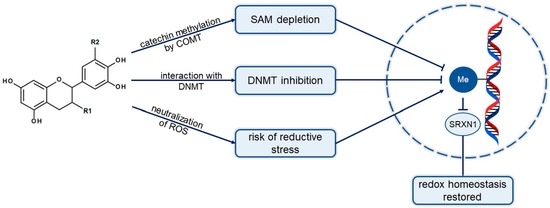
Beyond Antioxidant Activity: Redox Properties of Catechins May Affect Changes in the DNA Methylation Profile—The Example of SRXN1 Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selected Redox-Active Compounds
2.2. Cell Culture
2.3. Treatment of Cells with Selected Redox-Active Compounds
2.4. Total RNA Isolation
2.5. Microarray Analysis
2.6. Genomic DNA Isolation
2.7. Bisulfite Conversion
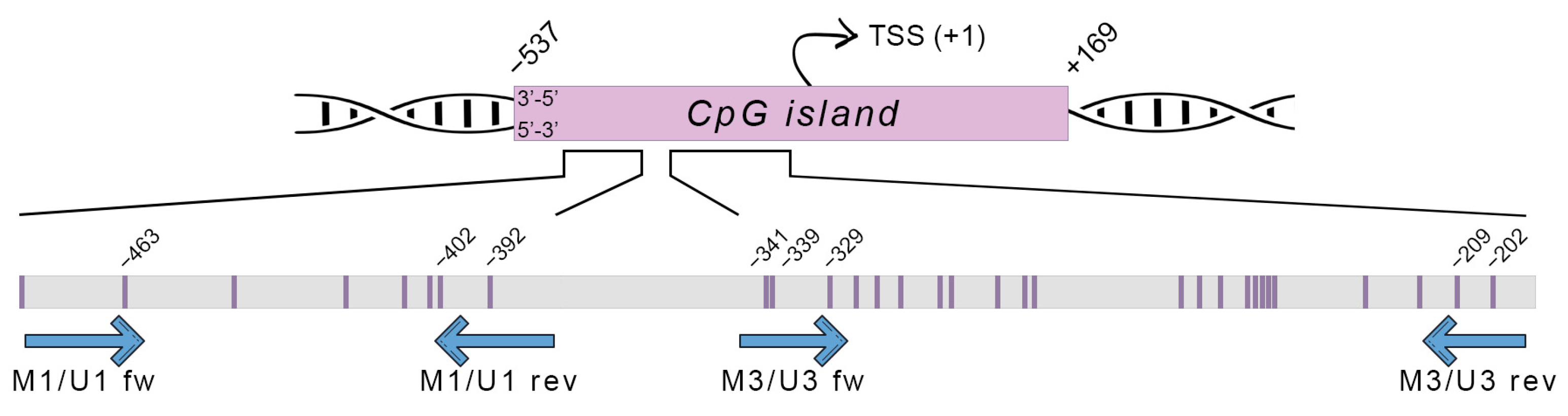
2.8. Prediction of CpG Islands and Primer Design for Methylation Analysis
2.9. Methylation-Specific PCR (MSP)
2.10. Methylation-Sensitive High-Resolution Melting (MS-HRM)
2.11. Quantitative Reverse Transcription PCR (RT-qPCR)
2.12. Statistical Analysis
3. Results
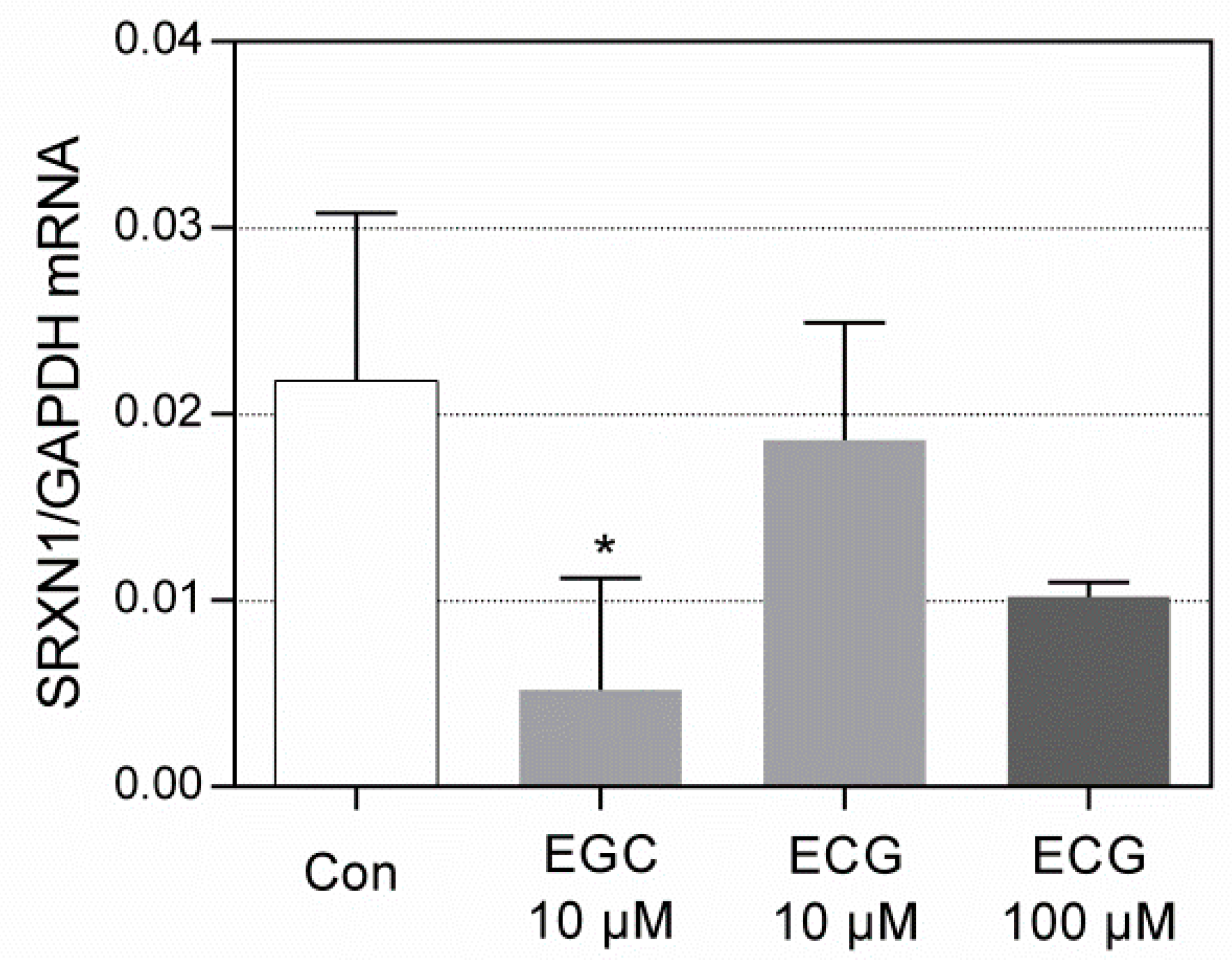
3.1. The Impact of Catechins on SRXN1 Expression
3.2. Methylation of SRXN1 Promoter
3.2.1. DNA Methylation within M1/U1 Region of SRXN1 Promoter Analyzed by MSP
3.2.2. DNA Methylation in Both M1/U1 and M3/U3 Regions of SRXN1 Promoter Analyzed by MS-HRM
3.3. RT-qPCR Analysis of SRXN1 Expression
3.4. The Search for Correlations between Investigated Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Durazzo, A.; Lucarini, M.; Souto, E.B.; Cicala, C.; Caiazzo, E.; Izzo, A.A.; Novellino, E.; Santini, A. Polyphenols: A Concise Overview on the Chemistry, Occurrence, and Human Health. Phyther. Res. 2019, 33, 2221–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grzesik, M.; Naparło, K.; Bartosz, G.; Sadowska-Bartosz, I. Antioxidant Properties of Catechins: Comparison with Other Antioxidants. Food Chem. 2018, 241, 480–492. [Google Scholar] [CrossRef]
- Yang, C.S.; Wang, X.; Lu, G.; Picinich, S.C. Cancer Prevention by Tea: Animal Studies, Molecular Mechanisms and Human Relevance. Nat. Rev. Cancer 2009, 9, 429–439. [Google Scholar] [CrossRef] [Green Version]
- Maiti, S.; Nazmeen, A.; Medda, N.; Patra, R.; Ghosh, T.K. Flavonoids Green Tea against Oxidant Stress and Inflammation with Related Human Diseases. Clin. Nutr. Exp. 2019, 24, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Reygaert, W.C. Green Tea Catechins: Their Use in Treating and Preventing Infectious Diseases. BioMed Res. Int. 2018, 2018, 9105261. [Google Scholar] [CrossRef]
