Revised Aspects into the Molecular Bases of Hydroxycinnamic Acid Metabolism in Lactobacilli
Abstract
:1. Introduction
Antimicrobial Stress on Lactobacilli Associated with Hydroxycinnamic Acids
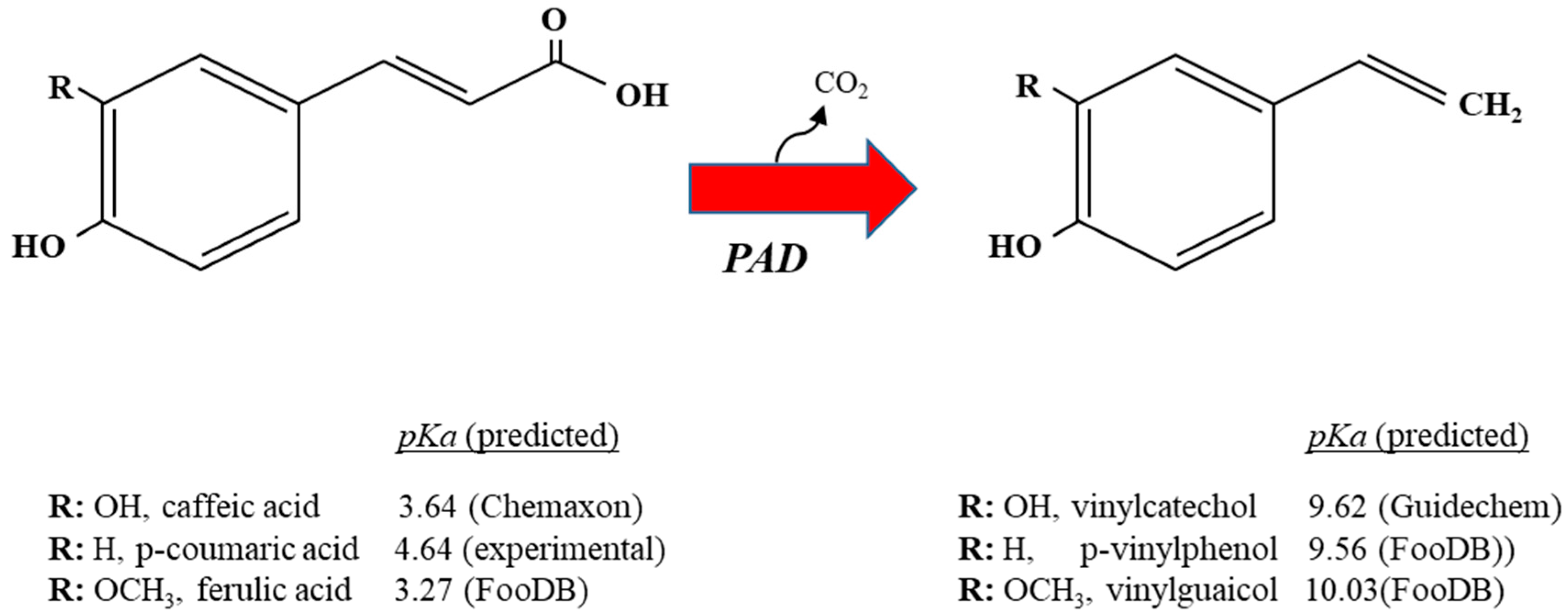
2. Decarboxylation of Hydroxycinnamic Acids
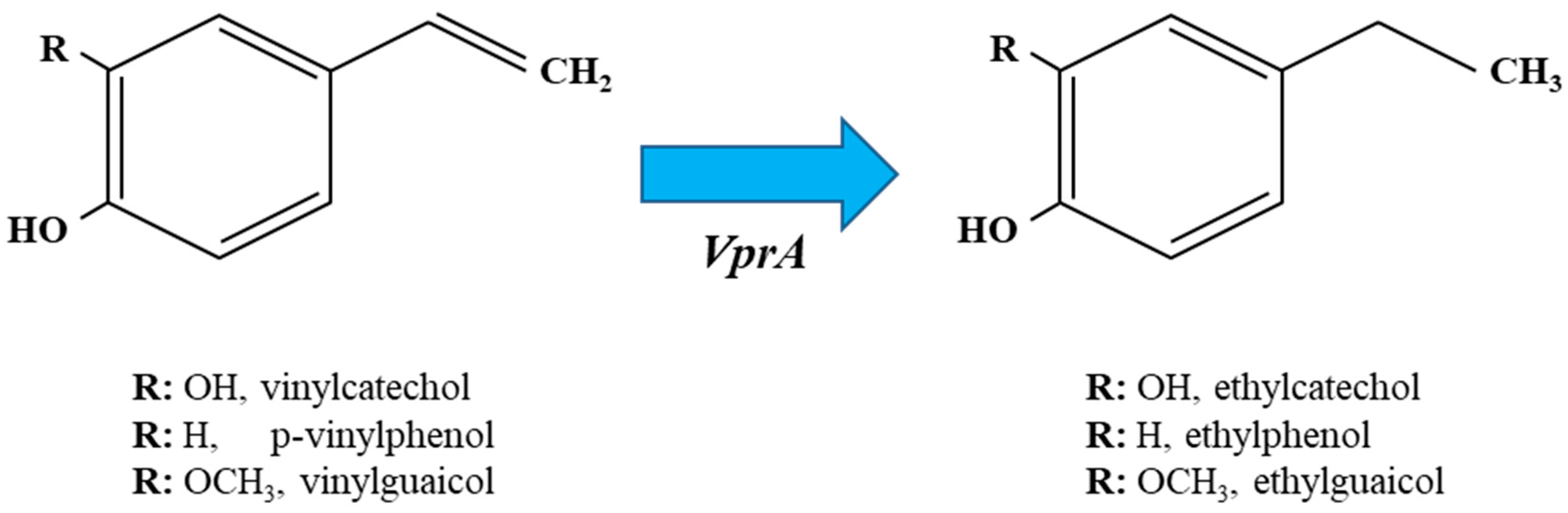
3. Reduction of Vinyl Derivatives of Hydroxycinnamic Acids into Ethyl Derivatives
4. Reduction of Hydroxycinnamic Acids
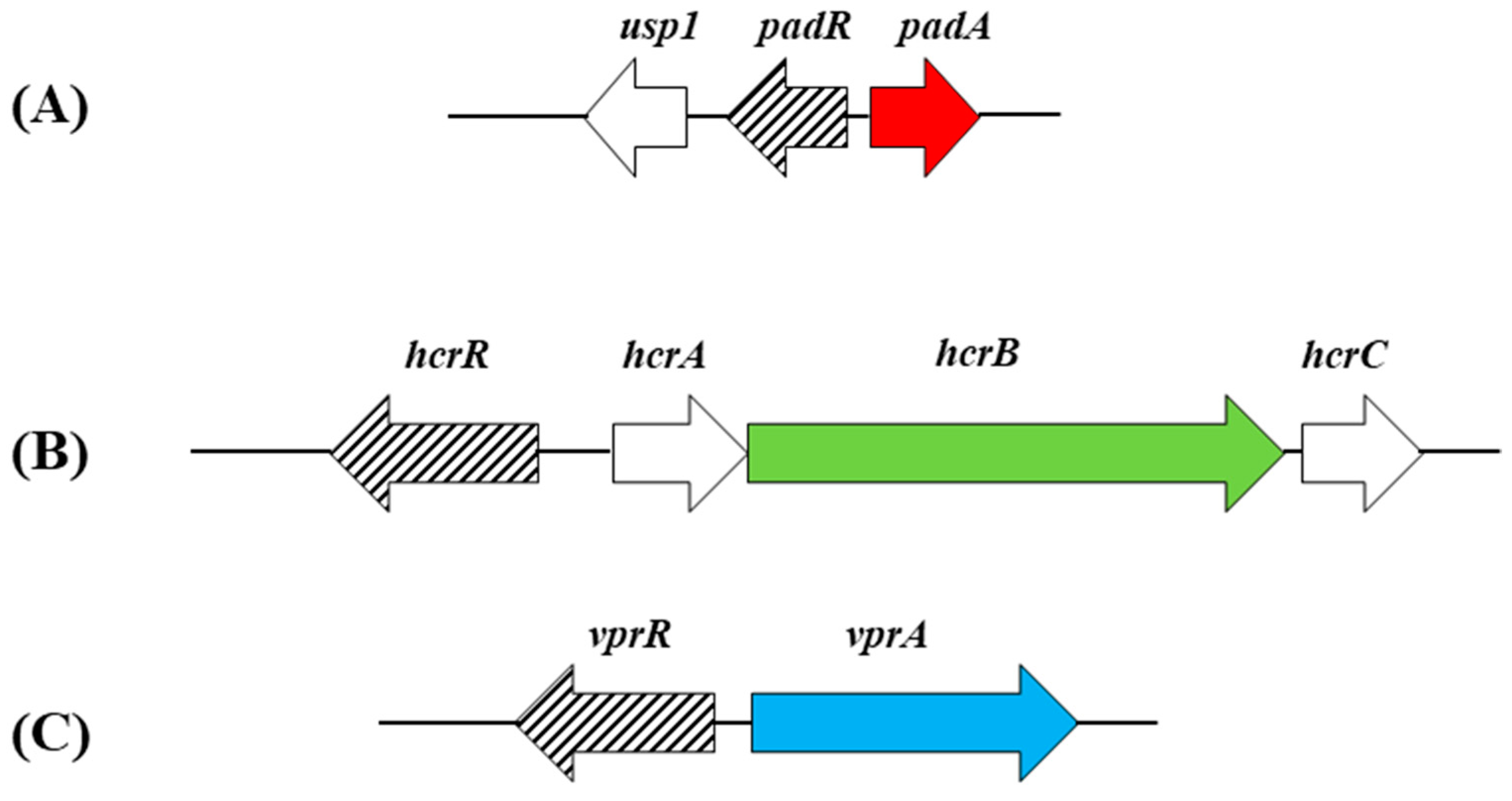
4.1. Variability in the HCA Reduction and Decarboxylation Metabolisms across Lactobacillus spp.
4.2. HrcAB Reductase: Open Questions
5. Concluding Remarks
Funding
Conflicts of Interest
References
- Clifford, M.N. Chlorogenic acids and other cinnamates–nature, occurrence and dietary burden. J. Sci. Food Agric. 1999, 79, 362–372. [Google Scholar] [CrossRef]
- Nguyen, T.; Sherratt, P.J.; Pickett, C.B. Regulatory Mechanisms Controlling Gene Expression Mediated by the Antioxidant Response Element. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 233–260. [Google Scholar] [CrossRef]
- Teixeira, J.; Gaspar, A.; Garrido, E.M.; Garrido, J.; Borges, F. Hydroxycinnamic Acid Antioxidants: An Electrochemical Overview. BioMed Res. Int. 2013, 2013, 251754. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Lobiuc, A.; Pavăl, N.-E.; Mangalagiu, I.I.; Gheorghiță, R.; Teliban, G.-C.; Amăriucăi-Mantu, D.; Stoleru, V. Future Antimicrobials: Natural and Functionalized Phenolics. Molecules 2023, 28, 1114. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Corcoran, B.M.; Stanton, C.; Fitzgerald, G.F.; Ross, R.P. Survival of Probiotic Lactobacilli in Acidic Environments Is Enhanced in the Presence of Metabolizable Sugars. Appl. Environ. Microbiol. 2005, 71, 3060–3067. [Google Scholar] [CrossRef] [Green Version]
- Reverón, I.; de las Rivas, B.; Muñoz, R.; López de Felipe, F. Genome-wide transcriptomic responses of a human isolate of Lactobacillus plantarum exposed to p-coumaric acid stress. Mol. Nutr. Food Res. 2012, 56, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Kao, K.C. Transcriptional Analysis of Lactobacillus brevis to N-Butanol and Ferulic Acid Stress Responses. PLoS ONE 2011, 6, e21438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivas-Sendra, A.; Landete, J.; Alcántara, C.; Zúñiga, M. Response of Lactobacillus casei BL23 to phenolic compounds. J. Appl. Microbiol. 