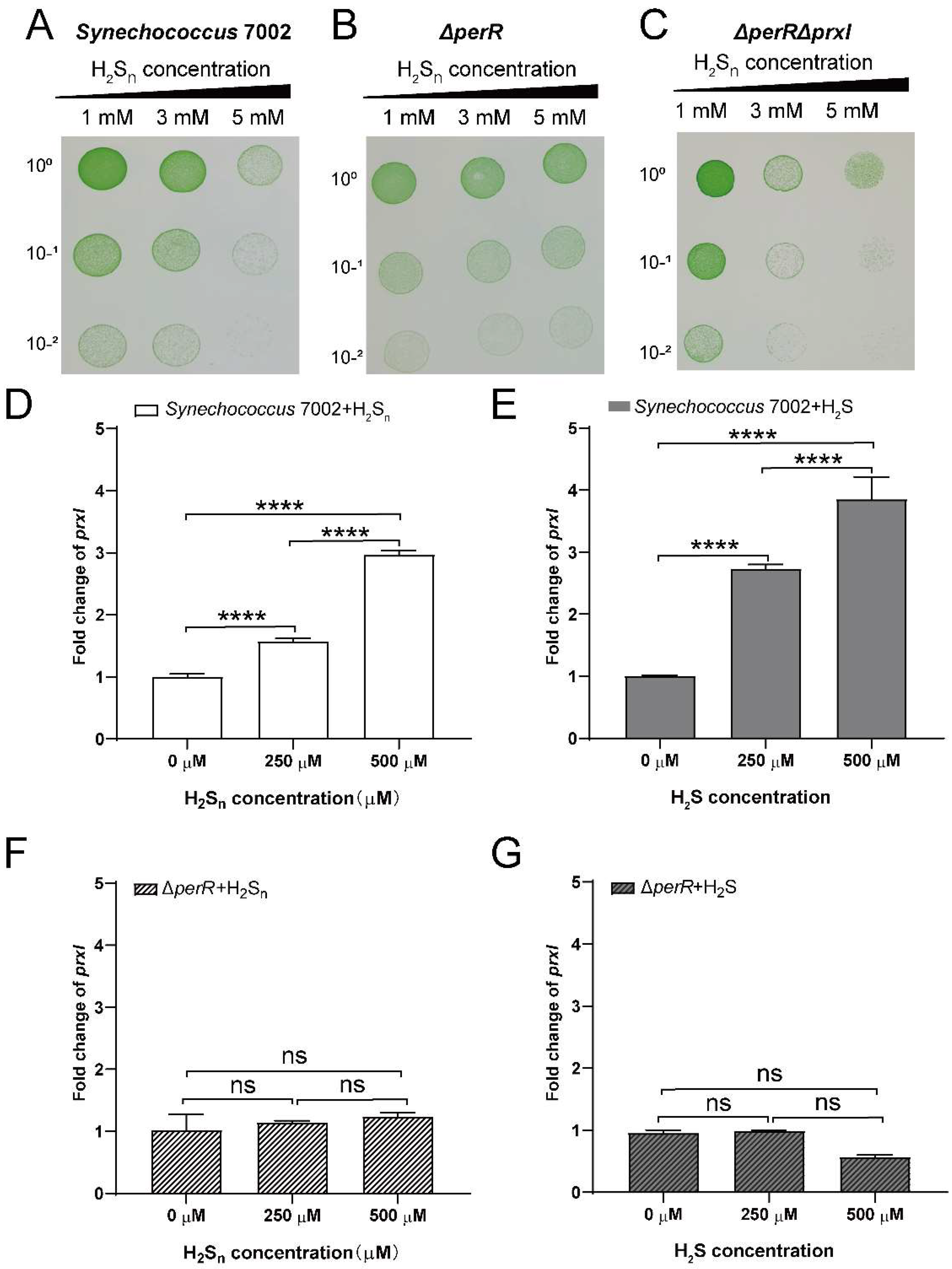
2.3. The Toxicity of H2Sn against PCC7002, PCC7002ΔperR, and PCC7002ΔprxIΔperR
2.4. Induction, RNA Extraction, and qRT-PCR Analysis
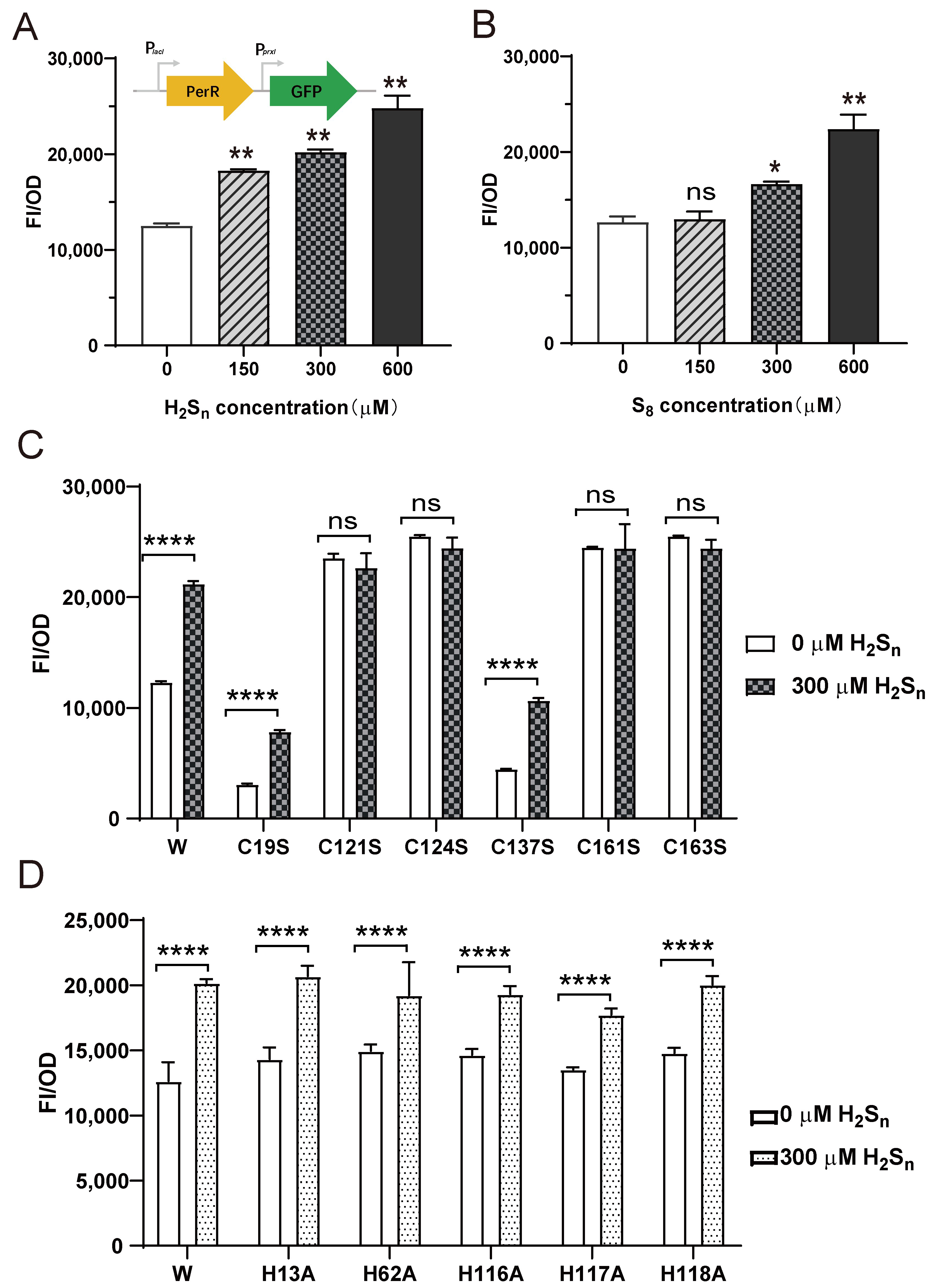
2.5. Construction of the perR-Repressed Reporter System
2.6. Construction, Overexpression, and Purification of PerR
2.7. Zn2+ Release Assay
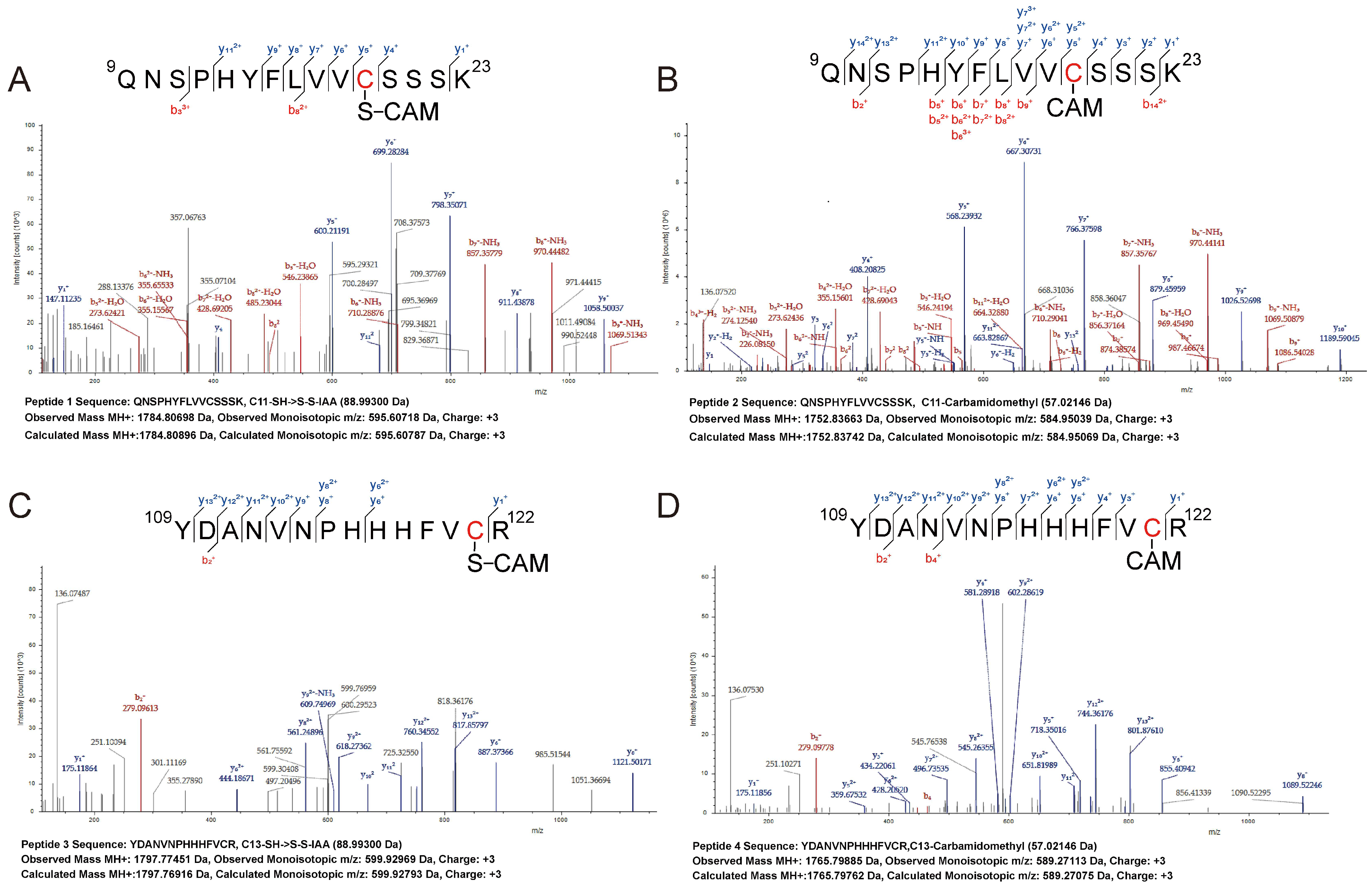
2.8. LC-MS/MS Analysis of PerR
2.9. Phylogenetic Analysis
3. Results
3.1. Phylogenetic Analysis of PerR in Cyanobacteria
3.2. PerR Deletion Increases the Tolerance of PCC7002 to High H2Sn
3.3. PerR Senses H2Sn and Regulates the Expression of prxI
3.4. Sulfane Sulfur Acts on the Cys4:Zn2+ Site of PerR
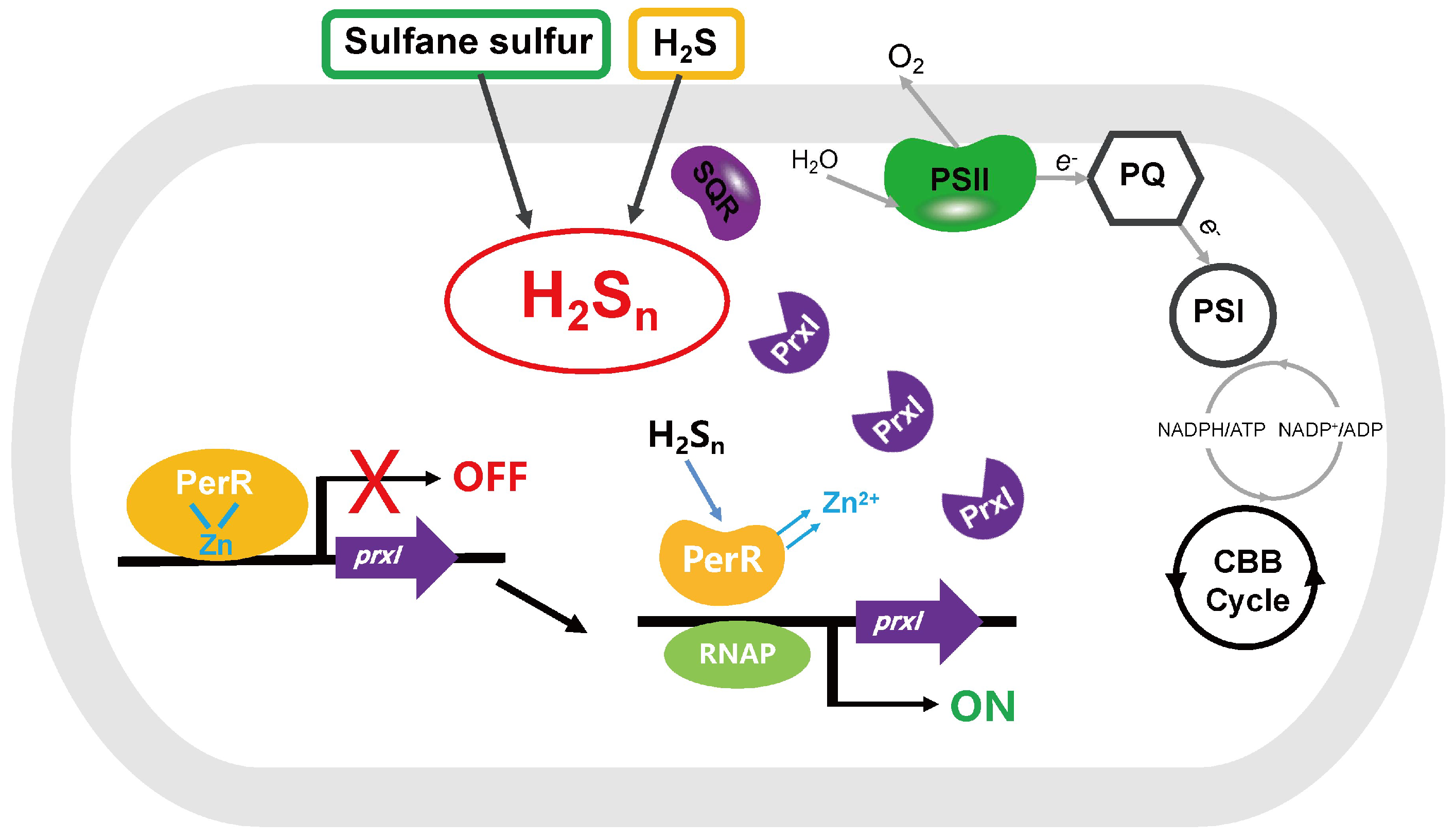
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schopf, J.W. Geological evidence of oxygenic photosynthesis and the biotic response to the 2400–2200 ma “great oxidation event”. Biochem. Biokhimiia 2014, 79, 165–177. [Google Scholar] [CrossRef]
- Soo, R.M.; Hemp, J.; Parks, D.H.; Fischer, W.W.; Hugenholtz, P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 2017, 355, 1436–1440. [Google Scholar] [CrossRef]
- Olson, K.R. Reactive oxygen species or reactive sulfur species: Why we should consider the latter. J. Exp. Biol. 2020, 223, 196352. [Google Scholar] [CrossRef]
- Johnston, D.T.; Wolfe-Simon, F.; Pearson, A.; Knoll, A.H. Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth’s middle age. Proc. Natl. Acad. Sci. USA 2009, 106, 16925–16929. [Google Scholar] [CrossRef]
- Buick, R. When did oxygenic photosynthesis evolve? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2008, 363, 2731–2743. [Google Scholar] [CrossRef]
- Oschlies, A. A committed fourfold increase in ocean oxygen loss. Nat. Commun. 2021, 12, 2307. [Google Scholar] [CrossRef]
- Haas, S.; de Beer, D.; Klatt, J.M.; Fink, A.; Rench, R.M.; Hamilton, T.L.; Meyer, V.; Kakuk, B.; Macalady, J.L. Low-light anoxygenic photosynthesis and Fe-S-Biogeochemistry in a microbial mat. Front. Microbiol. 2018, 9, 858. [Google Scholar] [CrossRef]
