Overexpression of Mitochondrial Catalase within Adipose Tissue Does Not Confer Systemic Metabolic Protection against Diet-Induced Obesity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA Isolation
2.3. RT Profiler qPCR Arrays
2.4. Quantitative PCR
2.5. Insulin Stimulation of Adipose Tissue
2.6. Protein Isolation
2.7. ELISA
2.8. Catalase Activity Assay
2.9. Glucose Tolerance Test
2.10. Insulin Tolerance Test
2.11. Triglyceride Assay
2.12. H & E Staining
2.13. Adipogenic Profiling
2.13.1. Isolation of Bone Marrow Cells
2.13.2. Adipogenesis Assay
2.14. Statistical Analysis
3. Results
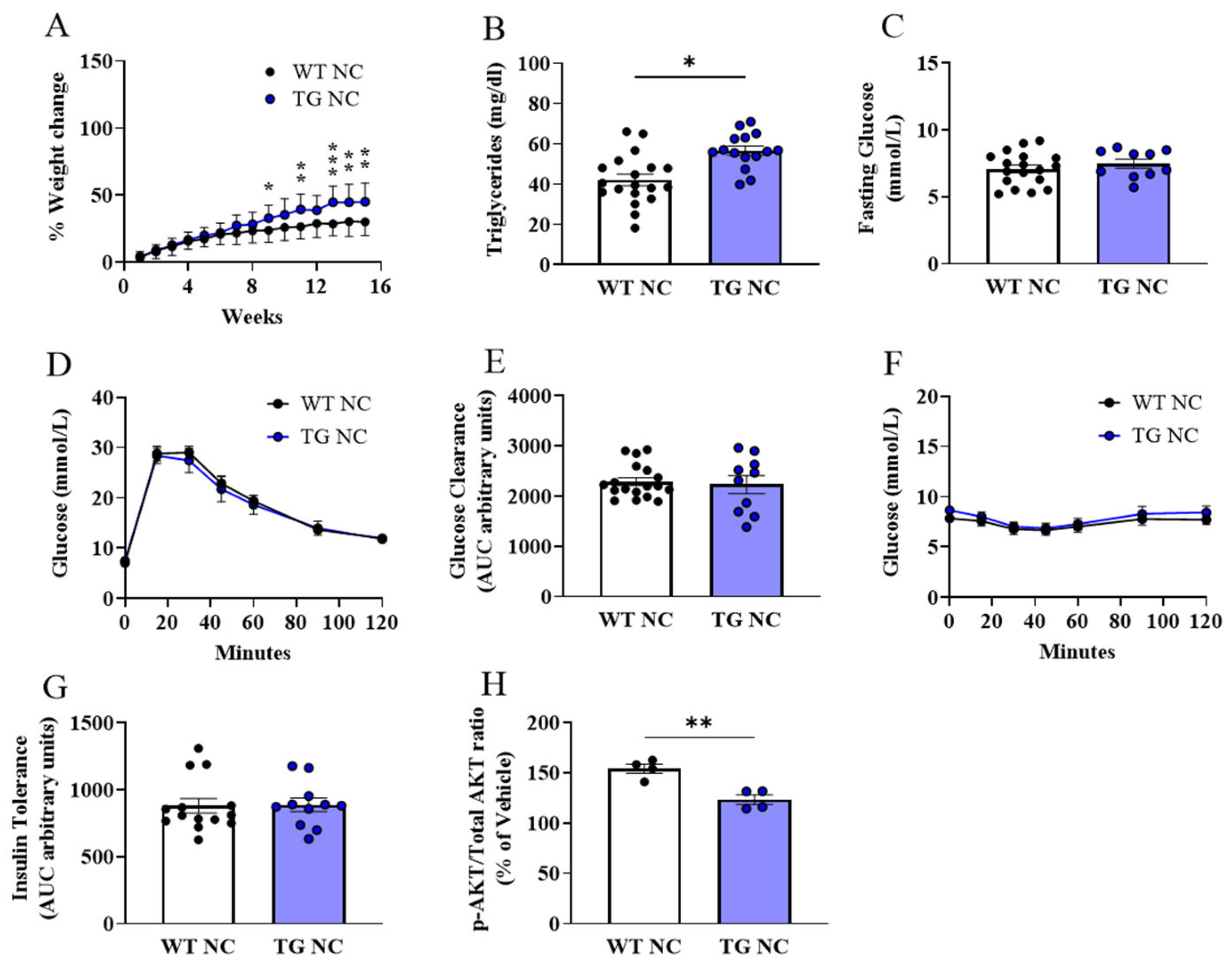
3.1. Metabolic Characterisation of AdipoQ-mCAT TG Mice
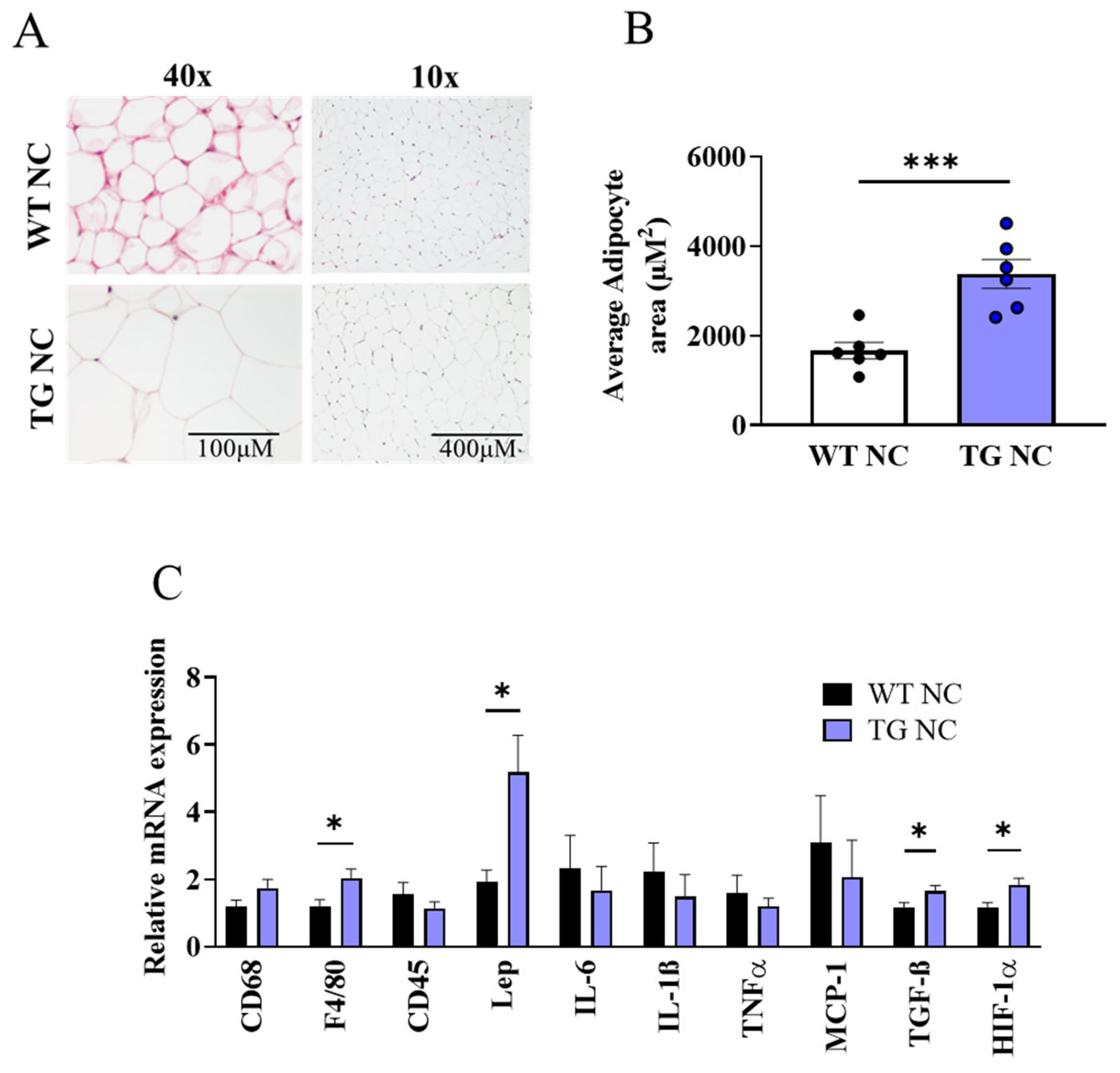
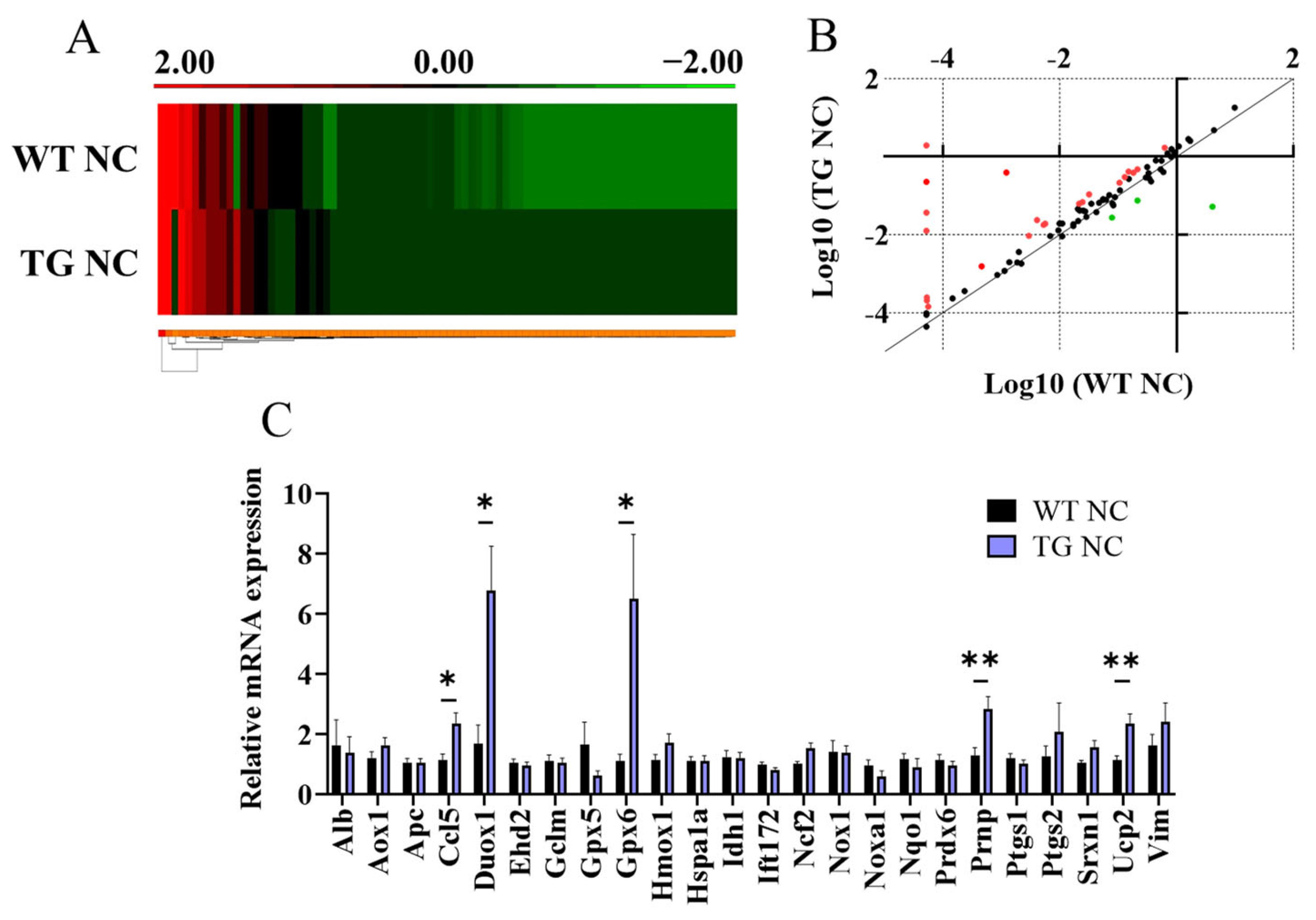
3.2. Characterisation of Adipose Tissue Changes in AdipoQ-mCAT TG vs. WT Mice Fed Normal Chow
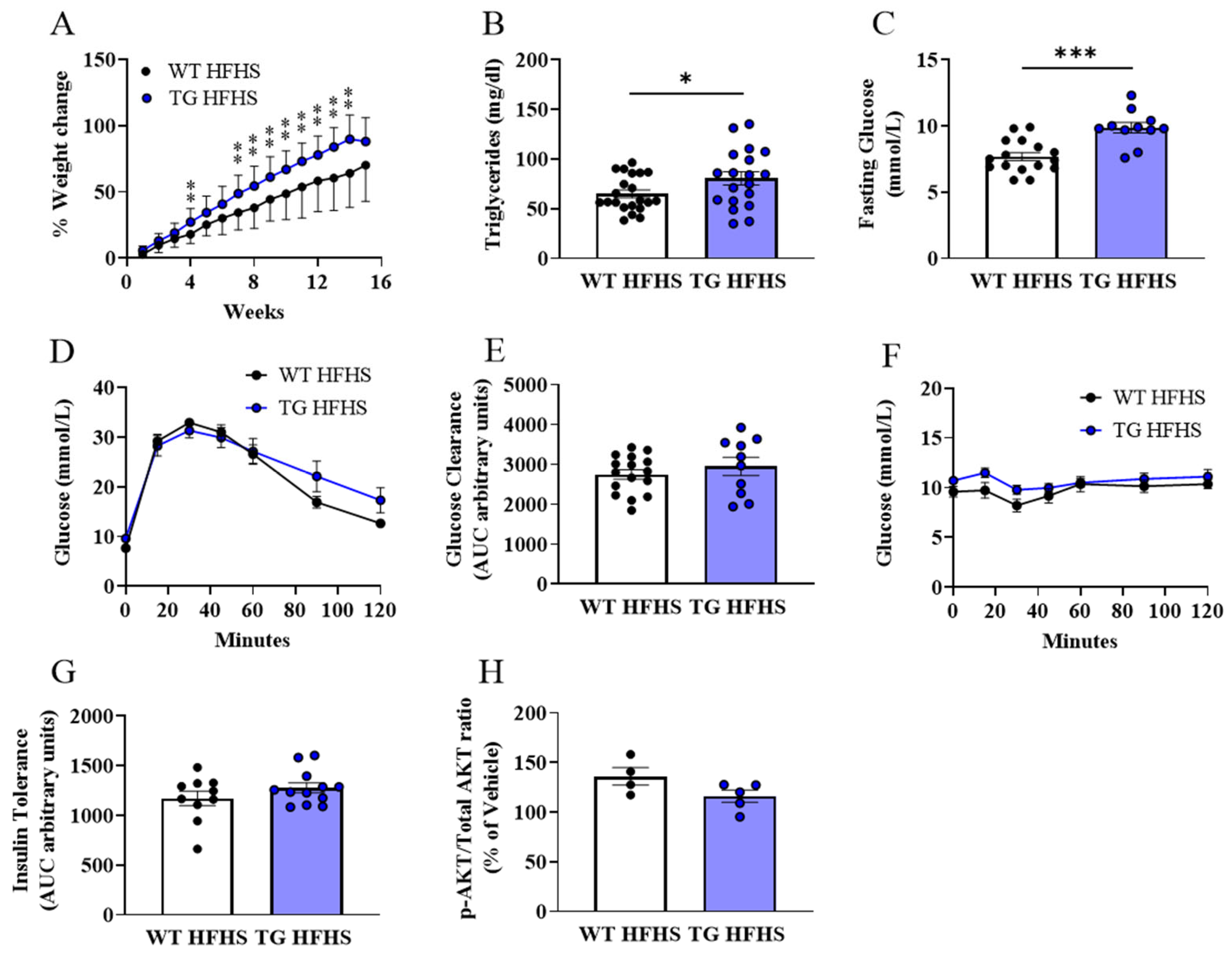
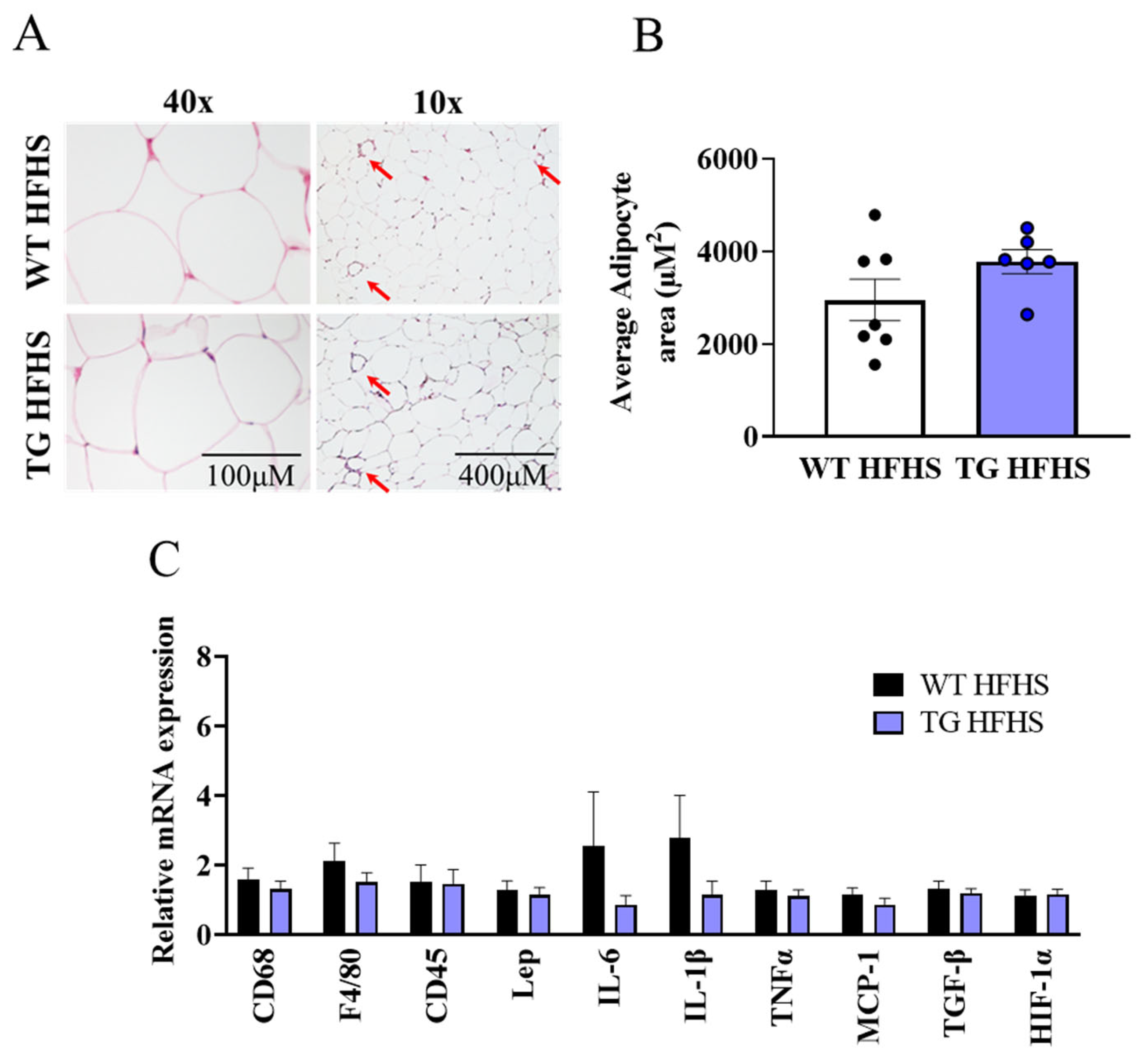
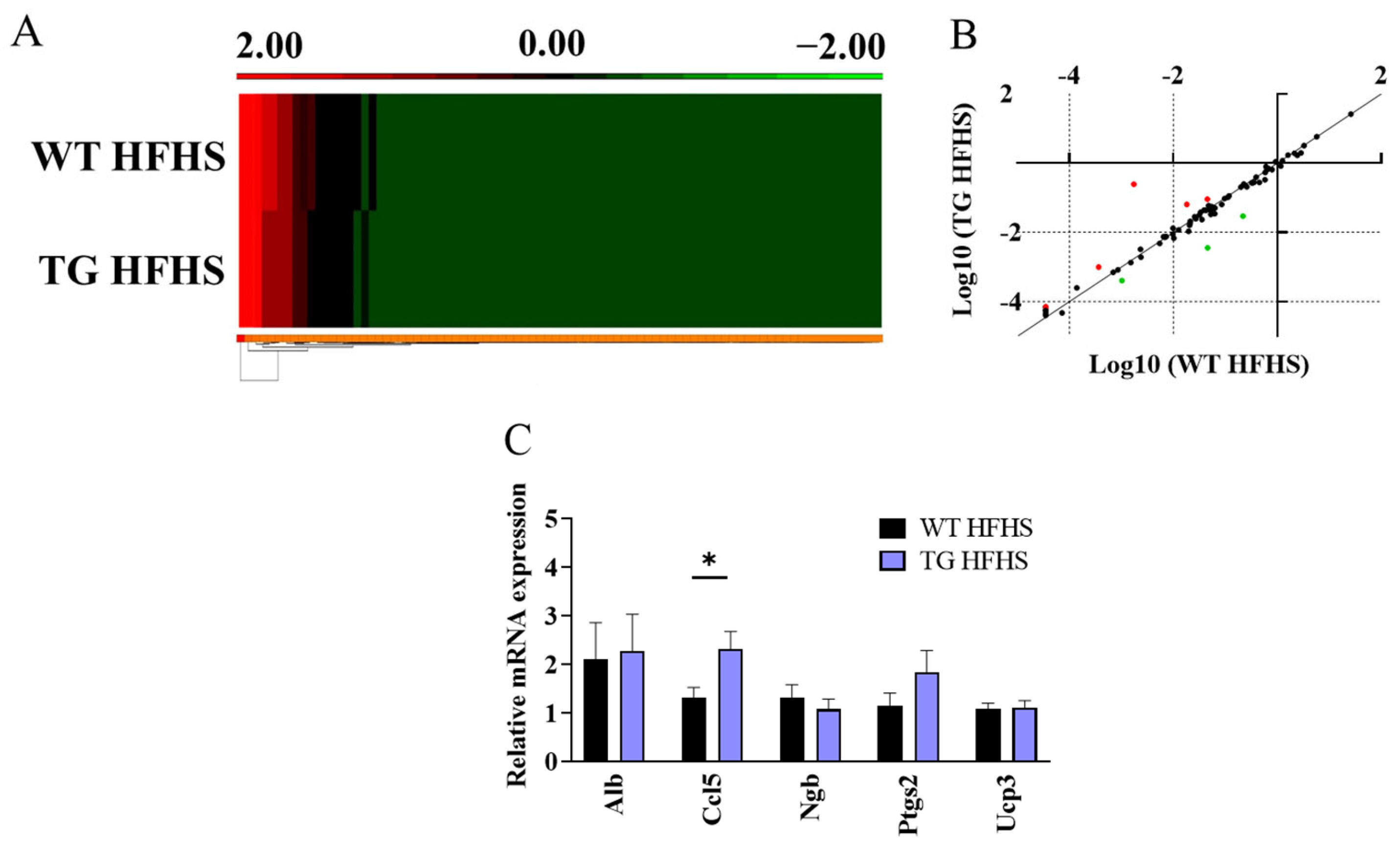
3.3. Obesogenic Diet-Induced Adipose Tissue Changes in AdipoQ-mCAT TG vs. WT Mice Fed HFHS Diet
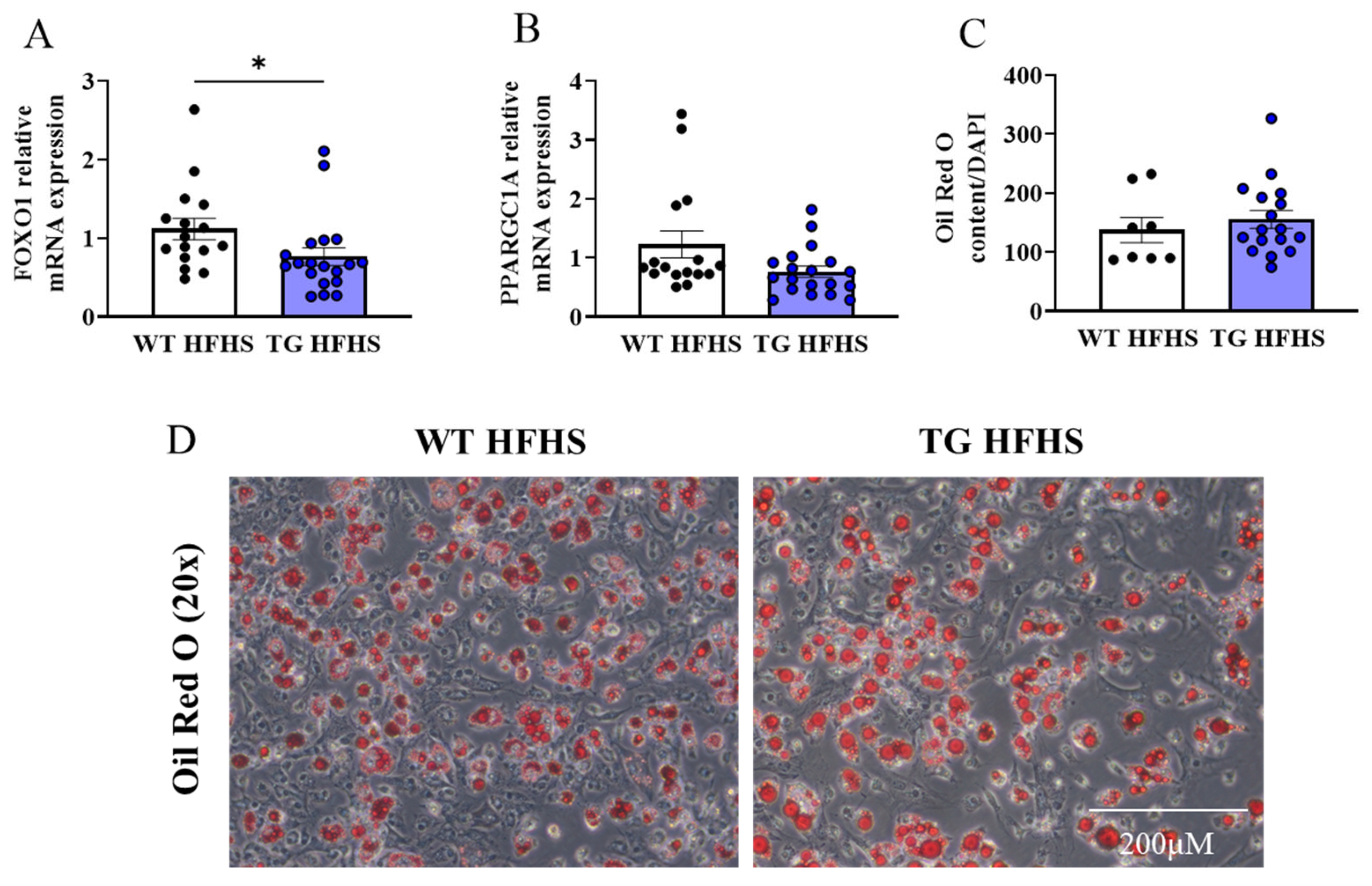
3.4. Adipogenic Profiling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 22 October 2022).
- Abbasi, F.; Brown, B.W., Jr.; Lamendola, C.; McLaughlin, T.; Reaven, G.M. Relationship between obesity, insulin resistance, and coronary heart disease risk. J. Am. Coll. Cardiol. 2002, 40, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Guh, D.P.; Zhang, W.; Bansback, N.; Amarsi, Z.; Birmingham, C.L.; Anis, A.H. The incidence of co-morbidities related to obesity and overweight: A systematic review and meta-analysis. BMC Public Health 2009, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.S.; Ning, H.; Wilkins, J.T.; Allen, N.; Carnethon, M.; Berry, J.D.; Sweis, R.N.; Lloyd-Jones, D.M. Association of Body Mass Index With Lifetime Risk of Cardiovascular Disease and Compression of Morbidity. JAMA Cardiol. 2018, 3, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Batchelor, B. Adipose tissue cellularity in human obesity. Clin. Endocrinol. Metab. 