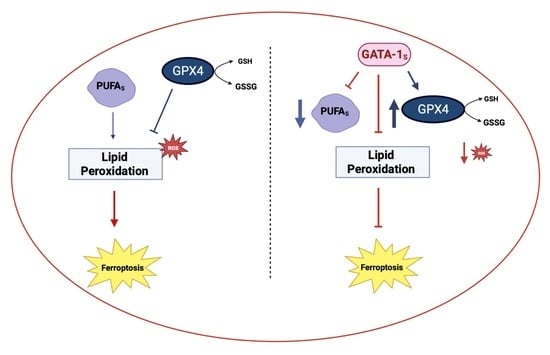
Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transient Transfection
2.3. Infrared Spectroscopy Analysis
2.4. Fatty Acids Composition Analysis by Gas Chromatography–Mass Spectrometry
2.5. Multivariate Data Analysis of FAMEs
2.6. Protein Extraction
2.7. Western Blot Analysis
2.8. Total RNA Extraction
2.9. Quantitative Real-Time PCR Analysis
2.10. Lipid Peroxidation Assay
2.11. Cell Viability and Cell Death Assays
2.12. Statistical Analysis
3. Results
3.1. Infrared Spectroscopy Analysis
3.2. Fatty Acids Composition
3.3. Correlation between GATA-1 Isoforms Expression and Lipid Peroxidation
3.4. Correlation between GATA-1 Isoforms and Expression Levels of Glutathione Peroxidase 4 (GPX4)
3.5. Effects of GPX4 Inhibition on Lipid Peroxidation in Cells Expressing GATA-1 Isoforms
3.6. Effects of GPX4 Inhibition on Cell Viability and Ferroptotic-Mediated Cell Death in Cells Overexpressing GATA-1 Isoforms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AA | Arachidonic (20:4) polyunsaturated omega-6 fatty acids |
| AdA | Adrenic (22:4) polyunsaturated omega-6 fatty acids |
| ASCA | ANOVA-simultaneous component analysis |
| CT value | Cycle threshold value |
| FAMEs | Fatty Acid Methyl Esters |
| FBS | Fetal bovine serum |
| FITC | Fluorescein Isothiocyanate |
| FLAG | DYKDDDDK tag peptide |
| GAPDH | Glyceraldehyde-3-phosphate Dehydrogenase |
| GATA-1 | Erythroid transcription factor |
| GATA-1FL | Full length isoform of the erythroid transcription factor GATA-1 |
| GATA-1S | Short length isoform of the erythroid transcription factor GATA-1 |
| GC- TOF-MS | Gas Chromatography–Time of Flight–Mass Spectrometry |
| GC-MS | Gas Chromatography–Mass Spectrometry |
| GPX4 | Glutathione peroxidase 4 |
| Hoechst 33342 | Trihydrochloride, Trihydrate solution |
| IR | Infrared spectroscopy analysis |
| LPO | Lipid hydroperoxides |
| LR | Lower Right Quadrant |
| MFAs | Monounsaturated fatty acids |
| MS | Mass Spectrometry |
| MTT assay | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide |
| Opti-MEM | Reduced-Serum Medium |
| p3XFLAG-CMV | Vector for expression of 3xFLAG-taged proteins |
| p53 | Cellular tumor antigen p5 |
| PAGE | Page PolyAcrylamide Gel Electrophoresis |
| PBS | Phosphate Buffer Saline |
| PCA | Principal Component Analysis |
| PCs | Principal Components |
| PFTBA | Perfluorotributylamine |
| PL-OH | Lipid alcohols |
| PL-OOH | Phospholipid peroxides |
| PUFA-PLs | Polyunsaturated fatty acids-containing phospholipids |
| qRT-PCR | Quantitative Real-time PCR |
| ROS | Reactive Oxygen Species |
| RPMI 1640 | Roswell Park Memorial Institute Culture Medium |
| RT-PCR | Real-time PCR analysis |
| SDHC | Subunit C of the Succinate Dehydrogenase Complex II of the respiratory chain |
| SDS | Sodium Dodecyl Sulphate |
| SFAs | Saturated fatty acids |
| UL | Upper Left Quadrant |
| UR | Upper Right Quadrant |
| 1R,3R-RSL3 | (1R,3R)-Methyl 2-(2-chloroacetyl)-2,3,4,9-tetrahydro-1-[4-(methoxycarbonyl)phenyl]-1H-pyrido[3,4-b]indole-3-carboxylate |
| 1S,3R-RSL3 | (1S,3R)-Methyl-2-(2-chloroacetyl)-2,3,4,9-tetrahydro-1-[4-(methoxycarbonyl)phenyl]-1H-pyrido[3,4-b]indole-3-carboxylate |
References
- Li, D.; Li, Y. The Interaction between Ferroptosis and Lipid Metabolism in Cancer. Sig. Transduct. Target Ther. 2020, 5, 108. [Google Scholar] [CrossRef]