- Klatt, J.M.; Al-Najjar, M.A.; Yilmaz, P.; Lavik, G.; de Beer, D.; Polerecky, L. Anoxygenic photosynthesis controls oxygenic photosynthesis in a cyanobacterium from a sulfidic spring. Appl. Environ. Microbiol. 2015, 81, 2025–2031. [Google Scholar] [CrossRef]
- Li, H.; Singh, A.K.; McIntyre, L.M.; Sherman, L.A. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 2004, 186, 3331–3345. [Google Scholar] [CrossRef]
- Kanesaki, Y.; Yamamoto, H.; Paithoonrangsarid, K.; Shoumskaya, M.; Suzuki, I.; Hayashi, H.; Murata, N. Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. Cell Mol. Biol. 2007, 49, 313–324. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ishizuka, T.; Katayama, M.; Kanehisa, M.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B.; Ikeuchi, M. Response to Oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2004, 45, 290–299. [Google Scholar] [CrossRef]
- Latifi, A.; Ruiz, M.; Zhang, C.C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 2009, 33, 258–278. [Google Scholar] [CrossRef]
- Xia, Y.; Lü, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017, 11, 2754–2766. [Google Scholar] [CrossRef]
- Li, K.; Xin, Y.; Xuan, G.; Zhao, R.; Liu, H.; Xia, Y.; Xun, L. Escherichia coli uses separate enzymes to produce H2S and Reactive Sulfane Sulfur from L-cysteine. Front. Microbiol. 2019, 10, 298. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, H.; Cui, F.; Liu, H.; Xun, L. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ. Microbiol. 2016, 18, 5123–5136. [Google Scholar] [CrossRef]
- Wang, R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol. Rev. 2012, 92, 791–896. [Google Scholar] [CrossRef]
- Xuan, G.; Lü, C.; Xu, H.; Chen, Z.; Li, K.; Liu, H.; Liu, H.; Xia, Y.; Xun, L. Sulfane Sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa. Mol. Microbiol. 2020, 114, 1038–1048. [Google Scholar] [CrossRef]
- Xuan, G.; Lv, C.; Xu, H.; Li, K.; Liu, H.; Xia, Y.; Xun, L. Sulfane sulfur regulates LasR-mediated quorum sensing and virulence in Pseudomonas aeruginosa PAO1. Antioxidants 2021, 10, 1498. [Google Scholar] [CrossRef]