1976, 5, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Knittle, J.L. Cellularity of obese and nonobese human adipose tissue. Fed. Proc. 1970, 29, 1516–1521. [Google Scholar]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Strissel, K.J.; Stancheva, Z.; Miyoshi, H.; Perfield, J.W., 2nd; DeFuria, J.; Jick, Z.; Greenberg, A.S.; Obin, M.S. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes 2007, 56, 2910–2918. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2017, 114, 1752–1761. [Google Scholar] [CrossRef]
- Vávrová, L.; Kodydková, J.; Zeman, M.; Dušejovská, M.; Macášek, J.; Staňková, B.; Tvrzická, E.; Žák, A. Altered Activities of Antioxidant Enzymes in Patients with Metabolic Syndrome. Obes. Facts 2013, 6, 39–47. [Google Scholar] [CrossRef]
- Albuali, W.H. Evaluation of oxidant-antioxidant status in overweight and morbidly obese Saudi children. World J. Clin. Pediatr. 2014, 3, 6–13. [Google Scholar] [CrossRef]
- Mohseni, R.; Sadeghabadi, Z.A.; Goodarzi, M.T.; Teimouri, M.; Nourbakhsh, M.; Azar, M.R. Evaluation of Mn-superoxide dismutase and catalase gene expression in childhood obesity: Its association with insulin resistance. J. Pediatr. Endocrinol. Metab. 2018, 31, 727–732. [Google Scholar] [CrossRef]
- Kataja-Tuomola, M.; Sundell, J.R.; Männistö, S.; Virtanen, M.J.; Kontto, J.; Albanes, D.; Virtamo, J. Effect of alpha-tocopherol and beta-carotene supplementation on the incidence of type 2 diabetes. Diabetologia 2008, 51, 47–53. [Google Scholar] [CrossRef]
- Song, Y.; Cook, N.R.; Albert, C.M.; Van Denburgh, M.; Manson, J.E. Effects of vitamins C and E and beta-carotene on the risk of type 2 diabetes in women at high risk of cardiovascular disease: A randomized controlled trial. Am. J. Clin. Nutr. 2009, 90, 429–437. [Google Scholar] [CrossRef]
- Wilson-Fritch, L.; Nicoloro, S.; Chouinard, M.; Lazar, M.A.; Chui, P.C.; Leszyk, J.; Straubhaar, J.; Czech, M.P.; Corvera, S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004, 114, 1281–1289. [Google Scholar] [CrossRef]
