Glutamine Starvation Affects Cell Cycle, Oxidative Homeostasis and Metabolism in Colorectal Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Growth Curves
2.3. Colony Forming Assay
2.4. Fluorescence-Activated Cell Sorting (FACS) Analysis
2.5. Sample Preparation for Metabolomics Analysis
2.6. Gas Chromatography-Mass Spectrometry Analysis
2.7. Glucose Uptake Assay
2.8. GLUT1 Protein Expression by Immunofluorescence
2.9. 2-Deoxyglucose Treatment and MTT Viability Test
2.10. Determination of Intracellular Aminothyol Levels
2.11. Statistical Analysis
3. Results
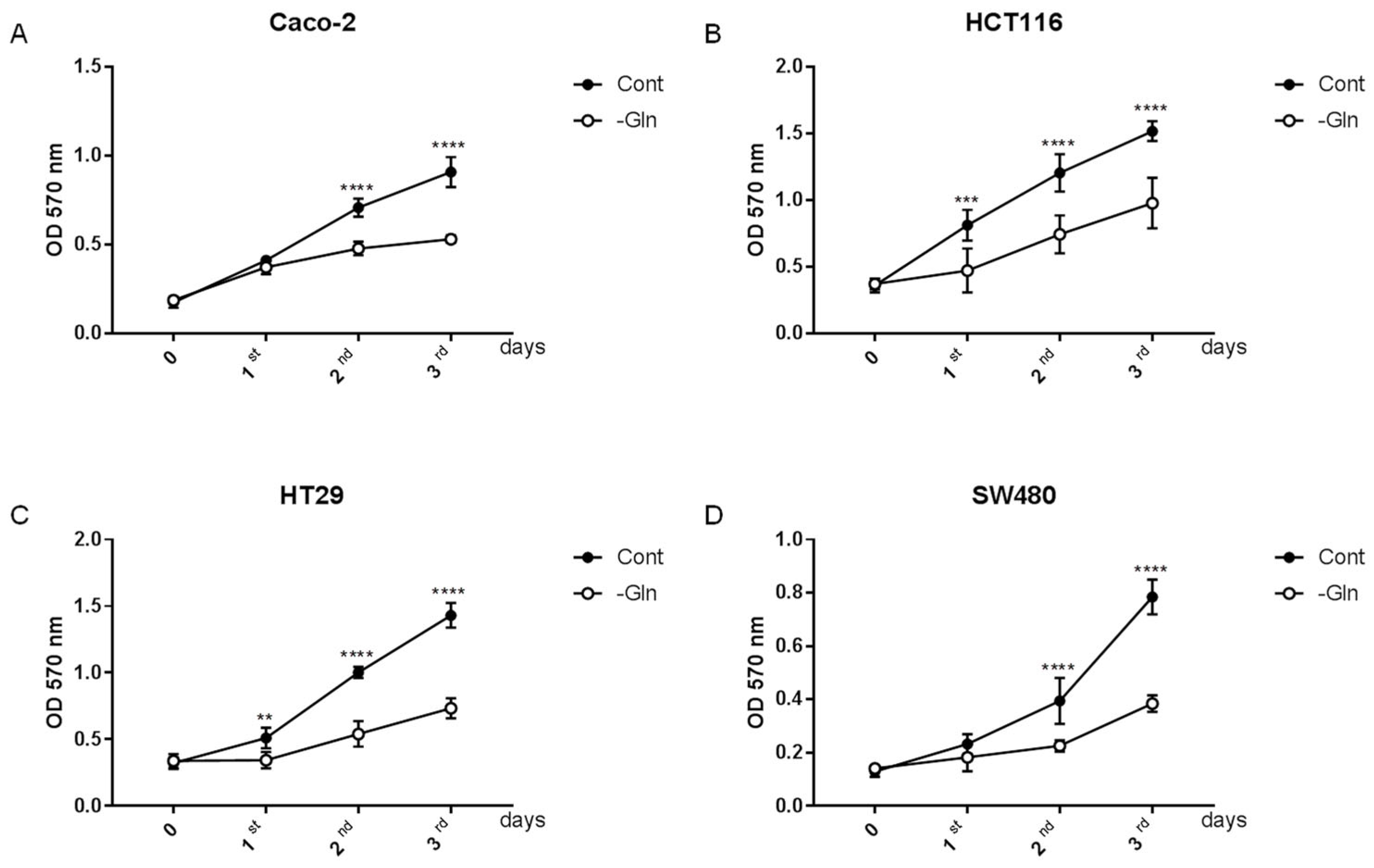
3.1. Glutamine Starvation Alters Growth Rate in Colorectal Cancer Cells
3.2. Glutamine Starvation Reduces Proliferative Capacity in Colorectal Cancer Cells
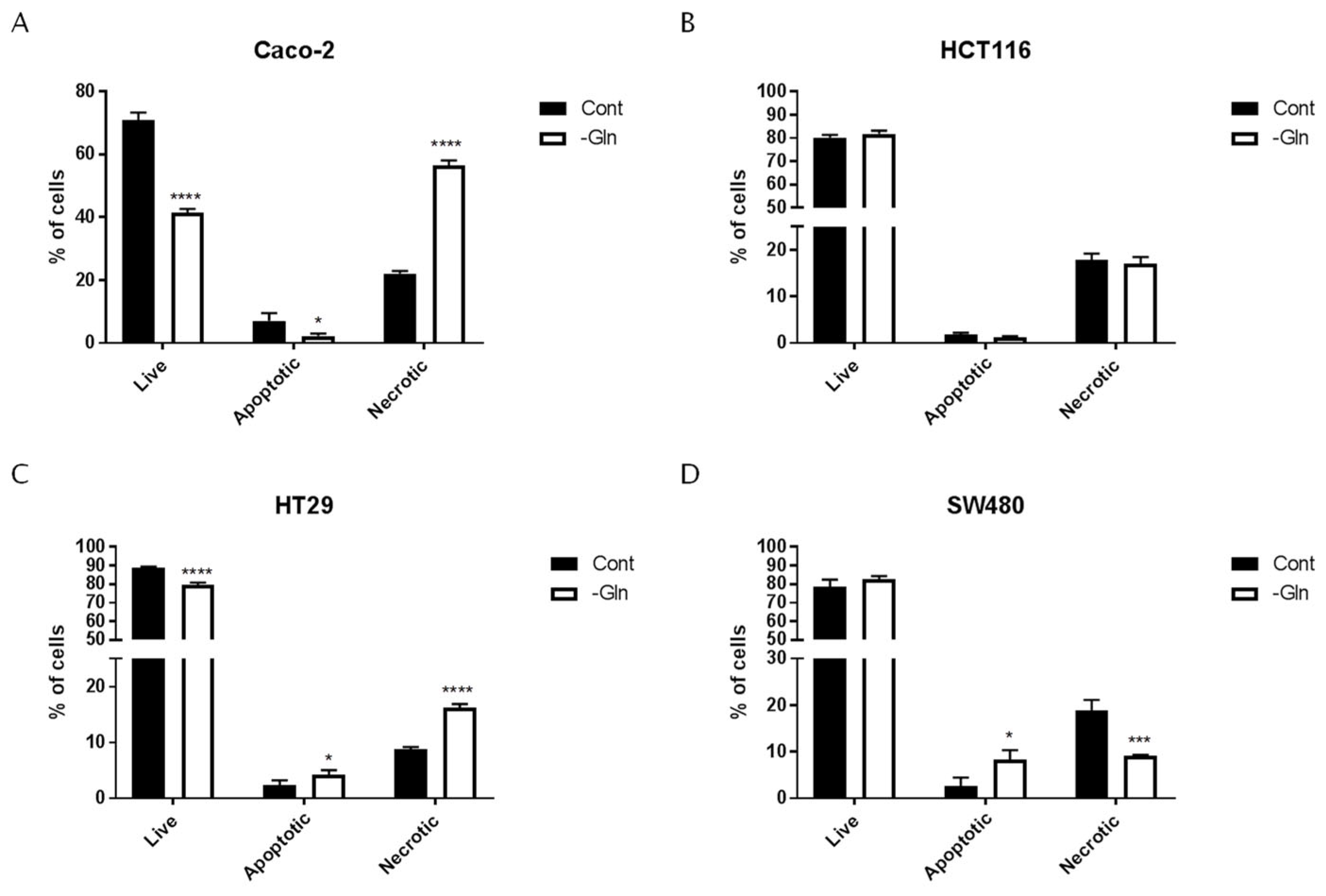
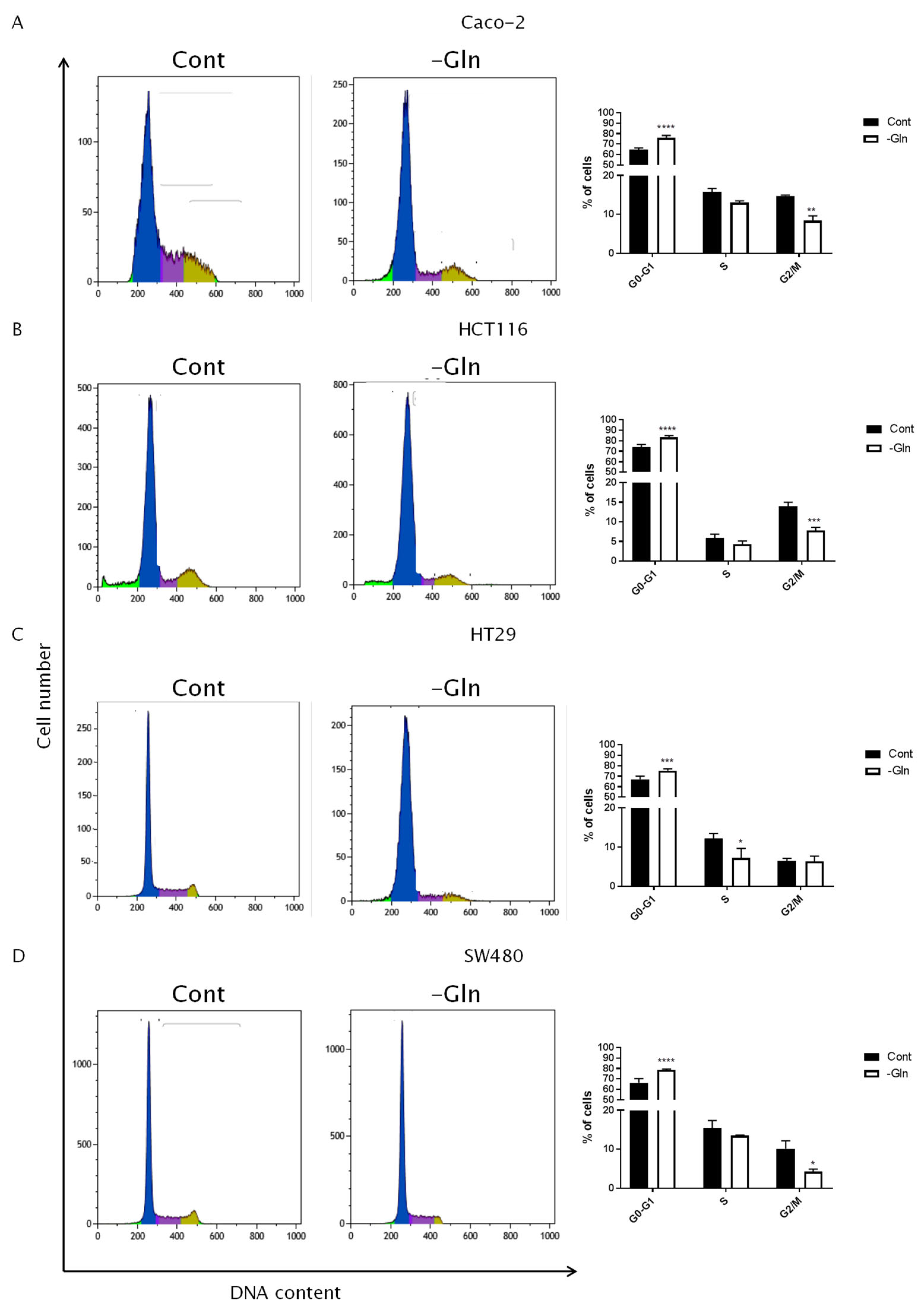
3.3. Glutamine Starvation Triggers Cell Death and G0/G1 Cell Cycle Arrest
3.4. Metabolomic Alterations under Glutamine Deprivation
3.5. Glutamine Deprivation Altered Glucose Uptake and GLUT1 Expression
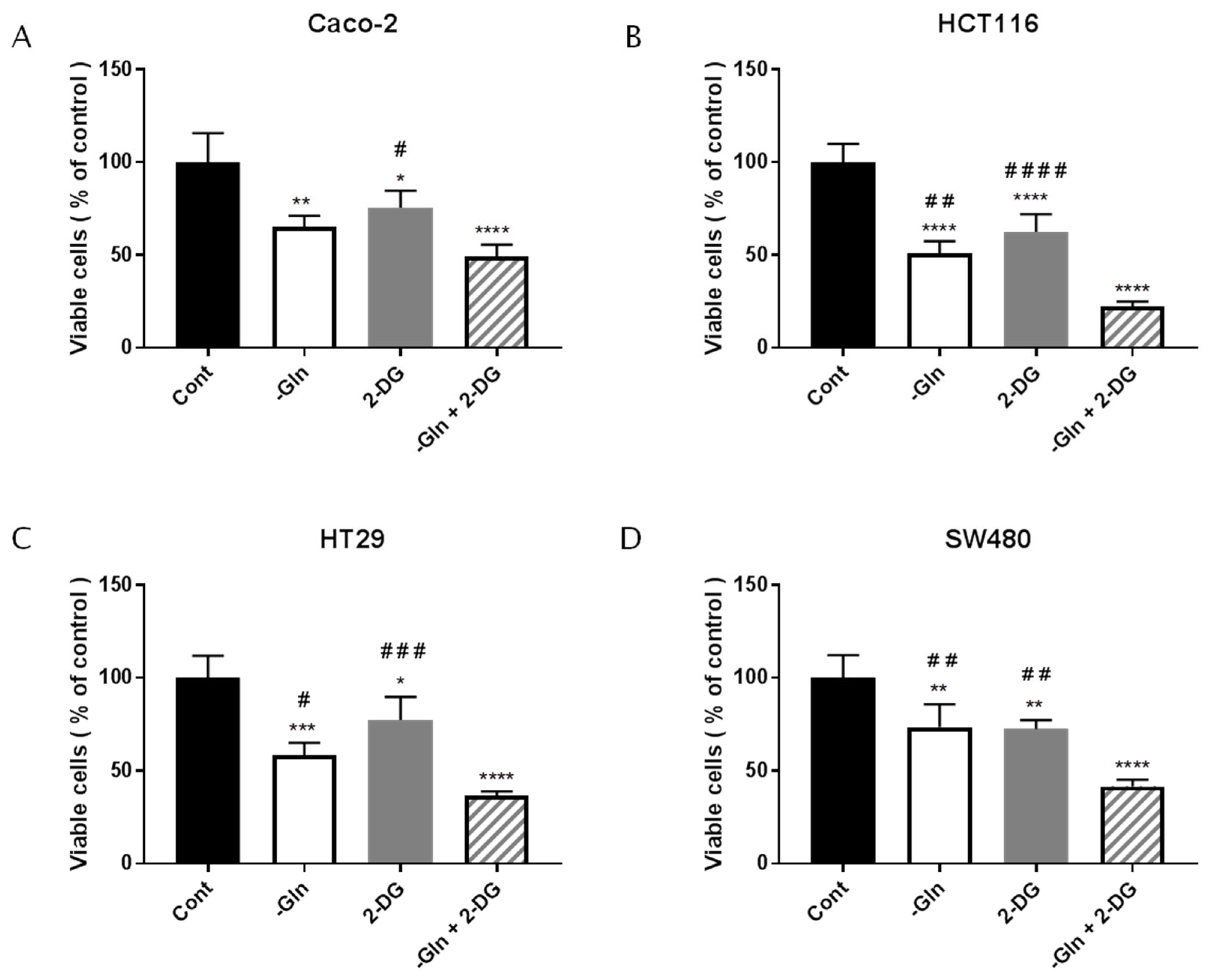
3.6. The Combined Treatment with Glutamine Deprivation and 2-Deoxy-D-Glucose Affected CRC Survival more than Glutamine Deprivation
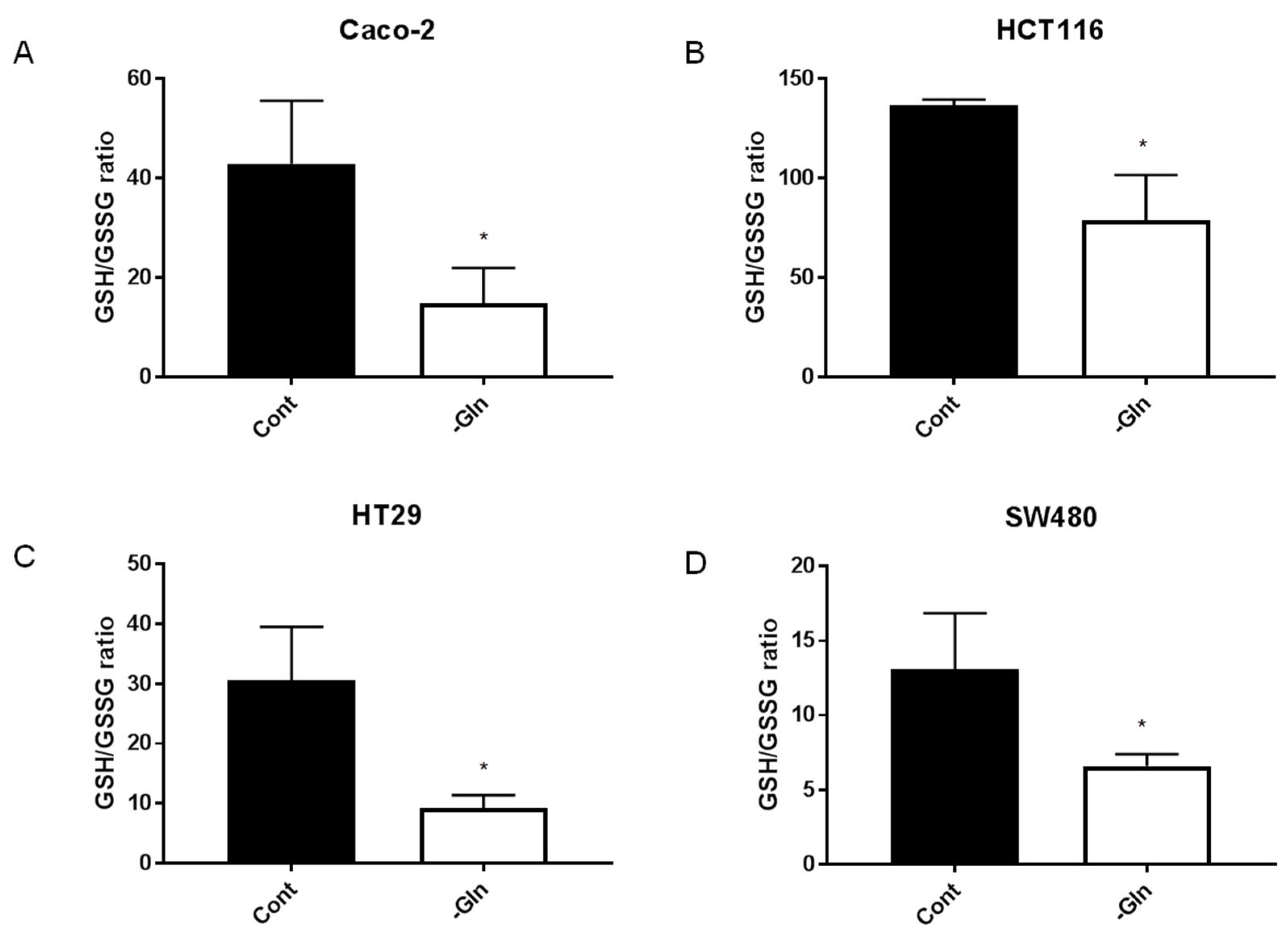
3.7. Glutamine-Deprivation-Induced Reduction in Antioxidant Defenses in CRC Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, J.; Prenen, H. Molecular genetics of colorectal cancer. Ann. Gastroenterol. 2014, 27, 9–14. [Google Scholar] [PubMed]