- Xun, H.; Xuan, G.; Liu, H.; Xia, Y.; Xun, L. Sulfane sulfur is a strong inducer of the multiple antibiotic resistance regulator MarR in Escherichia coli. Antioxidants 2021, 10, 1778. [Google Scholar] [CrossRef]
- Iciek, M.; Kowalczyk-Pachel, D.; Bilska-Wilkosz, A.; Kwiecien, I.; Gorny, M.; Wlodek, L. S-sulfhydration as a cellular redox regulation. Biosci. Rep. 2015, 36, e00304. [Google Scholar] [CrossRef]
- Yan, F.; Fojtikova, V.; Man, P.; Stranava, M.; Martínková, M.; Du, Y.; Huang, D.; Shimizu, T. Catalytic enhancement of the heme-based oxygen-sensing phosphodiesterase EcDOS by hydrogen sulfide is caused by changes in heme coordination structure. Biomet. Int. J. Role Met. Ions Biol. Biochem. Med. 2015, 28, 637–652. [Google Scholar] [CrossRef]
- Zivanovic, J.; Kouroussis, E.; Kohl, J.B.; Adhikari, B.; Bursac, B.; Schott-Roux, S.; Petrovic, D.; Miljkovic, J.L.; Thomas-Lopez, D.; Jung, Y.; et al. Selective persulfide detection reveals evolutionarily conserved antiaging effects of s-sulfhydration. Cell Metab. 2019, 30, 1152–1170.e13. [Google Scholar] [CrossRef]
- Bhatwalkar, S.B.; Mondal, R.; Krishna, S.B.N.; Adam, J.K.; Govender, P.; Anupam, R. Antibacterial properties of organosulfur compounds of Garlic (Allium sativum). Front. Microbiol. 2021, 12, 613077. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, J.; Lü, C.; Xia, Y.; Liu, H.; Jiao, N.; Xun, L.; Liu, J. Synechococcus sp. strain PCC7002 uses Sulfide:Quinone Oxidoreductase to detoxify exogenous sulfide and to convert endogenous sulfide to cellular sulfane sulfur. mBio 2020, 11, e03420–e03519. [Google Scholar] [CrossRef]
- Liu, H.; Xin, Y.; Xun, L. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl. Environ. Microbiol. 2014, 80, 1799–1806. [Google Scholar] [CrossRef]
- Lü, C.; Xia, Y.; Liu, D.; Zhao, R.; Gao, R.; Liu, H.; Xun, L. Cupriavidus necator H16 uses flavocytochrome c sulfide dehydrogenase to oxidize self-produced and added sulfide. Appl. Environ. Microbiol. 2017, 83, e01610–e01617. [Google Scholar] [CrossRef]
- Harada, M.; Akiyama, A.; Furukawa, R.; Yokobori, S.I.; Tajika, E.; Yamagishi, A. Evolution of superoxide dismutases and catalases in cyanobacteria: Occurrence of the antioxidant enzyme genes before the rise of atmospheric oxygen. J. Mol. Evol. 2021, 89, 527–543. [Google Scholar] [CrossRef]