- Vernochet, C.; Damilano, F.; Mourier, A.; Bezy, O.; Mori, M.A.; Smyth, G.; Rosenzweig, A.; Larsson, N.G.; Kahn, C.R. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014, 28, 4408–4419. [Google Scholar] [CrossRef]
- Schriner, S.E.; Linford, N.J.; Martin, G.M.; Treuting, P.; Ogburn, C.E.; Emond, M.; Coskun, P.E.; Ladiges, W.; Wolf, N.; Van Remmen, H.; et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 2005, 308, 1909–1911. [Google Scholar] [CrossRef]
- Dai, D.-F.; Santana, L.F.; Vermulst, M.; Tomazela, D.M.; Emond, M.J.; MacCoss, M.J.; Gollahon, K.; Martin, G.M.; Loeb, L.A.; Ladiges, W.C.; et al. Overexpression of Catalase Targeted to Mitochondria Attenuates Murine Cardiac Aging. Circulation 2009, 119, 2789–2797. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, J.S.; Alves, T.; Ladiges, W.; Rabinovitch, P.S.; Jurczak, M.J.; Choi, C.S.; Shulman, G.I.; Samuel, V.T. Mitochondrial-Targeted Catalase Protects Against High-Fat Diet-Induced Muscle Insulin Resistance by Decreasing Intramuscular Lipid Accumulation. Diabetes 2017, 66, 2072–2081. [Google Scholar] [CrossRef]
- Sverdlov, A.L.; Elezaby, A.; Behring, J.B.; Bachschmid, M.M.; Luptak, I.; Tu, V.H.; Siwik, D.A.; Miller, E.J.; Liesa, M.; Shirihai, O.S.; et al. High fat, high sucrose diet causes cardiac mitochondrial dysfunction due in part to oxidative post-translational modification of mitochondrial complex II. J. Mol. Cell. Cardiol. 2015, 78, 165–173. [Google Scholar] [CrossRef]
- Sverdlov, A.L.; Elezaby, A.; Qin, F.; Behring, J.B.; Luptak, I.; Calamaras, T.D.; Siwik, D.A.; Miller, E.J.; Liesa, M.; Shirihai, O.S.; et al. Mitochondrial Reactive Oxygen Species Mediate Cardiac Structural, Functional, and Mitochondrial Consequences of Diet-Induced Metabolic Heart Disease. J. Am. Heart Assoc. 2016, 5, e002555. [Google Scholar] [CrossRef] [PubMed]
- Galarraga, M.; Campión, J.; Muñoz-Barrutia, A.; Boqué, N.; Moreno, H.; Martínez, J.A.; Milagro, F.; Ortiz-de-Solórzano, C. Adiposoft: Automated software for the analysis of white adipose tissue cellularity in histological sections. J. Lipid Res. 2012, 53, 2791–2796. [Google Scholar] [CrossRef] [PubMed]