- Nagao-Kitamoto, H.; Kitamoto, S.; Kamada, N. Inflammatory bowel disease and carcinogenesis. Cancer Metastasis Rev. 2022, 41, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Peeters, P.J.; Bazelier, M.T.; Leufkens, H.G.; de Vries, F.; De Bruin, M.L. The Risk of Colorectal Cancer in Patients with Type 2 Diabetes: Associations with Treatment Stage and Obesity. Diabetes Care 2014, 38, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Yang, Y.; Wang, F.; Zhang, P.; Shi, C.; Zou, Y.; Qin, H. Obesity and Risk of Colorectal Cancer: A Systematic Review of Prospective Studies. PLoS ONE 2013, 8, e53916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, P.S.; Chen, T.-Y.; Giovannucci, E. Cigarette smoking and colorectal cancer incidence and mortality: Systematic review and meta-analysis. Int. J. Cancer 2009, 124, 2406–2415. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Mao, W.; Liu, T.; Lin, Q.; Lu, X.; Wang, Q.; Lin, F.; Ekelund, U.; Wijndaele, K. Sedentary Behavior and Incident Cancer: A Meta-Analysis of Prospective Studies. PLoS ONE 2014, 9, e105709. [Google Scholar] [CrossRef] [Green Version]
- Chan, D.S.M.; Lau, R.; Aune, D.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and Processed Meat and Colorectal Cancer Incidence: Meta-Analysis of Prospective Studies. PLoS ONE 2011, 6, e20456. [Google Scholar] [CrossRef] [Green Version]