- Kang, R.; Kroemer, G.; Tang, D. The Tumor Suppressor Protein P53 and the Ferroptosis Network. Free Radic. Biol. Med. 2019, 133, 162–168. [Google Scholar] [CrossRef]
- Yuan, H.; Li, X.; Zhang, X.; Kang, R.; Tang, D. Identification of ACSL4 as a Biomarker and Contributor of Ferroptosis. Biochem. Biophys. Res. Commun. 2016, 478, 1338–1343. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, X.; Jin, S.; Chen, Y.; Guo, R. Ferroptosis in Cancer Therapy: A Novel Approach to Reversing Drug Resistance. Mol. Cancer 2022, 21, 47. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cao, L.; Zhang, X.; Chu, B. Targeting Ferroptosis as a Vulnerability in Pulmonary Diseases. Cell Death Dis. 2022, 13, 649. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xie, Q.; Liu, X.; Wan, C.; Wu, W.; Fang, K.; Yao, Y.; Cheng, P.; Deng, D.; Liu, Z. Identification the Prognostic Value of Glutathione Peroxidases Expression Levels in Acute Myeloid Leukemia. Ann. Transl. Med. 2020, 8, 678. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, Z.; Peng, H. Molecular Mechanisms of Ferroptosis and Its Roles in Hematologic Malignancies. Front. Oncol. 2021, 11, 743006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Li, Q.; Xu, A.; Hu, Y.; Sun, C. Ferroptosis in Hematological Malignancies and Its Potential Network with Abnormal Tumor Metabolism. Biomed. Pharmacother. 2022, 148, 112747. [Google Scholar] [CrossRef]
- Lan, H.; Gao, Y.; Zhao, Z.; Mei, Z.; Wang, F. Ferroptosis: Redox Imbalance and Hematological Tumorigenesis. Front. Oncol. 2022, 12, 834681. [Google Scholar] [CrossRef]
- Trombetti, S.; Cesaro, E.; Catapano, R.; Sessa, R.; Lo Bianco, A.; Izzo, P.; Grosso, M. Oxidative Stress and ROS-Mediated Signaling in Leukemia: Novel Promising Perspectives to Eradicate Chemoresistant Cells in Myeloid Leukemia. IJMS 2021, 22, 2470. [Google Scholar] [CrossRef]
- Riccio, P.; Sessa, R.; de Nicola, S.; Petruzziello, F.; Trombetti, S.; Menna, G.; Pepe, G.; Maddalena, P.; Izzo, P.; Grosso, M. GATA-1 Isoforms Differently Contribute to the Production and Compartmentation of Reactive Oxygen Species in the Myeloid Leukemia Cell Line K562. J. Cell. Physiol. 2019, 234, 20829–20846. [Google Scholar] [CrossRef]
- Trombetti, S.; Sessa, R.; Catapano, R.; Rinaldi, L.; Lo Bianco, A.; Feliciello, A.; Izzo, P.; Grosso, M. Exploring the Leukemogenic Potential of GATA-1S, the Shorter Isoform of GATA-1: Novel Insights into Mechanisms Hampering Respiratory Chain Complex II Activity and Limiting Oxidative Phosphorylation Efficiency. Antioxidants 2021, 10, 1603. [Google Scholar] [CrossRef]
- Sarnelli, G.; Grosso, M.; Palumbo, I.; Pesce, M.; D’Alessandro, A.; Zaninotto, G.; Annese, V.; Petruzzelli, R.; Izzo, P.; Sepulveres, R.; et al. Allele-specific Transcriptional Activity of the Variable Number of Tandem Repeats of the Inducible Nitric Oxide Synthase Gene Is Associated with Idiopathic Achalasia. United Eur. Gastroenterol. J. 2017, 5, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.O.; Costa, H.; Lima, R.; Amaral, J.S. Simple Methodology for the Quantitative Analysis of Fatty Acids in Human Red Blood Cells. Chromatographia 2015, 78, 1271–1281. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.