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H.; Arden, K.C.; Accili, D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 2003, 4, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Bhandari, C.; Agnihotri, N. Dietary n-3 and n-6 polyunsaturated fatty acids differentially modulate the adiponectin and leptin mediated major signaling pathways in visceral and subcutaneous white adipose tissue in high fat diet induced obesity in Wistar rats. Nutr. Res. 2023, 110, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef]
- Han, Y.H.; Buffolo, M.; Pires, K.M.; Pei, S.; Scherer, P.E.; Boudina, S. Adipocyte-Specific Deletion of Manganese Superoxide Dismutase Protects From Diet-Induced Obesity Through Increased Mitochondrial Uncoupling and Biogenesis. Diabetes 2016, 65, 2639–2651. [Google Scholar] [CrossRef]
- Choi, M.J.; Jung, S.-B.; Lee, S.E.; Kang, S.G.; Lee, J.H.; Ryu, M.J.; Chung, H.K.; Chang, J.Y.; Kim, Y.K.; Hong, H.J.; et al. An adipocyte-specific defect in oxidative phosphorylation increases systemic energy expenditure and protects against diet-induced obesity in mouse models. Diabetologia 2020, 63, 837–852. [Google Scholar] [CrossRef]
- Petrus, P.; Fernandez, T.L.; Kwon, M.M.; Huang, J.L.; Lei, V.; Safikhan, N.S.; Karunakaran, S.; O’Shannessy, D.J.; Zheng, X.; Catrina, S.-B.; et al. Specific loss of adipocyte CD248 improves metabolic health via reduced white adipose tissue hypoxia, fibrosis and inflammation. EBioMedicine 2019, 44, 489–501. [Google Scholar] [CrossRef]
- Franz, M.J.; Boucher, J.L.; Rutten-Ramos, S.; VanWormer, J.J. Lifestyle Weight-Loss Intervention Outcomes in Overweight and Obese Adults with Type 2 Diabetes: A Systematic Review and Meta-Analysis of Randomized Clinical Trials. J. Acad. Nutr. Diet. 2015, 115, 1447–1463. [Google Scholar] [CrossRef]
- Swinburn, B.A.; Metcalf, P.A.; Ley, S.J. Long-Term (5-Year) Effects of a Reduced-Fat Diet Intervention in Individuals With Glucose Intolerance. Diabetes Care 2001, 24, 619–624. [Google Scholar] [CrossRef]