- Olson, K.R.; Gao, Y.; DeLeon, E.R.; Arif, M.; Arif, F.; Arora, N.; Straub, K.D. Catalase as a sulfide-sulfur oxido-reductase: An ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol. 2017, 12, 325–339. [Google Scholar] [CrossRef]
- Olson, K.R.; Gao, Y.; Arif, F.; Arora, K.; Patel, S.; DeLeon, E.R.; Sutton, T.R.; Feelisch, M.; Cortese-Krott, M.M.; Straub, K.D. Metabolism of hydrogen sulfide (H2S) and Production of Reactive Sulfur Species (RSS) by superoxide dismutase. Redox Biol. 2018, 15, 74–85. [Google Scholar] [CrossRef]
- Hou, N.; Yan, Z.; Fan, K.; Li, H.; Zhao, R.; Xia, Y.; Xun, L.; Liu, H. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol. 2019, 26, 101293. [Google Scholar] [CrossRef]
- Liu, D.; Chen, J.; Wang, Y.; Meng, Y.; Li, Y.; Huang, R.; Xia, Y.; Liu, H.; Jiao, N.; Xun, L.; et al. Synechococcus sp. PCC7002 uses peroxiredoxin to cope with reactive sulfur species stress. mBio 2022, 13, e0103922. [Google Scholar] [CrossRef] [PubMed]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Luebke, J.L.; Shen, J.; Bruce, K.E.; Kehl-Fie, T.E.; Peng, H.; Skaar, E.P.; Giedroc, D.P. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 2014, 94, 1343–1360. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, B.G.; Barbosa, R.L.; Soprano, A.S.; Campos, B.M.; de Souza, T.A.; Tonoli, C.C.C.; Leme, A.F.P.; Murakami, M.T.; Benedetti, C.E. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 2011, 286, 26148–26157. [Google Scholar] [CrossRef]
- Shimizu, T.; Shen, J.; Fang, M.; Zhang, Y.; Hori, K.; Trinidad, J.C.; Bauer, C.E.; Giedroc, D.P.; Masuda, S. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 2355–2360. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Lu, C.; Xia, Y.; Xin, Y.; Liu, H.; Xun, L.; Liu, H. FisR activates sigma(54)-dependent transcription of sulfide-oxidizing genes in Cupriavidus pinatubonensis JMP134. Mol. Microbiol. 2017, 105, 373–384. [Google Scholar] [CrossRef]
- Jo, I.; Chung, I.Y.; Bae, H.W.; Kim, J.S.; Song, S.; Cho, Y.H.; Ha, N.C. Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. USA 2015, 112, 6443–6448. [Google Scholar] [CrossRef]