-J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-Simultaneous Component Analysis (ASCA): A New Tool for Analyzing Designed Metabolomics Data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, N.; Khakimov, B.; Mikkelsen, M.S.; Nielsen, T.S.; Jensen, M.G.; Randazzo, A.; Engelsen, S.B. Structurally Different Mixed Linkage β-Glucan Supplements Differentially Increase Secondary Bile Acid Excretion in Hypercholesterolaemic Rat Faeces. Food Funct. 2020, 11, 514–523. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Bellisola, G.; Sorio, C. Infrared Spectroscopy and Microscopy in Cancer Research and Diagnosis. Am. J. Cancer Res. 2011, 2, 1–21. [Google Scholar]
- Dos Santos, A.F.; Inague, A.; Arini, G.S.; Terra, L.F.; Wailemann, R.A.M.; Pimentel, A.C.; Yoshinaga, M.Y.; Silva, R.R.; Severino, D.; de Almeida, D.R.Q.; et al. Distinct Photo-Oxidation-Induced Cell Death Pathways Lead to Selective Killing of Human Breast Cancer Cells. Cell Death Dis. 2020, 11, 1070. [Google Scholar] [CrossRef]
- Qin, S.; Wang, Y.; Li, L.; Liu, J.; Xiao, C.; Duan, D.; Hao, W.; Qin, C.; Chen, J.; Yao, L.; et al. Early-Life Vitamin B12 Orchestrates Lipid Peroxidation to Ensure Reproductive Success via SBP-1/SREBP1 in Caenorhabditis Elegans. Cell Rep. 2022, 40, 111381. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Palte, M.J.; Deik, A.A.; Li, H.; Eaton, J.K.; Wang, W.; Tseng, Y.-Y.; Deasy, R.; Kost-Alimova, M.; Dančík, V.; et al. A GPX4-Dependent Cancer Cell State Underlies the Clear-Cell Morphology and Confers Sensitivity to Ferroptosis. Nat. Commun. 2019, 10, 1617. [Google Scholar] [CrossRef] [PubMed]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in Ferroptosis and Its Pharmacological Implication. Free Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ Oxidoreductase FSP1 Acts Parallel to GPX4 to Inhibit Ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Shin, D.; Kim, E.H.; Lee, J.; Roh, J.-L. Nrf2 Inhibition Reverses Resistance to GPX4 Inhibitor-Induced Ferroptosis in Head and Neck Cancer. Free. Radic. Biol. Med. 2018, 129, 454–462. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Liu, S.; Duan, T.; Zhai, L.; Zhang, M.; Han, X.; Xiang, Y.; Huang, X.; Lin, H.; et al. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front. Pharmacol. 2018, 9, 1371. [Google Scholar] [CrossRef]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. IJMS 2021, 22, 12827. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, J.; Li, X.; Wu, Y.; Shi, H.; Chen, Y.; Lu, G.; Shen, H.; Lu, G.; Zhou, J. Quercetin Induces P53-independent Cancer Cell Death through Lysosome Activation by the Transcription Factor EB and Reactive Oxygen Species-dependent Ferroptosis. Br. J. Pharmacol. 2021, 178, 1133–1148. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Jiang, X.; Gu, W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol. 2020, 30, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tao, J.; Yang, Y.; Tan, S.; Liu, H.; Jiang, J.; Zheng, F.; Wu, B. Ferroptosis Involves in Intestinal Epithelial Cell Death in Ulcerative Colitis. Cell Death Dis. 2020, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Homma, T.; Kobayashi, S.; Conrad, M.; Konno, H.; Yokoyama, C.; Fujii, J. Nitric Oxide Protects against Ferroptosis by Aborting the Lipid Peroxidation Chain Reaction. Nitric Oxide 2021, 115, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Nag, P.; Grivei, A.; Giuliani, K.T.K.; Wang, X.; Diwan, V.; Hoy, W.; Healy, H.; Gobe, G.; Kassianos, A.J. Adenine Overload Induces Ferroptosis in Human Primary Proximal Tubular Epithelial Cells. Cell Death Dis. 2022, 13, 104. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Kim, W.K.; Bae, K.-H.; Lee, S.C.; Lee, E.-W. Lipid Metabolism and Ferroptosis. Biology 2021, 10, 184. [Google Scholar] [CrossRef]