- Mangels, D.R.; Mohler, E.R. Catechins as Potential Mediators of Cardiovascular Health. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Si, H.; Jia, Z.; Liu, D. Dietary Anti-Aging Polyphenols and Potential Mechanisms. Antioxidants 2021, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Kerimi, A.; Williamson, G. Differential Impact of Flavonoids on Redox Modulation, Bioenergetics, and Cell Signaling in Normal and Tumor Cells: A Comprehensive Review. Antioxidants Redox Signal. 2018, 29, 1633–1659. [Google Scholar] [CrossRef]
- Baranowska, M.; Suliborska, K.; Chrzanowski, W.; Kusznierewicz, B.; Namieśnik, J.; Bartoszek, A. The Relationship between Standard Reduction Potentials of Catechins and Biological Activities Involved in Redox Control. Redox Biol. 2018, 17, 355–366. [Google Scholar] [CrossRef]
- Yiannakopoulou, E.C. Targeting DNA Methylation with Green Tea Catechins. Pharmacology 2015, 95, 111–116. [Google Scholar] [CrossRef]
- Vidakovic, M.; Marinello, J.; Lahtela-Kakkonen, M.; Matulis, D.; Linkuvienė, V.; Michel, B.Y.; Navakauskienė, R.; Christodoulou, M.; Passarella, D.; Klimasauskas, S.; et al. New Insights into the Epigenetic Activities of Natural Compounds. OBM Genet. 2018, 131, 3022–3041. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kwak, J.; Choi, H.K.; Choi, K.C.; Kim, S.; Lee, J.; Jun, W.; Park, H.J.; Yoon, H.G. EGCG Suppresses Prostate Cancer Cell Growth Modulating Acetylation of Androgen Receptor by Anti-Histone Acetyltransferase Activity. Int. J. Mol. Med. 2012, 30, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (-)-Epigallocatechin-3-Gallate Reactivates Silenced Tumor Suppressor Genes, Cip1/P21 and P16INK4a, by Reducing DNA Methylation and Increasing Histones Acetylation in Human Skin Cancer Cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Cyr, A.R.; Domann, F.E. The Redox Basis of Epigenetic Modifications: From Mechanisms to Functional Consequences. Antioxid. Redox Signal. 2011, 15, 551–589. [Google Scholar] [CrossRef] [Green Version]
- Vanden Berghe, W. Epigenetic Impact of Dietary Polyphenols in Cancer Chemoprevention: Lifelong Remodeling of Our Epigenomes. Pharmacol. Res. 2012, 65, 565–576. [Google Scholar] [CrossRef]
- Lu, H.; Meng, X.; Yang, C.S. Enzymology of Methylation of Tea Catechins and Inhibition of Catechol-O-Methyltransferase by (-)-Epigallocatechin Gallate. Drug Metab. Dispos. 2003, 31, 572–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, J.L.; Shim, J.Y.; Zhu, B.T. Mechanisms for the Inhibition of DNA Methyltransferases by Tea Catechins and Bioflavonoids. Mol. Pharmacol. 