- Islami, F.; Goding Sauer, A.; Miller, K.D.; Siegel, R.L.; Fedewa, S.A.; Jacobs, E.J.; McCullough, M.L.; Patel, A.V.; Ma, J.; Soerjomataram, I.; et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA A Cancer J. Clin. 2017, 68, 31–54. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Lui, R.N.; Tsoi, K.K.; Ho, J.M.; Lo, C.; Chan, F.C.; Kyaw, M.H.; Sung, J.J. Global Increasing Incidence of Young-Onset Colorectal Cancer Across 5 Continents: A Joinpoint Regression Analysis of 1,922,167 Cases. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1275–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meester, R.G.S.; Mannalithara, A.; Lansdorp-Vogelaar, I.; Ladabaum, U. Trends in Incidence and Stage at Diagnosis of Colorectal Cancer in Adults Aged 40 Through 49 Years, 1975–2015. JAMA 2019, 321, 1933–1934. [Google Scholar] [CrossRef] [PubMed]
- El Din, K.S.; Loree, J.; Sayre, E.C.; Gill, S.; Brown, C.; Dau, H.; De Vera, M.A. Trends in the epidemiology of young-onset colorectal cancer: A worldwide systematic review. BMC Cancer 2020, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernfeld, E.; Foster, D.A. Glutamine as an Essential Amino Acid for KRas-Driven Cancer Cells. Trends Endocrinol. Metab. 2019, 30, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Samuels, Y.; Li, Q.; Krokowski, D.; Guan, B.