- Ngo, D.T.M.; Sverdlov, A.L.; Karki, S.; Macartney-Coxson, D.; Stubbs, R.S.; Farb, M.G.; Carmine, B.; Hess, D.T.; Colucci, W.S.; Gokce, N. Oxidative modifications of mitochondrial complex II are associated with insulin resistance of visceral fat in obesity. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E168–E177. [Google Scholar] [CrossRef]
- Anderson, E.J.; Lustig, M.E.; Boyle, K.E.; Woodlief, T.L.; Kane, D.A.; Lin, C.-T.; Price, J.W., III; Kang, L.; Rabinovitch, P.S.; Szeto, H.H.; et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Investig. 2009, 119, 573–581. [Google Scholar] [CrossRef]
- Lark, D.S.; Kang, L.; Lustig, M.E.; Bonner, J.S.; James, F.D.; Neufer, P.D.; Wasserman, D.H. Enhanced mitochondrial superoxide scavenging does not improve muscle insulin action in the high fat-fed mouse. PLoS ONE 2015, 10, e0126732. [Google Scholar] [CrossRef]
- Paglialunga, S.; Ludzki, A.; Root-McCaig, J.; Holloway, G.P. In adipose tissue, increased mitochondrial emission of reactive oxygen species is important for short-term high-fat diet-induced insulin resistance in mice. Diabetologia 2015, 58, 1071–1080. [Google Scholar] [CrossRef]
- Coudray, C.; Fouret, G.; Lambert, K.; Ferreri, C.; Rieusset, J.; Blachnio-Zabielska, A.; Lecomte, J.; Ebabe Elle, R.; Badia, E.; Murphy, M.P.; et al. A mitochondrial-targeted ubiquinone modulates muscle lipid profile and improves mitochondrial respiration in obesogenic diet-fed rats. Br. J. Nutr. 2016, 115, 1155–1166. [Google Scholar] [CrossRef]
- Marín-Royo, G.; Rodríguez, C.; Le Pape, A.; Jurado-López, R.; Luaces, M.; Antequera, A.; Martínez-González, J.; Souza-Neto, F.V.; Nieto, M.L.; Martínez-Martínez, E.; et al. The role of mitochondrial oxidative stress in the metabolic alterations in diet-induced obesity in rats. FASEB J. 2019, 33, 12060–12072. [Google Scholar] [CrossRef]
- Jeong, E.M.; Chung, J.; Liu, H.; Go, Y.; Gladstein, S.; Farzaneh-Far, A.; Lewandowski, E.D.; Dudley, S.C., Jr. Role of Mitochondrial Oxidative Stress in Glucose Tolerance, Insulin Resistance, and Cardiac Diastolic Dysfunction. J. Am. Heart Assoc. 2016, 5, e003046. [Google Scholar] [CrossRef]