2005, 68, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Liu, Y. DNA Methylation in Human Diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for Current Nutrigenetic, Nutrigenomic, and Nutriepigenetic Approaches for Precision Nutrition Involving the Prevention and Management of Chronic Diseases Associated with Obesity. J. Nutrigenet. Nutrigenomics 2017, 10, 43–62. [Google Scholar] [CrossRef]
- Jacob, C. Redox Signalling via the Cellular Thiolstat. Biochem. Soc. Trans. 2011, 39, 1247–1253. [Google Scholar] [CrossRef]
- Cox, A.G.; Winterbourn, C.C.; Hampton, M.B. Mitochondrial Peroxiredoxin Involvement in Antioxidant Defence and Redox Signalling. Biochem. J. 2010, 425, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczyńska, M.; Jakubek, P.; Bartoszek, A. More than Just Antioxidants: Redox-Active Components and Mechanisms Shaping Redox Signalling Network. Antioxidants 2022, 11, 2403. [Google Scholar] [CrossRef] [PubMed]
- Baranowska, M.; Koziara, Z.; Suliborska, K.; Chrzanowski, W.; Wormstone, M.; Namieśnik, J.; Bartoszek, A. Interactions between Polyphenolic Antioxidants Quercetin and Naringenin Dictate the Distinctive Redox-Related Chemical and Biological Behaviour of Their Mixtures. Sci. Rep. 2021, 11, 12282. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicka, G.T.; Hagen, T.M.; Tones, D.P. Glutathione in Food. J. Food Compos. Anal. 1989, 2, 327–337. [Google Scholar] [CrossRef]
- Jakubek, P.; Rajić, J.; Baranowska, M.; Vidaković, M.; Bartoszek, A.; Namieśnik, J. DNA Methylation Changes Induced by Redox-Active Compounds—Choosing the Right PCR-Based Method. Proceedings 2019, 11, 20. [Google Scholar] [CrossRef] [Green Version]
- Rajić, J.; Inic-Kanada, A.; Stein, E.; Dinić, S.; Schuerer, N.; Uskoković, A.; Ghasemian, E.; Mihailović, M.; Vidaković, M.; Grdović, N.; et al. Chlamydia Trachomatis Infection Is Associated with E-Cadherin Promoter Methylation, Downregulation of E-Cadherin Expression, and Increased Expression of Fibronectin and α-SMA—Implications for Epithelial-Mesenchymal Transition. Front. Cell. Infect. Microbiol. 2017, 7, 253. [Google Scholar] [CrossRef] [Green Version]
- Krzystek-Korpacka, M.; Hotowy, K.; Czapinska, E.; Podkowik, M.; Bania, J.; Gamian, A.; Bednarz-Misa, I. Serum Availability Affects Expression of Common House-Keeping Genes in Colon Adenocarcinoma Cell Lines: Implications for Quantitative Real-Time PCR Studies. Cytotechnology 2016, 68, 2503–2517. [Google Scholar] [CrossRef] [Green Version]