-J.; Wang, C.; Jin, Z.; Dong, B.; Cao, B.; Feng, X.; et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016, 7, 11971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, D.R.; DeBerardinis, R.J.; Mancuso, A.; Sayed, N.; Zhang, X.-Y.; Pfeiffer, H.K.; Nissim, I.; Daikhin, E.; Yudkoff, M.; McMahon, S.B.; et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA 2008, 105, 18782–18787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [Green Version]
- Bott, A.J.; Maimouni, S.; Zong, W.-X. The Pleiotropic Effects of Glutamine Metabolism in Cancer. Cancers 2019, 11, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhao, X.; Chen, V.; Feng, Y.; Wang, L.; Croniger, C.; Conlon, R.A.; Markowitz, S.; Fearon, E.; Puchowicz, M.; et al. Colorectal cancers utilize glutamine as an anaplerotic substrate of the TCA cycle in vivo. Sci. Rep. 2019, 9, 19180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Li, J.; Sun, X.; Guo, Y.; Chu, D.; Wei, L.; Li, X.; Yang, G.; Liu, X.; Yao, L.; et al. Tumor suppressor NDRG2 inhibits glycolysis and glutaminolysis in colorectal cancer cells by repressing c-Myc expression. Oncotarget 2015, 6, 26161–26176. [Google Scholar] [CrossRef] [PubMed]
- Vermeersch, K.A.; Styczynski, M.P. Applications of metabolomics in cancer research. J. Carcinog. 2013, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D. Metabolomics and the Multi-Omics View of Cancer. Metabolites 2022, 12, 154. [Google Scholar] [CrossRef]
- ATCC. Available online: https://www.atcc.org/ (accessed on 2 March 2023).