- Lee, J.W.; Helmann, J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 2006, 440, 363–367. [Google Scholar] [CrossRef]
- Lee, J.W.; Helmann, J.D. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 2006, 281, 23567–23578. [Google Scholar] [CrossRef]
- Ludwig, M.; Chua, T.T.; Chew, C.Y.; Bryant, D.A. Fur-type transcriptional repressors and metal homeostasis in the cyanobacterium Synechococcus sp. PCC 7002. Front. Microbiol. 2015, 6, 1217. [Google Scholar] [CrossRef] [Green Version]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef]
- Stevens, S.E.; Porter, R.D. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 1980, 77, 6052–6056. [Google Scholar] [CrossRef]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]
- Nagy, P.; Pálinkás, Z.; Nagy, A.; Budai, B.; Tóth, I.; Vasas, A. Chemical aspects of hydrogen sulfide measurements in physiological samples. Biochim. Biophys. Acta 2014, 1840, 876–891. [Google Scholar] [CrossRef]
- Szekeres, E.; Sicora, C.; Dragos, N.; Druga, B. Selection of proper reference genes for the cyanobacterium Synechococcus PCC 7002 using real-time quantitative PCR. FEMS Microbiol. Lett. 2014, 359, 102–109. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xia, Y.; Xun, L. Revised mechanism and improved efficiency of the quikchange site-directed mutagenesis method. Methods Mol. Biol. 2017, 1498, 367–374. [Google Scholar] [CrossRef]
- Waugh, D.S. The remarkable solubility-enhancing power of Escherichia coli maltose-binding protein. Postep. Biochem. 2016, 62, 377–382. [Google Scholar] [CrossRef]
- Lu, T.; Cao, Q.; Pang, X.; Xia, Y.; Xun, L.; Liu, H. Sulfane sulfur-activated actinorhodin production and sporulation is maintained by a natural gene circuit in Streptomyces coelicolor. Microb. Biotechnol. 2020, 13, 1917–1932. [Google Scholar] [CrossRef]
- Doka, E.; Pader, I.; Biro, A.; Johansson, K.; Cheng, Q.; Ballago, K.; Prigge, J.R.; Pastor-Flores, D.; Dick, T.P.; Schmidt, E.E.; et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016, 2, e1500968. [Google Scholar] [CrossRef] [Green Version]