- Reuland, D.J.; Khademi, S.; Castle, C.J.; Irwin, D.C.; McCord, J.M.; Miller, B.F.; Hamilton, K.L. Upregulation of Phase II Enzymes through Phytochemical Activation of Nrf2 Protects Cardiomyocytes against Oxidant Stress. Free Radic. Biol. Med. 2013, 56, 102–111. [Google Scholar] [CrossRef]
- Suliborska, K. Określenie Mocy Przeciwutleniającej Wybranych Przeciwutleniaczy z Wykorzystaniem Metod Elektrochemicznych; Gdańsk University of Technology: Gdańsk, Poland, 2020. [Google Scholar]
- Martin-Benlloch, X.; Novodomska, A.; Jacquemin, D.; Davioud-Charvet, E.; Elhabiri, M. Iron(Iii) Coordination Properties of Ladanein, a Flavone Lead with a Broad-Spectrum Antiviral Activity. New J. Chem. 2018, 42, 8074–8087. [Google Scholar] [CrossRef]
- Friso, S.; Udali, S.; De Santis, D.; Choi, S.W. One-Carbon Metabolism and Epigenetics. Mol. Aspects Med. 2017, 54, 28–36. [Google Scholar] [CrossRef]
- Bheemanaik, S.; Reddy, Y.V.R.; Rao, D.N. Structure, Function and Mechanism of Exocyclic DNA Methyltransferases. Biochem. J. 2006, 399, 177–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients That Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Perán, E.; Cabezas-Herrera, J.; del Campo, L.S.; Rodríguez-López, J.N. Effects of Folate Cycle Disruption by the Green Tea Polyphenol Epigallocatechin-3-Gallate. Int. J. Biochem. Cell Biol. 2007, 39, 2215–2225. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Ferk, F.; Sterneder, S.; Setayesh, T.; Roth, S.; Kepcija, T.; Noorizadeh, R.; Rebhan, I.; Greunz, M.; Beckmann, J.; et al. EGCG Prevents High Fat Diet-Induced Changes in Gut Microbiota, Decreases of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Oxid. Med. Cell. Longev. 2017, 2017, 3079148. [Google Scholar] [CrossRef] [Green Version]
- Jyothidasan, A.; Sunny, S.; Murugesan, S.; Quiles, J.M.; Challa, A.K.; Dalley, B.; Cinghu, S.K.; Nanda, V.; Rajasekaran, N.S. Transgenic Expression of Nrf2 Induces a Pro-Reductive Stress and Adaptive Cardiac Remodeling in the Mouse. Genes 2022, 13, 1514. [Google Scholar] [CrossRef] [PubMed]
- Talebi, M.; Talebi, M.; Farkhondeh, T.; Mishra, G.; İlgün, S.; Samarghandian, S. New Insights into the Role of the Nrf2 Signaling Pathway in Green Tea Catechin Applications. Phyther. Res. 2021, 35, 3078–3112. [Google Scholar] [CrossRef]