- Fink, B.; Coppey, L.; Davidson, E.; Shevalye, H.; Obrosov, A.; Chheda, P.R.; Kerns, R.; Sivitz, W.; Yorek, M. Effect of mitoquinone (Mito-Q) on neuropathic endpoints in an obese and type 2 diabetic rat model. Free Radic. Res. 2020, 54, 311–318. [Google Scholar] [CrossRef]
- Rindler, P.M.; Cacciola, A.; Kinter, M.; Szweda, L.I. Catalase-dependent H2O2 consumption by cardiac mitochondria and redox-mediated loss in insulin signaling. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H1091–H1096. [Google Scholar] [CrossRef]
- Merry, T.L.; Tran, M.; Stathopoulos, M.; Wiede, F.; Fam, B.C.; Dodd, G.T.; Clarke, I.; Watt, M.J.; Andrikopoulos, S.; Tiganis, T. High-fat-fed obese glutathione peroxidase 1-deficient mice exhibit defective insulin secretion but protection from hepatic steatosis and liver damage. Antioxid. Redox Signal. 2014, 20, 2114–2129. [Google Scholar] [CrossRef]
- Okuno, Y.; Fukuhara, A.; Hashimoto, E.; Kobayashi, H.; Kobayashi, S.; Otsuki, M.; Shimomura, I. Oxidative Stress Inhibits Healthy Adipose Expansion Through Suppression of SREBF1-Mediated Lipogenic Pathway. Diabetes 2018, 67, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Townsend, L.K.; Weber, A.J.; Barbeau, P.A.; Holloway, G.P.; Wright, D.C. Reactive oxygen species-dependent regulation of pyruvate dehydrogenase kinase-4 in white adipose tissue. Am. J. Physiol. Cell Physiol. 2020, 318, C137–C149. [Google Scholar] [CrossRef] [PubMed]
- May, J.M.; de Haën, C. The insulin-like effect of hydrogen peroxide on pathways of lipid synthesis in rat adipocytes. J. Biol. Chem. 1979, 254, 9017–9021. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Tanaka, M.; Takemura, Y.; Matsuzaki, H.; Ono, Y.; Kikkawa, U.; Nishizuka, Y. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc. Natl. Acad. Sci. USA 1997, 94, 11233–11237. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, M.; Yu, Z.X.; Ferrans, V.J.; Irani, K.; Finkel, T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 1995, 270, 296–299. [Google Scholar] [CrossRef]