- Fan, K.; Chen, Z.; Liu, H. Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity. Sci. Adv. 2020, 6, eabb6477. [Google Scholar] [CrossRef]
- Dóka, É.; Arnér, E.S.J.; Schmidt, E.E.; Dick, T.P.; van der Vliet, A.; Yang, J.; Szatmári, R.; Ditrói, T.; Wallace, J.L.; Cirino, G.; et al. Comment on “Evidence that the ProPerDP method is inadequate for protein persulfidation detection due to lack of specificity”. Sci. Adv. 2021, 7, abe7006. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Sollitto, M.; Pallavicini, A.; Castellano, I. The complex evolutionary history of sulfoxide synthase in ovothiol biosynthesis. Proc. Biol. Sci. 2019, 286, 20191812. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Naser-Khdour, S.; Minh, B.Q.; Lanfear, R. Assessing confidence in root placement on phylogenies: An empirical study using non-reversible models for mammals. Syst. Biol. 2021, 71, 959–972. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Näär, A.M.; Ryu, S.; Tjian, R. Cofactor requirements for transcriptional activation by Sp1. Cold Spring Harb. Symp. Quant. Biol. 1998, 63, 189–199. [Google Scholar] [CrossRef]
- Urnov, F.D. A feel for the template: Zinc finger protein transcription factors and chromatin. Biochem. Cell Biol. 2002, 80, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Andrianopoulos, A. Evolution of a fungal regulatory gene family: The Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 1997, 21, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.O.; Merchant, K.; Nudelman, R.; Beyer, W.F.; Keng, T.; DeAngelo, J.; Hausladen, A.; Stamler, J.S. OxyR: A molecular code for redox-related signaling. Cell 2002, 109, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-J.; Kim, S.-J.; Mukhopadhyay, P.; Cho, S.; Woo, J.-R.; Storz, G.; Ryu, S.-E. Structural basis of the redox switch in the OxyR transcription factor. Cell 2001, 105, 103–113. [Google Scholar] [CrossRef]
- Storz, G.; Tartaglia, L.A.; Ames, B.N. Transcriptional regulator of oxidative stress-inducible genes: Direct activation by oxidation. Science 1990, 248, 189–194. [Google Scholar] [CrossRef]