- Chen, L.; Cao, H.; Xiao, J. Polyphenols: Absorption, Bioavailability, and Metabolomics; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 10, ISBN 9780128135723. [Google Scholar]
- Stalmach, A.; Mullen, W.; Steiling, H.; Williamson, G.; Lean, M.E.J.; Crozier, A. Absorption, Metabolism, and Excretion of Green Tea Flavan-3-Ols in Humans with an Ileostomy. Mol. Nutr. Food Res. 2010, 54, 323–334. [Google Scholar] [CrossRef]
- Yashin, A.; Nemzer, B.; Yashin, Y. Bioavailability of Tea Components. J. Food Res. 2012, 1, 281–290. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Romá-Mateo, C.; Pérez-Machado, G.; Peiró-Chova, L.; Pallardó, F.V. Role of Glutathione in the Regulation of Epigenetic Mechanisms in Disease. Free Radic. Biol. Med. 2017, 112, 36–48. [Google Scholar] [CrossRef]
- Forshaw, T.E.; Reisz, J.A.; Nelson, K.J.; Gumpena, R.; Lawson, J.R.; Jönsson, T.J.; Wu, H.; Clodfelter, J.E.; Johnson, L.C.; Furdui, C.M.; et al. Specificity of Human Sulfiredoxin for Reductant and Peroxiredoxin Oligomeric State. Antioxidants 2021, 10, 946. [Google Scholar] [CrossRef]
- Chan, K.Y.; Zhang, L.; Zuo, Z. Intestinal Efflux Transport Kinetics of Green Tea Catechins in Caco-2 Monolayer Model. J. Pharm. Pharmacol. 2010, 59, 395–400. [Google Scholar] [CrossRef]
- Stalmach, A.; Troufflard, S.; Serafini, M.; Crozier, A. Absorption, Metabolism and Excretion of Choladi Green Tea Flavan-3-Ols by Humans. Mol. Nutr. Food Res. 2009, 53, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ni, X. ROS-Mediated DNA Methylation Pattern Alterations in Carcinogenesis. Curr. Drug Targets 2015, 16, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Fu, L.; Jung, Y.; Lo Conte, M.; Lawson, J.R.; Lowther, W.T.; Sun, R.; Liu, K.; Yang, J.; Carroll, K.S. Chemical Proteomics Reveals New Targets of Cysteine Sulfinic Acid Reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [Google Scholar] [CrossRef] [PubMed]





| Compound | GSH | C | EC | EGC | ECG | EGCG | Ref. |
|---|---|---|---|---|---|---|---|
| Structure | Tripeptide | Parent compound | Epimer of C | Epimer of GC | Ester of EC with gallate | Ester of EGC with gallate | [9] |
| Catechol moiety | 0 | 1 | 1 | 0 | 1 | 0 | |
| Pyrogallol moiety | 0 | 0 | 0 | 1 | 0 | 1 | |
| Galloyl moiety | 0 | 0 | 0 | 0 | 1 | 1 | |
| Cell-free systems | |||||||
| Electrochemical parameters | |||||||
| E0 at 37 °C (V) | 0.310 ± 0.003 | 0.281 ± 0.008 | 0.277 ± 0.005 | 0.287 ± 0.003 | 0.098 ± 0.002 | 0.104 ± 0.002 | [9] |