- Mouradov, D.; Sloggett, C.; Jorissen, R.N.; Love, C.G.; Li, S.; Burgess, A.W.; Arango, D.; Strausberg, R.L.; Buchanan, D.; Wormald, S.; et al. Colorectal Cancer Cell Lines Are Representative Models of the Main Molecular Subtypes of Primary Cancer. Cancer Res. 2014, 74, 3238–3247. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, M.; Tomlinson, I.P.M.; Rowan, A.; Pignatelli, M.; Bodmer, W.F. β-Catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. USA 1997, 94, 10330–10334. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Bodmer, W.F. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc. Natl. Acad. Sci. USA 2006, 103, 976–981. [Google Scholar] [CrossRef] [Green Version]
- Fujinaga, T.; Kumamaru, W.; Sugiura, T.; Kobayashi, Y.; Ohyama, Y.; Ikari, T.; Onimaru, M.; Akimoto, N.; Jogo, R.; Mori, Y. Biological characterization and analysis of metastasis-related genes in cell lines derived from the primary lesion and lymph node metastasis of a squamous cell carcinoma arising in the mandibular gingiva. Int. J. Oncol. 2014, 44, 1614–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tronci, L.; Serreli, G.; Piras, C.; Frau, D.; Dettori, T.; Deiana, M.; Murgia, F.; Santoru, M.; Spada, M.; Leoni, V.; et al. Vitamin C Cytotoxicity and Its Effects in Redox Homeostasis and Energetic Metabolism in Papillary Thyroid Carcinoma Cell Lines. Antioxidants 2021, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.-H.; Li, L.-L.; Wu, G.-G.; Xie, Y.-C.; Zhang, G.-X.; Chen, W.; Yang, H.-F.; Liu, Q.-L.; Li, W.-H.; He, W.-G.; et al. Upregulation of CPE promotes cell proliferation and tumorigenicity in colorectal cancer. BMC Cancer 2013, 13, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, Y.; Gao, W.; Li, F.; Bo, Y.; Zhu, M.; Fu, R.; Liu, Q.; Wen, S.; Wang, B. Identification and characterization of CD133+CD44+ cancer stem cells from human laryngeal squamous cell carcinoma cell lines. J. Cancer 2017, 8, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettori, T.; Sanna, G.; Cocco, A.; Serreli, G.; Deiana, M.; Palmas, V.; Onnis, V.; Pilia, L.; Melis, N.; Moi, D.; et al. Synthesis and Antiproliferative Effect of Halogenated Coumarin Derivatives. Molecules 2022, 27, 8897. [Google Scholar] [CrossRef]
- Santoru, M.L.; Piras, C.; Murgia, F.; Spada, M.; Tronci, L.; Leoni, V.P.; Serreli, G.; Deiana, M.; Atzori, L. Modulatory Effect of Nicotinic Acid on the Metabolism of Caco-2 Cells Exposed to IL-1β and LPS. Metabolites 2020, 10, 204. [Google Scholar] [CrossRef]
- Caria, P.; Pillai, R.; Dettori, T.; Frau, D.V.; Zavattari, P.; Riva, G.; Romano, G.; Pani, F.; Bentivegna, A.; Giovannoni, R.; et al. Thyrospheres from B-CPAP Cell Line with BRAF and TERT Promoter Mutations have Different Functional and Molecular Features than Parental Cells. J. Cancer 2017, 8, 1629–1639. [Google Scholar] [CrossRef] [Green Version]
- Incani, A.; Serra, G.; Atzeri, A.; Melis, M.P.; Serreli, G.; Bandino, G.; Sedda, P.; Campus, M.; Tuberoso, C.I.; Deiana, M. Extra virgin olive oil phenolic extracts counteract the pro-oxidant effect of dietary oxidized lipids in human intestinal cells. Food Chem. Toxicol. 2016, 90, 171–180. [Google Scholar] [CrossRef]
- Gwangwa, M.V.; Joubert, A.M.; Visagie, M.H. Effects of glutamine deprivation on oxidative stress and cell survival in breast cell lines. Biol. Res. 2019, 52, 15. [Google Scholar] [CrossRef]
- Lee, P.; Malik, D.; Perkons, N.; Huangyang, P.; Khare, S.; Rhoades, S.; Gong, Y.-Y.; Burrows, M.; Finan, J.M.; Nissim, I.; et al. Targeting glutamine metabolism slows soft tissue sarcoma growth. Nat. Commun. 2020, 11, 498. [Google Scholar] [CrossRef] [Green Version]
- Márquez, J.; Alonso, F.J.; Matés, J.M.; Segura, J.A.; Martín-Rufián, M.; Campos-Sandoval, J.A. Glutamine Addiction in Gliomas. Neurochem. Res. 2017, 42, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Ambs, S. Metabolic signatures of human breast cancer. Mol. Cell. Oncol. 2015, 2, e992217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Wei, B.; Lu, C.; Li, P.; Chen, L. Glutaminase sustains cell survival via the regulation of glycolysis and glutaminolysis in colorectal cancer. Oncol. Lett. 2017, 14, 3117–3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhove, K.; Derveaux, E.; Graulus, G.-J.; Mesotten, L.; Thomeer, M.; Noben, J.-P.; Guedens, W.; Adriaensens, P. Glutamine Addiction and Therapeutic Strategies in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandian, S.R.K.; Deepak, V.; Nellaiah, H.; Sundar, K. PEG–PHB-glutaminase nanoparticle inhibits cancer cell proliferation in vitro through glutamine deprivation. Vitr. Cell. Dev. Biol.–Anim. 2014, 51, 372–380. [Google Scholar] [CrossRef]
- Sun, N.; Liang, Y.; Chen, Y.; Wang, L.; Li, D.; Liang, Z.; Sun, L.; Wang, Y.; Niu, H. Glutamine affects T24 bladder cancer cell proliferation by activating STAT3 through ROS and glutaminolysis. Int. J. Mol. Med. 2019, 44, 2189–2200. [Google Scholar] [CrossRef] [Green Version]