- Tseng, H.J.; McEwan, A.G.; Apicella, M.A.; Jennings, M.P. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 2003, 71, 550–556. [Google Scholar] [CrossRef]
- Wu, H.J.; Seib, K.L.; Srikhanta, Y.N.; Kidd, S.P.; Edwards, J.L.; Maguire, T.L.; Grimmond, S.M.; Apicella, M.A.; McEwan, A.G.; Jennings, M.P. PerR controls Mn-dependent resistance to oxidative stress in Neisseria gonorrhoeae. Mol. Microbiol. 2006, 60, 401–416. [Google Scholar] [CrossRef]
- Ashby, M.K.; Houmard, J. Cyanobacterial two-component proteins: Structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 2006, 70, 472–509. [Google Scholar] [CrossRef]
- Koskinen, S.; Hakkila, K.; Kurkela, J.; Tyystjärvi, E.; Tyystjärvi, T. Inactivation of group 2 σ factors upregulates production of transcription and translation machineries in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2018, 8, 10305. [Google Scholar] [CrossRef]
- Grettenberger, C.L. Novel Gloeobacterales spp. from diverse environments across the globe. mSphere 2021, 6, e0006121. [Google Scholar] [CrossRef]
- Briand, J.-F.; Robillot, C.; Quiblier, C.; Bernard, C. A perennial bloom of planktothrix agardhii (cyanobacteria) in a shallow eutrophic french lake: Limnological and microcystin production studies. Arch. Fur Hydrobiol. 2002, 153, 605–622. [Google Scholar] [CrossRef]
- Schunck, H.; Lavik, G.; Desai, D.K.; Grosskopf, T.; Kalvelage, T.; Loscher, C.R.; Paulmier, A.; Contreras, S.; Siegel, H.; Holtappels, M.; et al. Giant hydrogen sulfide plume in the oxygen minimum zone off Peru supports chemolithoautotrophy. PLoS ONE 2013, 8, e68661. [Google Scholar] [CrossRef]
- Klatt, J.M.; Gomez-Saez, G.V.; Meyer, S.; Ristova, P.P.; Yilmaz, P.; Granitsiotis, M.S.; Macalady, J.L.; Lavik, G.; Polerecky, L.; Bühring, S.I. Versatile cyanobacteria control the timing and extent of sulfide production in a Proterozoic analog microbial mat. ISME J. 2020, 14, 3024–3037. [Google Scholar] [CrossRef] [PubMed]
- Stal, L.J.; Moezelaar, R. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 1997, 21, 179–211. [Google Scholar] [CrossRef]