2011, 111, 1473–1481. [Google Scholar] [CrossRef] [Green Version]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 781–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damasceno, S.S.; Dantas, B.B.; Ribeiro-Filho, J.; Araújo, A.M.; Da Costa, J.G.M. Chemical Properties of Caffeic and Ferulic Acids in Biological System: Implications in Cancer Therapy. A Review. Curr. Pharm. Des. 2017, 23, 3015–3023. [Google Scholar] [CrossRef]
- Song, Y.; Buettner, G.R. Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free. Radic. Biol. Med. 2010, 15, 919–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Felipe, F.L.; Rivas, B.D.L.; Muñoz, R. Molecular Responses of Lactobacilli to Plant Phenolic Compounds: A Comparative Review of the Mechanisms Involved. Antioxidants 2021, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; van der Veen, S.; Nakajima, H.; Abee, T. Effect of respiration and manganese on oxidative stress resistance of Lactobacillus plantarum WCFS1. Microbiology 2012, 158, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Segura-Aguilar, J.; Lind, C. On the mechanism of the Mn3+-induced neurotoxicity of dopamine: Prevention of quinone-derived oxygen toxicity by DT diaphorase and superoxide dismutase. Chem. Interact. 1989, 72, 309–324. [Google Scholar] [CrossRef]
- Gaur, G.; Oh, J.-H.; Filannino, P.; Gobbetti, M.; van Pijkeren, J.-P.; Gänzle, M.G. Genetic Determinants of Hydroxycinnamic Acid Metabolism in Heterofermentative Lactobacilli. Appl. Environ. Microbiol. 2020, 86, e02461-19. [Google Scholar] [CrossRef]
- Carrasco, J.A.; Lucena-Padrós, H.; Brenes, M.; Ruiz-Barba, J.L. Expression of genes involved in metabolism of phenolic compounds by Lactobacillus pentosus and its relevance for table-olive fermentations. Food Microbiol. 2018, 76, 382–389. [Google Scholar] [CrossRef] [Green Version]
- Barthelmebs, L.; Divies, C.; Cavin, J.F. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl. Environ. Microbiol. 2000, 66, 3368–3375. [Google Scholar] [CrossRef] [Green Version]