| Ep,a + ɛ at 25 °C (V) | 0.984 ± 0.000 | 0.273 ± 0.000 | 0.260 ± 0.007 | 0.140 ± 0.007 | 0.230 ± 0.007 | 0.120 ± 0.009 | [29] |
| Ip,a at 25 °C (µA) | 2.60 ± 0.17 | 5.40 ± 0.38 | 8.10 ± 0.28 | 12.60 ± 0.26 | 5.70 ± 1.06 | 6.30 ± 0.63 | |
| Qox. at 25 °C (µC·cm−2) | 2.6 ± 0.08 | 5.20 ± 0.05 | 6.70 ± 0.10 | 3.90 ± 0.04 | 3.70 ± 0.65 | 4.70 ± 0.35 | |
| AOE (µJ·cm−2) | 233.3 ± 3.50 | 184.40 ± 4.42 | 227.00 ± 1.40 | 65.50 ± 0.32 | 113.80 ± 13.83 | 119.30 ± 7.86 | |
| t (s) | 6.6 ± 0.0 | 4.0 ± 0.0 | 4.2 ± 0.0 | 2.9 ± 0.0 | 3.9 ± 0.0 | 4.8 ± 0.0 | |
| AOP (µW·cm−2) | 35.4 ± 0.53 | 46.10 ± 1.10 | 54.10 ± 0.34 | 22.60 ± 0.11 | 29.20 ± 3.55 | 24.90 ± 1.64 | |
| Antioxidant activity by spectrophotometric tests | |||||||
| Fe(III) chelation, pFe pH 7.0 | - | 17.6 * | - | 23.1 | 23.2 | 23.4 | [30] |
| FRAP (mol TE/mol), pH 3.6 | 0.01 ± 0.00 | 0.79 ± 0.00 | 0.92 ± 0.00 | 1.03 ± 0.01 | 2.34 ± 0.01 | 2.21 ± 0.01 | [2] |
| Spectro. test | Antioxidant activity, n10 value at 37 °C | ||||||
| ABTS | 1.52 ± 0.02 | 5.44 ± 0.07 | 5.44 ± 0.11 | 5.98 ± 0.10 | 7.76 ± 0.11 | 8.33 ± 0.09 | [9] |
| DPPH | 0.01 | 2.15 ± 0.03 | 2.20 ± 0.03 | 3.02 ± 0.03 | 5.48 ± 0.07 | 6.16 ± 0.08 | |
| FC | 0.39 | 1.96 ± 0.02 | 1.83 ± 0.03 | 1.50 ± 0.02 | 2.66 ± 0.02 | 2.31 ± 0.03 | |
| Protection against oxidation a/fluorescein bleaching b expressed as IC50 (µM) | |||||||
| DHR123 a | 14.07 ± 0.36 | 0.81 ± 0.07 | 1.36 ± 0.04 | 1.21 ± 0.05 | 1.08 ± 0.04 | 1.15 ± 0.00 | [2] |
| NaOCl b | 3.52 ± 0.05 | 0.34 ± 0.00 | 0.51 ± 0.03 | 0.25 ± 0.03 | 0.31 ± 0.01 | 0.36 ± 0.01 | |
| AAPH b | 15.44 ± 0.01 | 0.67 ± 0.04 | 1.08 ± 0.06 | 0.66 ± 0.03 | 1.87 ± 0.07 | 2.16 ± 0.17 | |
| Cell culture | |||||||
| Conc. | Cell growth (% of control) at 37 °C, 24 h | ||||||
| 1 µM | 94.3 ± 5.8 | 109.7 ± 10.8 | 127.8 ± 10.2 | 97.7 ± 9.3 | 110.5 ± 11.3 | 94.3 ± 5.8 | [9] |
| 10 µM | 90.8 ± 4.3 | 96.1 ± 9.9 | 106.5 ± 7.8 | 92.9 ± 10.4 | 101.2 ± 10.6 | 90.8 ± 4.3 | |
| Conc. | Cellular antioxidant activity, CAA value at 37 °C, 1 h | ||||||
| 1 µM | 7 ± 7 | 11 ± 8 | 23 ± 3 | 17 ± 5 | 35 ± 9 | 22 ± 10 | [9] |
| 10 µM | 22 ± 6 | 21 ± 10 | 20 ± 7 | 23 ± 9 | 35 ± 9 | 25 ± 10 | |
| Conc. | Genotoxicity % at 37 °C, 24 h | ||||||
| 1 µM | 6.75 ± 0.88 | 3.58 ± 0.29 | 3.07 ± 0.23 | 3.54 ± 0.05 | 2.92 ± 0.69 | 2.59 ± 0.81 | [9] |
| 10 µM | 6.35 ± 2.01 | 3.77 ± 0.62 | 3.04 ± 1.00 | 3.02 ± 0.37 | 2.55 ± 0.35 | 2.66 ± 0.63 | |