- Willems, L.; Jacque, N.; Jacquel, A.; Neveux, N.; Maciel, T.T.; Lambert, M.; Schmitt, A.; Poulain, L.; Green, A.S.; Uzunov, M.; et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 2013, 122, 3521–3532. [Google Scholar] [CrossRef] [Green Version]
- Fumarola, C.; Zerbini, A.; Guidotti, G.G. Glutamine deprivation-mediated cell shrinkage induces ligand-independent CD95 receptor signaling and apoptosis. Cell Death Differ. 2001, 8, 1004–1013. [Google Scholar] [CrossRef]
- Ortega, A.L.; Carretero, J.; Obrador, E.; Gambini, J.; Asensi, M.; Rodilla, V.; Estrela, J.M. Tumor Cytotoxicity by Endothelial Cells. J. Biol. Chem. 2003, 278, 13888–13897. [Google Scholar] [CrossRef] [Green Version]
- Yuneva, M.; Zamboni, N.; Oefner, P.; Sachidanandam, R.; Lazebnik, Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007, 178, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Shanware, N.P.; Bray, K.; Eng, C.H.; Wang, F.; Follettie, M.; Myers, J.; Fantin, V.R.; Abraham, R.T. Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat. Commun. 2014, 5, 4900. [Google Scholar] [CrossRef] [Green Version]
- Chiodi, I.; Picco, G.; Martino, C.; Mondello, C. Cellular response to glutamine and/or glucose deprivation in in vitro transformed human fibroblasts. Oncol. Rep. 2019, 41, 3555–3564. [Google Scholar] [CrossRef]
- Polat, I.H.; Tarrado-Castellarnau, M.; Benito, A.; Hernandez-Carro, C.; Centelles, J.; Marin, S.; Cascante, M. Glutamine Modulates Expression and Function of Glucose 6-Phosphate Dehydrogenase via NRF2 in Colon Cancer Cells. Antioxidants 2021, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Saqcena, M.; Menon, D.; Patel, D.; Mukhopadhyay, S.; Chow, V.; Foster, D.A. Amino Acids and mTOR Mediate Distinct Metabolic Checkpoints in Mammalian G1 Cell Cycle. PLoS ONE 2013, 8, e74157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, T.Q.; Lowman, X.H.; A Reid, M.; Mendez-Dorantes, C.; Pan, M.; Yang, Y.; Kong, M. Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction. Oncogene 2016, 36, 1991–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, S.L.; Palacios-Callender, M.; Frakich, N.; Carcamo, S.; Kovacs, I.; Tudzarova, S.; Moncada, S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl. Acad. Sci. USA 2011, 108, 21069–21074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estévez-García, I.O.; Cordoba-Gonzalez, V.; Lara-Padilla, E.; Fuentes-Toledo, A.; Falfán-Valencia, R.; Campos-Rodríguez, R.; Abarca-Rojano, E. Glucose and glutamine metabolism control by APC and SCF during the G1-to-S phase transition of the cell cycle. J. Physiol. Biochem. 2014, 70, 569–581. [Google Scholar] [CrossRef]
- Kuo, C.-C.; Ling, H.-H.; Chiang, M.-C.; Chung, C.-H.; Lee, W.-Y.; Chu, C.-Y.; Wu, Y.-C.; Chen, C.-H.; Lai, Y.-W.; Tsai, I.-L.; et al. Metastatic Colorectal Cancer Rewrites Metabolic Program Through a Glut3-YAP-dependent Signaling Circuit. Theranostics 2019, 9, 2526–2540. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Lee, G.; Oh, T.-I.; Kim, B.M.; Shim, D.-W.; Lee, K.-H.; Kim, Y.J.; Lim, B.O.; Lim, J.-H. Inhibition of glutamine utilization sensitizes lung cancer cells to apigenin-induced apoptosis resulting from metabolic and oxidative stress. Int. J. Oncol. 2015, 48, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of Glucose Transporters and Glutaminase Synergistically Impairs Tumor Cell Growth. Cell Chem. Biol. 2019, 26, 1214–1228.e25. [Google Scholar] [CrossRef]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cetinbas, N.M.; Sudderth, J.; Harris, R.C.; Cebeci, A.; Negri, G.L.; Yılmaz, H.; DeBerardinis, R.J.; Sorensen, P.H. Glucose-dependent anaplerosis in cancer cells is required for cellular redox balance in the absence of glutamine. Sci. Rep. 2016, 6, 32606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, L.; Shine, A.; Hewage, C.; Malthouse, J.; Brindle, K.; McClenaghan, N.; Flatt, P.; Newsholme, P. A Nuclear Magnetic Resonance-Based Demonstration of Substantial Oxidative L-Alanine Metabolism and L-Alanine-Enhanced Glucose Metabolism in a Clonal Pancreatic Beta-Cell Line–Metabolism of L-Alanine Is Important to the Regulation of Insulin Secretion. Diabetes 2002, 51, 1714–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlessi, R.; Rowlands, J.; Ellison, G.; de Oliveira Alves, H.H.; Newsholme, P.; Mamotte, C. Glutamine deprivation induces metabolic adaptations associated with beta cell dysfunction and exacerbate lipotoxicity. Mol. Cell. Endocrinol. 2019, 491, 110433. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Mou, J.; Shao, B.; Wei, Y.; Liang, H.; Takano, N.; Semenza, G.L.; Xie, G. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis. 2019, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zou, Y.; Mao, D.; Sun, D.; Gao, G.; Shi, J.; Liu, X.; Zhu, C.; Yang, M.; Ye, W.; et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 2014, 206, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Sasayama, T.; Nagashima, H.; Irino, Y.; Takahashi, M.; Izumi, Y.; Uno, T.; Satoh, N.; Kitta, A.; Kyotani, K.; et al. Glioma cells require one-carbon metabolism to survive glutamine starvation. Acta Neuropathol. Commun. 2021, 9, 16. [Google Scholar] [CrossRef]