- Kalinowska, M.; Laderiere, B.; Champagne, P.; Kowczyk-Sadowy, M.; Lewandowski, W. Mn(II), Cu(II) and Cd(II) p-coumarates: FT-IR, FT-Raman, ¹H and ¹³C NMR and thermogravimetric studies. Spectrochim. Acta A 2013, 103, 264–271. [Google Scholar] [CrossRef]
- Garrido, J.; Gaspar, A.; Garrido, E.M.; Miri, R.; Tavakkoli, M.; Pourali, S.; Saso, L.; Borges, F.; Firuzi, O. Alkyl esters of hydroxycinnamic acids with improved antioxidant activity and lipophilicity protect PC12 cells against oxidative stress. Biochimie 2012, 94, 961–967. [Google Scholar] [CrossRef]
- Kortenska, V.D.; Velikova, M.P.; Yanishlieva, N.V.; Totzeva, I.R.; Bankova, V.S.; Marcucci, M.C. Kinetics of lipid oxidation in the presence of cinnamic acid derivatives. Eur. J. Lipid Sci. Technol. 2002, 104, 19–28. [Google Scholar] [CrossRef]
- Medina, I.; Undeland, I.; Larsson, K.; Storrø, I.; Rustad, T.; Jacobsen, C.; Kristinová, V.; Gallardo, J.M. Activity of caffeic acid in different fish lipid matrices: A review. Food Chem. 2012, 131, 730–740. [Google Scholar] [CrossRef]
- Rogozinska, M.; Korsak, D.; Mroczek, J.; Biesaga, M. Catabolism of hydroxycinnamic acids in contact with probiotic Lactobacillus. J. Appl. Microbiol. 2021, 131, 1464–1473. [Google Scholar] [CrossRef]
- Nguyen, T.K.C.; Tran, N.P.; Cavin, J.-F. Genetic and Biochemical Analysis of PadR- padC Promoter Interactions during the Phenolic Acid Stress Response in Bacillus subtilis 168. J. Bacteriol. 2011, 193, 4180–4191. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, H.; Landete, J.M.; Curiel, J.A.; de Las Rivas, B.; Mancheño, J.M.; Muñoz, R. Characterization of the p-Coumaric Acid Decarboxylase from Lactobacillus plantarum CECT 748T. J. Agric. Food Chem. 2008, 56, 3068–3072. [Google Scholar] [CrossRef]
- Cavin, J.F.; Barthelmebs, L.; Guzzo, J.; Van Beeumen, J.; Samyn, B.; Travers, J.F.; Diviès, C. Purification and characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum. FEMS Microbiol. Lett. 1997, 147, 291–295. [Google Scholar] [CrossRef]
- Landete, J.M.; Rodríguez, H.; Curiel, J.A.; Rivas, B.d.L.; Mancheño, J.M.; Muñoz, R. Gene cloning, expression, and characterization of phenolic acid decarboxylase from Lactobacillus brevis RM84. J. Ind. Microbiol. Biotechnol. 2010, 37, 617–624. [Google Scholar] [CrossRef] [Green Version]