- Yu, Y.; Xie, Y.; Cao, L.; Yang, L.; Yang, M.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. The Ferroptosis Inducer Erastin Enhances Sensitivity of Acute Myeloid Leukemia Cells to Chemotherapeutic Agents. Mol. Cell. Oncol. 2015, 2, e1054549. [Google Scholar] [CrossRef]
- Chlon, T.M.; McNulty, M.; Goldenson, B.; Rosinski, A.; Crispino, J.D. Global Transcriptome and Chromatin Occupancy Analysis Reveal the Short Isoform of GATA1 Is Deficient for Erythroid Specification and Gene Expression. Haematologica 2015, 100, 575–584. [Google Scholar] [CrossRef][Green Version]
- Doré, L.C.; Crispino, J.D. Transcription Factor Networks in Erythroid Cell and Megakaryocyte Development. Blood 2011, 118, 231–239. [Google Scholar] [CrossRef]
- Halsey, C.; Docherty, M.; McNeill, M.; Gilchrist, D.; Le Brocq, M.; Gibson, B.; Graham, G. The GATA1s Isoform Is Normally Down-Regulated during Terminal Haematopoietic Differentiation and over-Expression Leads to Failure to Repress MYB, CCND2 and SKI during Erythroid Differentiation of K562 Cells. J. Hematol. Oncol. 2012, 5, 45. [Google Scholar] [CrossRef]
- Lentjes, M.H.; Niessen, H.E.; Akiyama, Y.; de Bruïne, A.P.; Melotte, V.; van Engeland, M. The Emerging Role of GATA Transcription Factors in Development and Disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef]
- Xu, C.; Fu, H.; Gao, L.; Wang, L.; Wang, W.; Li, J.; Li, Y.; Dou, L.; Gao, X.; Luo, X.; et al. BCR-ABL/GATA1/MiR-138 Mini Circuitry Contributes to the Leukemogenesis of Chronic Myeloid Leukemia. Oncogene 2014, 33, 44–54. [Google Scholar] [CrossRef] [PubMed]











| Transcript | Accession Number | Primer | Sequence 5′-3′ | Amplicon Size |
|---|---|---|---|---|
| GPX4 | NM_002085.5 | For | CCTGGACAAGTACCGGGGC | 140 bp |
| Rev | CTTCGTTACTCCCTGGCTCCT | |||
| β-actin | NM_001101.5 | For | CGACAGGATGCAGAAGGAGA | 160 bp |
| Rev | CGTCATACTCCTGCTTGCTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trombetti, S.; Iaccarino, N.; Riccio, P.; Sessa, R.; Catapano, R.; Salvatore, M.; Luka, S.; de Nicola, S.; Izzo, P.; Roperto, S.; et al. Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation. Antioxidants 2023, 12, 537. https://doi.org/10.3390/antiox12030537
Trombetti S, Iaccarino N, Riccio P, Sessa R, Catapano R, Salvatore M, Luka S, de Nicola S, Izzo P, Roperto S, et al. Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation. Antioxidants. 2023; 12(3):537. https://doi.org/10.3390/antiox12030537
Chicago/Turabian StyleTrombetti, Silvia, Nunzia Iaccarino, Patrizia Riccio, Raffaele Sessa, Rosa Catapano, Marcella Salvatore, Stelina Luka, Sergio de Nicola, Paola Izzo, Sante Roperto, and et al. 2023. "Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation" Antioxidants 12, no. 3: 537. https://doi.org/10.3390/antiox12030537
APA StyleTrombetti, S., Iaccarino, N., Riccio, P., Sessa, R., Catapano, R., Salvatore, M., Luka, S., de Nicola, S., Izzo, P., Roperto, S., Maddalena, P., Randazzo, A., & Grosso, M. (2023). Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation. Antioxidants, 12(3), 537. https://doi.org/10.3390/antiox12030537