- Geck, R.; Toker, A. Nonessential amino acid metabolism in breast cancer. Adv. Biol. Regul. 2016, 62, 11–17. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [Green Version]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma—Deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Hutschenreuther, A.; Birkenmeier, G.; Bigl, M.; Krohn, K.; Birkemeyer, C. Glycerophosphoglycerol, Beta-Alanine, and Pantothenic Acid as Metabolic Companions of Glycolytic Activity and Cell Migration in Breast Cancer Cell Lines. Metabolites 2013, 3, 1084–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanware, N.P.; Mullen, A.R.; DeBerardinis, R.J.; Abraham, R.T. Glutamine: Pleiotropic roles in tumor growth and stress resistance. J. Mol. Med. 2011, 89, 229–236. [Google Scholar] [CrossRef]
- Caiola, E.; Colombo, M.; Sestito, G.; Lupi, M.; Marabese, M.; Pastorelli, R.; Broggini, M.; Brunelli, L. Glutaminase Inhibition on NSCLC Depends on Extracellular Alanine Exploitation. Cells 2020, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Sudderth, J.; Yang, C.; Mullen, A.R.; Jin, E.S.; Matés, J.M.; DeBerardinis, R.J. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8674–8679. [Google Scholar] [CrossRef] [Green Version]
- Singleton, D.C.; Dechaume, A.-L.; Murray, P.M.; Katt, W.P.; Baguley, B.C.; Leung, E.Y. Pyruvate anaplerosis is a mechanism of resistance to pharmacological glutaminase inhibition in triple-receptor negative breast cancer. BMC Cancer 2020, 20, 470. [Google Scholar] [CrossRef] [PubMed]
- Croze, M.L.; Soulage, C.O. Potential role and therapeutic interests of myo-inositol in metabolic diseases. Biochimie 2013, 95, 1811–1827. [Google Scholar] [CrossRef]
- Le, A.; Lane, A.N.; Hamaker, M.; Bose, S.; Gouw, A.; Barbi, J.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Zhang, H.; et al. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012, 15, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Otto, A.M. Metabolic Constants and Plasticity of Cancer Cells in a Limiting Glucose and Glutamine Microenvironment-A Pyruvate Perspective. Front. Oncol. 2020, 10, 596197. [Google Scholar] [CrossRef]
- Bansal, A.; Celeste Simon, M. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [Green Version]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of Glutathione in Cancer Progression and Chemoresistance. Oxid. Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [Green Version]
- Eklöf, V.; Wikberg, M.L.; Edin, S.; Dahlin, A.M.; Jonsson, B.-A.; Öberg, Å.; Rutegård, J.; Palmqvist, R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br. J. Cancer 2013, 108, 2153–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Cell line | Caco-2 | HCT116 | HT29 | SW480 | References |
|---|---|---|---|---|---|
| APC | c.4099C>T | Wild type | c.2557G>T | c.4012C>T | [27,28] |
| BRAF | Wild type | Wild type | c.1799T>A | Wild type | ATCC [27,29] |
| CTNNB1 | c.734G>C | c.133_ 135delTCT | Wild type | Wild type | [27,30] |
| KRAS | Wild type | c.38G>A | Wild type | c.35G>T | [27,28] |
| PIK3CA | Wild type | c.3140A>G | c.1345C>A | Wild type | [27] |
| SMAD4 | c.1051G>C | Wild type | c.931C>T | Wild type | [27,28] |
| TP53 | c.610G>T | Wild type | c.818G>A | c.818G>A | [27,31] |
| Histotype | Colorectal carcinoma | Colorectal carcinoma | Colorectal adenocarcinoma | Colorectal adenocarcinoma | [29] |
| Duke’s stage | / | D | C | B | [29] |
| Cell Lines | Cell Viability in -Gln Condition (% of Controls) | ±SD |
|---|---|---|
| Caco-2 | 55.02 | 5.04 |
| HCT116 | 62.01 | 3.25 |
| HT29 | 48.83 | 7.07 |
| SW480 | 49.34 | 0.57 |
| Cell Lines | Live | Apoptotic | Necrotic | ||||
|---|---|---|---|---|---|---|---|
| % of Cells ±SD | p-Value (vs. Cont) | % of Cells ±SD | p-Value (vs. Cont) | % of Cells ±SD | p-Value (vs. Cont) | ||
| Caco-2 | Cont | 70.93 ± 2.39 | 7.05 ± 2.48 | 22.02 ± 0.86 | |||
| -Gln | 41.36 ± 1.27 | <0.0001 | 2.20 ± 0.86 | 0.0025 | 56.44 ± 1.67 | <0.0001 | |
| HCT116 | Cont | 80.24 ± 1.15 | 1.84 ± 0.33 | 17.93 ± 1.28 | |||
| -Gln | 81.73 ± 1.46 | ns | 1.24 ± 0.21 | ns | 17.02 ± 1.48 | ns | |
| HT29 | Cont | 88.92 ± 0.49 | 2.32 ± 0.90 | 8.77 ± 0.40 | |||
| -Gln | 82.36 ± 0.56 | <0.0001 | 4.16 ± 0.88 | 0.0395 | 16.24 ± 0.63 | <0.0001 | |
| SW480 | Cont | 78.43 ± 4.00 | 2.66 ± 1.80 | 18.91 ± 2.22 | |||
| -Gln | 82.48 ± 2.60 | ns | 8.33 ± 2.83 | 0.0311 | 9.19 ± 0.23 | 0.0030 | |
| Cell Line | G0/G1 Phase | S Phase | G2/M Phase | PI | |||||
|---|---|---|---|---|---|---|---|---|---|
| % of Cells ±SD | p-Value (vs. Cont) | % of Cells ±SD | p-Value (vs. Cont) | % of Cells ±SD | p-Value (vs. Cont) | % of Cells ±SD | p-Value (vs. Cont) | ||
| Caco-2 | Cont | 64.71 ± 1.57 | 15.85 ± 0.84 | 14,76 ± 0.20 | 30.61 ± 1.04 | ||||
| -Gln | 76.12 ± 2.18 | <0.0001 | 13.06 ± 0.45 | ns | 8.41 ± 1.25 | 0.0017 | 21.46 ± 1.59 | 0.0059 | |
| HCT116 | Cont | 73.77 ± 2.71 | 5.79 ± 1.03 | 13.98 ± 1.01 | 19.77 ± 2.03 | ||||
| -Gln | 83.28 ± 1.75 | <0.0001 | 4.28 ± 0.79 | ns | 7.81 ± 0.79 | 0.0009 | 12.09 ± 0.31 | 0.0029 | |