- Santamaría, L.; Reverón, I.; de Felipe, F.L.; Rivas, B.D.L.; Muñoz, R. Ethylphenol Formation by Lactobacillus plantarum: Identification of the Enzyme Involved in the Reduction of Vinylphenols. Appl. Environ. Microbiol. 2018, 84, e01064-18. [Google Scholar] [CrossRef] [Green Version]
- Needham, B.D.; Funabashi, M.; Adame, M.D.; Wang, Z.; Boktor, J.C.; Haney, J.; Wu, W.-L.; Rabut, C.; Ladinsky, M.S.; Hwang, S.-J.; et al. A gut-derived metabolite alters brain activity and anxiety behaviour in mice. Nature 2022, 602, 647–653. [Google Scholar] [CrossRef]
- Tlais, A.Z.A.; Junior, W.J.F.L.; Filannino, P.; Campanaro, S.; Gobbetti, M.; Di Cagno, R. How Microbiome Composition Correlates with Biochemical Changes during Sauerkraut Fermentation: A Focus on Neglected Bacterial Players and Functionalities. Microbiol. Spectr. 2022, 10, e00168-22. [Google Scholar] [CrossRef]
- Kuji, M.; Itoh, N.; Ohba, Y.; Yamada, K.; Hashimoto, K. Inhibitory effect of 4-ethylcatechol on β-glucuronidase activity. Food Sci. Technol. Res. 2021, 27, 797–806. [Google Scholar] [CrossRef]
- Li, Z.-H.; Wang, Q.; Ruan, X.; Pan, C.-D.; Jiang, D.-A. Phenolics and Plant Allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, H.; Landete, J.M.; Rivas, B.d.L.; Muñoz, R. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem. 2008, 107, 1393–1398. [Google Scholar] [CrossRef] [Green Version]
- Filannino, P.; Di Cagno, R.; Crecchio, C.; De Virgilio, C.; De Angelis, M.; Gobbetti, M. Transcriptional reprogramming and phenotypic switching associated with the adaptation of Lactobacillus plantarum C2 to plant niches. Sci. Rep. 2016, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Wrolstad, R. Phenolic Composition of Authentic Pineapple Juice. J. Food Sci. 2002, 67, 155–161. [Google Scholar] [CrossRef]
- Santamaría, L.; Reverón, I.; de Felipe, F.L.; Rivas, B.D.L.; Muñoz, R. Unravelling the Reduction Pathway as an Alternative Metabolic Route to Hydroxycinnamate Decarboxylation in Lactobacillus plantarum. Appl. Environ. Microbiol. 2018, 84, e01123-18. [Google Scholar] [CrossRef] [Green Version]
- Hertzberger, R.; Arents, J.; Dekker, H.L.; Pridmore, R.D.; Gysler, C.; Kleerebezem, M.; de Mattos, M.J.T. H2O2 Production in Species of the Lactobacillus acidophilus Group: A Central Role for a Novel NADH-Dependent Flavin Reductase. Appl. Environ. Microbiol. 2014, 80, 2229–2239. [Google Scholar] [CrossRef] [Green Version]