- Mahadev, K.; Motoshima, H.; Wu, X.; Ruddy, J.M.; Arnold, R.S.; Cheng, G.; Lambeth, J.D.; Goldstein, B.J. The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol. Cell. Biol. 2004, 24, 1844–1854. [Google Scholar] [CrossRef]
- Lee, H.; Lee, Y.J.; Choi, H.; Ko, E.H.; Kim, J.-W. Reactive Oxygen Species Facilitate Adipocyte Differentiation by Accelerating Mitotic Clonal Expansion. J. Biol. Chem. 2009, 284, 10601–10609. [Google Scholar] [CrossRef]
- de Villiers, D.; Potgieter, M.; Ambele, M.A.; Adam, L.; Durandt, C.; Pepper, M.S. The Role of Reactive Oxygen Species in Adipogenic Differentiation. Adv. Exp. Med. Biol. 2018, 1083, 125–144. [Google Scholar]
- Munekata, K.; Sakamoto, K. Forkhead transcription factor Foxo1 is essential for adipocyte differentiation. In Vitro Cell. Dev. Biol. Anim. 2009, 45, 642–651. [Google Scholar] [CrossRef]
- Higuchi, M.; Dusting, G.J.; Peshavariya, H.; Jiang, F.; Hsiao, S.; Chan, E.C.; Liu, G.-S. Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and Forkhead box O1 mediated upregulation of antioxidant enzymes. Stem Cells Dev. 2013, 22, 878–888. [Google Scholar] [CrossRef]
- Tormos, K.V.; Anso, E.; Hamanaka, R.B.; Eisenbart, J.; Joseph, J.; Kalyanaraman, B.; Chandel, N.S. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011, 14, 537–544. [Google Scholar] [CrossRef]
- Kamata, H.; Honda, S.-I.; Maeda, S.; Chang, L.; Hirata, H.; Karin, M. Reactive Oxygen Species Promote TNFα-Induced Death and Sustained JNK Activation by Inhibiting MAP Kinase Phosphatases. Cell 2005, 120, 649–661. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Voigt, A.; Agnew, K.; van Schothorst, E.M.; Keijer, J.; Klaus, S. Short-term, high fat feeding-induced changes in white adipose tissue gene expression are highly predictive for long-term changes. Mol. Nutr. Food Res. 2013, 57, 1423–1434. [Google Scholar] [CrossRef]
- Bodur, A.; İnce, İ.; Kahraman, C.; Abidin, İ.; Aydin-Abidin, S.; Alver, A. Effect of a high sucrose and high fat diet in BDNF (+/−) mice on oxidative stress markers in adipose tissues. Arch. Biochem. Biophys. 2019, 665, 46–56. [Google Scholar] [CrossRef]