| Conc. | Fold regulation of gene expression at 37 °C, 24 h | ||||||
| 1 µM | 1.8 | 1.7 | −1.1 | −2.0 | 1.2 | 1.0 | [9] |
| 10 µM | 1.4 | 1.0 | −1.4 | −2.5 | −1.6 | −1.8 | |
| Conc. | Fold change of % DNA methylation at 37 °C, 24 h | ||||||
| MSP M1/U1 | Current study | ||||||
| 1 µM | 0.802 ± 0.136 | 1.005 ± 0.047 | 0.687 ± 0.053 | 1.139 ± 0.202 | 1.109 ± 0.317 | 0.671 ± 0.162 | |
| 10 µM | 1.159 ± 0.131 | 1.084 ± 0.069 | 0.900 ± 0.024 | 1.074 ± 0.213 | 1.204 ± 0.052 | 0.915 ± 0.269 | |
| Conc. | MS-HRM M1/U1 | ||||||
| 1 µM | 1.199 ± 0.471 | 0.498 ± 0.054 | 0.419 ± 0.051 | 0.547 ± 0.167 | 0.478 ± 0.032 | 0.569 ± 0.106 | |
| 10 µM | 1.009 ± 0.208 | 0.330 ± 0.088 | 0.236 ± 0.217 | 1.768 ± 0.171 | 1.889 ± 0.029 | 0.269 ± 0.126 | |
| Conc. | MS-HRM M3/U3 | ||||||
| 1 µM | 1.163 ± 0.080 | 0.811 ± 0.219 | 0.809 ± 0.034 | 0.918 ± 0.073 | 0.816 ± 0.084 | 0.882 ± 0.082 | |
| 10 µM | 1.191 ± 0.084 | 0.797 ± 0.070 | 0.685 ± 0.037 | 1.331 ± 0.130 | 1.410 ± 0.227 | 0.920 ± 0.236 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakubek, P.; Rajić, J.; Kuczyńska, M.; Suliborska, K.; Heldt, M.; Dziedziul, K.; Vidaković, M.; Namieśnik, J.; Bartoszek, A. Beyond Antioxidant Activity: Redox Properties of Catechins May Affect Changes in the DNA Methylation Profile—The Example of SRXN1 Gene. Antioxidants 2023, 12, 754. https://doi.org/10.3390/antiox12030754
Jakubek P, Rajić J, Kuczyńska M, Suliborska K, Heldt M, Dziedziul K, Vidaković M, Namieśnik J, Bartoszek A. Beyond Antioxidant Activity: Redox Properties of Catechins May Affect Changes in the DNA Methylation Profile—The Example of SRXN1 Gene. Antioxidants. 2023; 12(3):754. https://doi.org/10.3390/antiox12030754
Chicago/Turabian StyleJakubek, Patrycja, Jovana Rajić, Monika Kuczyńska, Klaudia Suliborska, Mateusz Heldt, Karol Dziedziul, Melita Vidaković, Jacek Namieśnik, and Agnieszka Bartoszek. 2023. "Beyond Antioxidant Activity: Redox Properties of Catechins May Affect Changes in the DNA Methylation Profile—The Example of SRXN1 Gene" Antioxidants 12, no. 3: 754. https://doi.org/10.3390/antiox12030754
APA StyleJakubek, P., Rajić, J., Kuczyńska, M., Suliborska, K., Heldt, M., Dziedziul, K., Vidaković, M., Namieśnik, J., & Bartoszek, A. (2023). Beyond Antioxidant Activity: Redox Properties of Catechins May Affect Changes in the DNA Methylation Profile—The Example of SRXN1 Gene. Antioxidants, 12(3), 754. https://doi.org/10.3390/antiox12030754