- Valladares, R.B.; Graves, C.; Wright, K.; Gardner, C.L.; Lorca, G.L.; Gonzalez, C.F. H2O2 production rate in Lactobacillus johnsonii is modulated via the interplay of a heterodimeric flavin oxidoreductase with a soluble 28 Kd PAS domain containing protein. Front. Microbiol. 2015, 6, 716. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Zetina, S.M.; González-Manzano, S.; Ayuda-Durán, B.; Santos-Buelga, C.; González-Paramás, A.M. Caffeic and Dihydrocaffeic Acids Promote Longevity and Increase Stress Resistance in Caenorhabditis elegans by Modulating Expression of Stress-Related Genes. Molecules 2021, 26, 1517. [Google Scholar] [CrossRef]
- Silva, F.A.M.; Borges, F.; Guimarães, C.; Lima, J.L.F.C.; Matos, C.; Reis, S. Phenolic Acids and Derivatives: Studies on the Relationship among Structure, Radical Scavenging Activity, and Physicochemical Parameters. J. Agric. Food Chem. 2000, 48, 2122–2126. [Google Scholar] [CrossRef] [PubMed]
- Esteves, M.; Siquet, C.; Gaspar, A.; Rio, V.; Sousa, J.B.; Reis, S.; Marques, M.P.M.; Borges, F. Antioxidant Versus Cytotoxic Properties of Hydroxycinnamic Acid Derivatives—A New Paradigm in Phenolic Research. Arch. Pharm. 2008, 341, 164–173. [Google Scholar] [CrossRef] [Green Version]
- Santamaría, L.; Reverón, I.; Plaza-Vinuesa, L.; Oliveros, J.C.; Rivas, B.D.L.; Muñoz, R.; De Felipe, F.L. Oleuropein Transcriptionally Primes Lactobacillus plantarum to Interact With Plant Hosts. Front. Microbiol. 2019, 10, 2177. [Google Scholar] [CrossRef] [PubMed]
- Reverón, I.; Rivas, B.D.L.; Matesanz, R.; Muñoz, R.; de Felipe, F.L. Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb. Cell Factories 2015, 14, 160. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Lin, Y.; Shen, X.; Jain, R.; Sun, X.; Yuan, Q.; Yan, Y. Aerobic biosynthesis of hydrocinnamic acids in Escherichia coli with a strictly oxygen-sensitive enoate reductase. Metab. Eng. 2016, 35, 75–82. [Google Scholar] [CrossRef]
- Aguirre, J.D.; Culotta, V.C. Battles with Iron: Manganese in Oxidative Stress Protection. J. Biol. Chem. 2012, 287, 13541–13548. [Google Scholar] [CrossRef] [Green Version]