- Klatt, J.M.; de Beer, D.; Hausler, S.; Polerecky, L. Cyanobacteria in sulfidic spring microbial mats can perform oxygenic and anoxygenic photosynthesis simultaneously during an entire diurnal period. Front. Microbiol. 2016, 7, 1973. [Google Scholar] [CrossRef]
- Liao, C.; Seebeck, F.P. Convergent evolution of ergothioneine biosynthesis in cyanobacteria. ChemBioChem 2017, 18, 2115–2118. [Google Scholar] [CrossRef]
- Brancaccio, M.; Tangherlini, M.; Danovaro, R.; Castellano, I. Metabolic adaptations to marine environments: Molecular diversity and evolution of ovothiol biosynthesis in bacteria. Genome Biol. Evol. 2021, 13, evab169. [Google Scholar] [CrossRef]
- Sevilla, E.; Sarasa-Buisan, C.; Gonzalez, A.; Cases, R.; Kufryk, G.; Peleato, M.L.; Fillat, M.F. Regulation by FurC in Anabaena links the oxidative stress response to photosynthetic metabolism. Plant Cell Physiol. 2019, 60, 1778–1789. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Song, H.; Li, Y.; Huang, R.; Liu, H.; Tang, K.; Jiao, N.; Liu, J. The Transcriptional Repressor PerR Senses Sulfane Sulfur by Cysteine Persulfidation at the Structural Zn2+ Site in Synechococcus sp. PCC7002. Antioxidants 2023, 12, 423. https://doi.org/10.3390/antiox12020423
Liu D, Song H, Li Y, Huang R, Liu H, Tang K, Jiao N, Liu J. The Transcriptional Repressor PerR Senses Sulfane Sulfur by Cysteine Persulfidation at the Structural Zn2+ Site in Synechococcus sp. PCC7002. Antioxidants. 2023; 12(2):423. https://doi.org/10.3390/antiox12020423
Chicago/Turabian StyleLiu, Daixi, Hui Song, Yuanning Li, Ranran Huang, Hongyue Liu, Kunxian Tang, Nianzhi Jiao, and Jihua Liu. 2023. "The Transcriptional Repressor PerR Senses Sulfane Sulfur by Cysteine Persulfidation at the Structural Zn2+ Site in Synechococcus sp. PCC7002" Antioxidants 12, no. 2: 423. https://doi.org/10.3390/antiox12020423
APA StyleLiu, D., Song, H., Li, Y., Huang, R., Liu, H., Tang, K., Jiao, N., & Liu, J. (2023). The Transcriptional Repressor PerR Senses Sulfane Sulfur by Cysteine Persulfidation at the Structural Zn2+ Site in Synechococcus sp. PCC7002. Antioxidants, 12(2), 423. https://doi.org/10.3390/antiox12020423