- Illesca, P.; Valenzuela, R.; Espinosa, A.; Echeverría, F.; Soto-Alarcón, S.; Ortiz, M.; Campos, C.; Vargas, R.; Videla, L.A. The metabolic dysfunction of white adipose tissue induced in mice by a high-fat diet is abrogated by co-administration of docosahexaenoic acid and hydroxytyrosol. Food Funct. 2020, 11, 9086–9102. [Google Scholar] [CrossRef]
- Luptak, I.; Qin, F.; Sverdlov, A.L.; Pimentel, D.R.; Panagia, M.; Croteau, D.; Siwik, D.A.; Bachschmid, M.M.; He, H.; Balschi, J.A.; et al. Energetic Dysfunction Is Mediated by Mitochondrial Reactive Oxygen Species and Precedes Structural Remodeling in Metabolic Heart Disease. Antioxid. Redox Signal. 2019, 31, 539–549. [Google Scholar] [CrossRef]
- Ryan, T.E.; Schmidt, C.A.; Green, T.D.; Spangenburg, E.E.; Neufer, P.D.; McClung, J.M. Targeted Expression of Catalase to Mitochondria Protects Against Ischemic Myopathy in High-Fat Diet-Fed Mice. Diabetes 2016, 65, 2553–2568. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Croft, A.J.; Kelly, C.; Chen, D.; Haw, T.J.; Sverdlov, A.L.; Ngo, D.T.M. Overexpression of Mitochondrial Catalase within Adipose Tissue Does Not Confer Systemic Metabolic Protection against Diet-Induced Obesity. Antioxidants 2023, 12, 1137. https://doi.org/10.3390/antiox12051137
Croft AJ, Kelly C, Chen D, Haw TJ, Sverdlov AL, Ngo DTM. Overexpression of Mitochondrial Catalase within Adipose Tissue Does Not Confer Systemic Metabolic Protection against Diet-Induced Obesity. Antioxidants. 2023; 12(5):1137. https://doi.org/10.3390/antiox12051137
Chicago/Turabian StyleCroft, Amanda J., Conagh Kelly, Dongqing Chen, Tatt Jhong Haw, Aaron L. Sverdlov, and Doan T. M. Ngo. 2023. "Overexpression of Mitochondrial Catalase within Adipose Tissue Does Not Confer Systemic Metabolic Protection against Diet-Induced Obesity" Antioxidants 12, no. 5: 1137. https://doi.org/10.3390/antiox12051137
APA StyleCroft, A. J., Kelly, C., Chen, D., Haw, T. J., Sverdlov, A. L., & Ngo, D. T. M. (2023). Overexpression of Mitochondrial Catalase within Adipose Tissue Does Not Confer Systemic Metabolic Protection against Diet-Induced Obesity. Antioxidants, 12(5), 1137. https://doi.org/10.3390/antiox12051137