- Dukan, S.; Nyström, T. Oxidative Stress Defense and Deterioration of Growth-arrested Escherichia coli Cells. J. Biol. Chem. 1999, 274, 26027–26032. [Google Scholar] [CrossRef] [Green Version]
- De Felipe, F.L.; Hugenholtz, J. Purification and characterisation of the water forming NADH-oxidase from Lactococcus lactis. Int. Dairy J. 2001, 11, 37–44. [Google Scholar] [CrossRef]
- Zhang, Y.-W.; Tiwari, M.K.; Gao, H.; Dhiman, S.S.; Jeya, M.; Lee, J.-K. Cloning and characterization of a thermostable H2O-forming NADH oxidase from Lactobacillus rhamnosus. Enzym. Microb. Technol. 2012, 50, 255–262. [Google Scholar] [CrossRef]
- Filannino, P.; Gobbetti, M.; De Angelis, M.; Di Cagno, R. Hydroxycinnamic Acids Used as External Acceptors of Electrons: An Energetic Advantage for Strictly Heterofermentative Lactic Acid Bacteria. Appl. Environ. Microbiol. 2014, 80, 7574–7582. [Google Scholar] [CrossRef] [Green Version]
- Filannino, P.; Di Cagno, R.; Addante, R.; Pontonio, E.; Gobbetti, M. Metabolism of Fructophilic Lactic Acid Bacteria Isolated from the Apis mellifera L. Bee Gut: Phenolic Acids as External Electron Acceptors. Appl. Environ. Microbiol. 2016, 82, 6899–6911. [Google Scholar] [CrossRef] [PubMed] [Green Version]

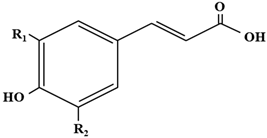 | |||
| Compound | R1 | R2 | Ep (mV) |
| p-Coumaric acid | OH | H | +736 |
| Caffeic acid | OH | H | +183 |
| Ferulic acid | OCH3 | H | +335 |
| Sinapic acid | OCH3 | OCH3 | +188 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López de Felipe, F. Revised Aspects into the Molecular Bases of Hydroxycinnamic Acid Metabolism in Lactobacilli. Antioxidants 2023, 12, 1294. https://doi.org/10.3390/antiox12061294
López de Felipe F. Revised Aspects into the Molecular Bases of Hydroxycinnamic Acid Metabolism in Lactobacilli. Antioxidants. 2023; 12(6):1294. https://doi.org/10.3390/antiox12061294
Chicago/Turabian StyleLópez de Felipe, Félix. 2023. "Revised Aspects into the Molecular Bases of Hydroxycinnamic Acid Metabolism in Lactobacilli" Antioxidants 12, no. 6: 1294. https://doi.org/10.3390/antiox12061294
APA StyleLópez de Felipe, F. (2023). Revised Aspects into the Molecular Bases of Hydroxycinnamic Acid Metabolism in Lactobacilli. Antioxidants, 12(6), 1294. https://doi.org/10.3390/antiox12061294