| HT29 | Cont | 67.10 ± 3.02 | 12.16 ± 1.35 | 6.51 ± 0.62 | 18.66 ± 1.95 | ||||
| -Gln | 75.45 ± 1.64 | 0.0005 | 7.22 ± 2.42 | 0.0243 | 6.31 ± 1.40 | ns | 13.54 ± 1.67 | 0.0258 | |
| SW480 | Cont | 66.21 ± 4.11 | 15.49 ± 1.87 | 10.12 ± 2.08 | 25.61 ± 3.62 | ||||
| -Gln | 78.61 ± 0.86 | <0.0001 | 13.50 ± 0.14 | ns | 4.26 ± 0.65 | 0.0143 | 17.77 ± 0.79 | 0.0215 | |
| Metabolites | Caco-2 | HCT116 | HT29 | SW480 | ||||
|---|---|---|---|---|---|---|---|---|
| Fold Change -Gln/Cont | p-Value | Fold Change -Gln/Cont | p-Value | Fold Change -Gln/Cont | p-Value | Fold Change -Gln/Cont | p-Value | |
| Adenosine monophosphate | 0.60 | 0.0135 | 0.40 | ns | 0.48 | 0.0017 | 0.82 | ns |
| Aminomalonc acid | 1.62 | 0.0003 | 4.04 | 0.0012 | - | - | 3.96 | 0.0154 |
| Beta-Alanine | 0.36 | 0.0007 | 0.51 | 0.0054 | 0.43 | <0.0001 | 0.51 | 0.0016 |
| Beta-Glycerophosphoric acid | 1.02 | ns | 1.23 | ns | - | - | - | - |
| Cholesterol | 1.56 | 0.0169 | 0.98 | ns | 0.56 | ns | 0.87 | ns |
| Citric acid | 0.36 | <0.0001 | 0.85 | ns | 0.69 | 0.0029 | 0.96 | ns |
| Creatinine | 1.67 | 0.0059 | 1.40 | 0.0342 | - | - | - | - |
| D-Fructose | 0.80 | 0.0010 | 0.93 | ns | 1.75 | 0.0062 | - | - |
| D-Galactose | 1.53 | 0.0004 | 2.57 | 0.0021 | 1.50 | 0.0065 | 3.82 | 0.0042 |
| D-Glucose | 1.66 | 0.0011 | 2.47 | 0.0039 | 1.51 | 0.0049 | 4.11 | 0.0030 |
| D-Glucose 6-phosphate | 1.42 | ns | 1.31 | ns | 0.92 | ns | 1.42 | 0.0036 |
| D-Ribose 5-phosphate | 0.29 | 0.0010 | 0.77 | ns | 0.49 | ns | 1.56 | ns |
| Fumaric acid | 0.05 | ns | 0.69 | 0.0022 | 0.14 | <0.0001 | 0.53 | 0.0381 |
| Gamma-Aminobutyric acid | 0.19 | 0.0025 | 0.36 | 0.0210 | 1.08 | ns | 0.28 | 0.0199 |
| Glycerol 3-phosphate | 1.37 | 0.0256 | 1.01 | ns | 1.58 | <0.0001 | 1.95 | 0.0008 |
| Glycine | 3.56 | <0.0001 | 3.87 | <0.0001 | 8.44 | <0.0001 | 3.68 | 0.0002 |
| L-Alanine | 0.13 | 0.0070 | 0.46 | ns | 0.20 | 0.0431 | 0.71 | ns |
| L-Aspartic acid | 0.27 | 0.0039 | 0.29 | 0.0050 | - | - | 0.16 | 0.0091 |
| L-Glutamic acid | 0.13 | 0.0021 | 0.72 | 0.0109 | 0.31 | 0.0012 | 0.26 | 0.0270 |
| L-Isoluecine | 1.08 | ns | 2.27 | 0.0068 | - | - | 2.20 | 0.0193 |
| L-Lactic acid | 0.67 | 0.0039 | 0.30 | <0.0001 | 0.54 | 0.0001 | 0.92 | ns |
| L-Leucine | 0.29 | ns | 2.71 | 0.0011 | - | - | 2.50 | 0.0024 |
| L-Phenylalanine | 2.00 | 0.0064 | 4.08 | <0.0001 | - | - | 3.53 | 0.0016 |
| L-Proline | 0.97 | ns | 0.41 | 0.0068 | - | - | 0.86 | ns |
| L-Sorbose | 0.83 | 0.0114 | 1.06 | ns | 1.93 | 0.0004 | - | - |
| L-Threonine | 2.31 | <0.0001 | 5.07 | <0.0001 | 2.63 | 0.0121 | 2.99 | <0.0001 |
| L-Tryptophan | 1.34 | ns | 2.14 | <0.0001 | - | - | 1.99 | 0.0030 |
| L-Tyrosine | 2.69 | 0.0007 | 3.15 | 0.0006 | 2.42 | ns | 3.16 | 0.0014 |
| L-Valine | 1.17 | 0.0242 | 2.92 | 0.0093 | 1.63 | ns | 1.74 | 0.0244 |
| Malic acid | 0.13 | <0.0001 | 0.79 | ns | 0.14 | <0.0001 | 0.51 | ns |
| Mannitol | 1.99 | 0.0022 | 0.54 | 0.0006 | - | - | 1.60 | 0.0095 |
| Mannose 6-phosphate | 0.48 | 0.001 | 1.38 | ns | 0.81 | ns | 1.36 | 0.0180 |
| Myo-Inositol | 0.78 | 0.0093 | 0.65 | 0.0010 | 0.63 | 0.0001 | 0.38 | <0.0001 |
| Myo-Inositol 1-phosphate | 1.03 | ns | 0.86 | ns | 1.07 | ns | 0.92 | ns |
| Niacinamide | - | - | 0.86 | ns | - | - | 0.98 | ns |
| Pantothenic acid | 0.92 | ns | 0.90 | ns | 0.92 | ns | 1.42 | 0.0112 |
| Pyroglutamic acid | 0.10 | 0.0022 | 0.12 | 0.0333 | 0.13 | <0.0001 | 0.16 | 0.0011 |
| Serine | 7.14 | <0.0001 | 7.94 | <0.0001 | 13.71 | 0.03052 | 2.46 | 0.0003 |
| Taurine | 2.21 | ns | 0.90 | ns | - | - | 1.10 | ns |
| Uridine 5-monophosphate | 1.09 | ns | 0.42 | 0.0001 | 0.40 | ns | - | - |
| 5′Methylthioadenosine | 0.32 | 0.0498 | 1.07 | ns | 0.94 | ns | 0.88 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spada, M.; Piras, C.; Diana, G.; Leoni, V.P.; Frau, D.V.; Serreli, G.; Simbula, G.; Loi, R.; Noto, A.; Murgia, F.; et al. Glutamine Starvation Affects Cell Cycle, Oxidative Homeostasis and Metabolism in Colorectal Cancer Cells. Antioxidants 2023, 12, 683. https://doi.org/10.3390/antiox12030683
Spada M, Piras C, Diana G, Leoni VP, Frau DV, Serreli G, Simbula G, Loi R, Noto A, Murgia F, et al. Glutamine Starvation Affects Cell Cycle, Oxidative Homeostasis and Metabolism in Colorectal Cancer Cells. Antioxidants. 2023; 12(3):683. https://doi.org/10.3390/antiox12030683
Chicago/Turabian StyleSpada, Martina, Cristina Piras, Giulia Diana, Vera Piera Leoni, Daniela Virginia Frau, Gabriele Serreli, Gabriella Simbula, Roberto Loi, Antonio Noto, Federica Murgia, and et al. 2023. "Glutamine Starvation Affects Cell Cycle, Oxidative Homeostasis and Metabolism in Colorectal Cancer Cells" Antioxidants 12, no. 3: 683. https://doi.org/10.3390/antiox12030683
APA StyleSpada, M., Piras, C., Diana, G., Leoni, V. P., Frau, D. V., Serreli, G., Simbula, G., Loi, R., Noto, A., Murgia, F., Caria, P., & Atzori, L. (2023). Glutamine Starvation Affects Cell Cycle, Oxidative Homeostasis and Metabolism in Colorectal Cancer Cells. Antioxidants, 12(3), 683. https://doi.org/10.3390/antiox12030683










