Influence of the Degree of Unsaturation in Fish Oil Supplements on Oxidative Stress and Protein Carbonylation in the Cerebral Cortex and Cerebellum of Healthy Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model and Experimental Design
2.2. Fatty Acid Extraction and Profile Analysis
2.3. Analysis of Lipid Peroxidation Products
2.4. Analysis of Protein Glutathionylation
2.5. Analysis of Total and Specific Protein Carbonylation
2.6. Carbonylated Proteome and Functional Enrichment Analysis of Rat Cerebral Cortex and Cerebellum
2.7. Antioxidant Status and Enzymatic Activities in Brain
2.8. Image Analysis and Densitometry Measures
2.9. Statistics
2.10. Materials and Reagents
3. Results and Discussion
3.1. Comparison of the Lipid Profile Between Cortex and Cerebellum and Response to the Degree of Unsaturation of Fatty Acids in Oils Administered to the Rats
3.2. Redox Status in the Cortex and the Cerebellum and Response to the Degree of Unsaturation of Fatty Acids in Oils Administered to the Rats
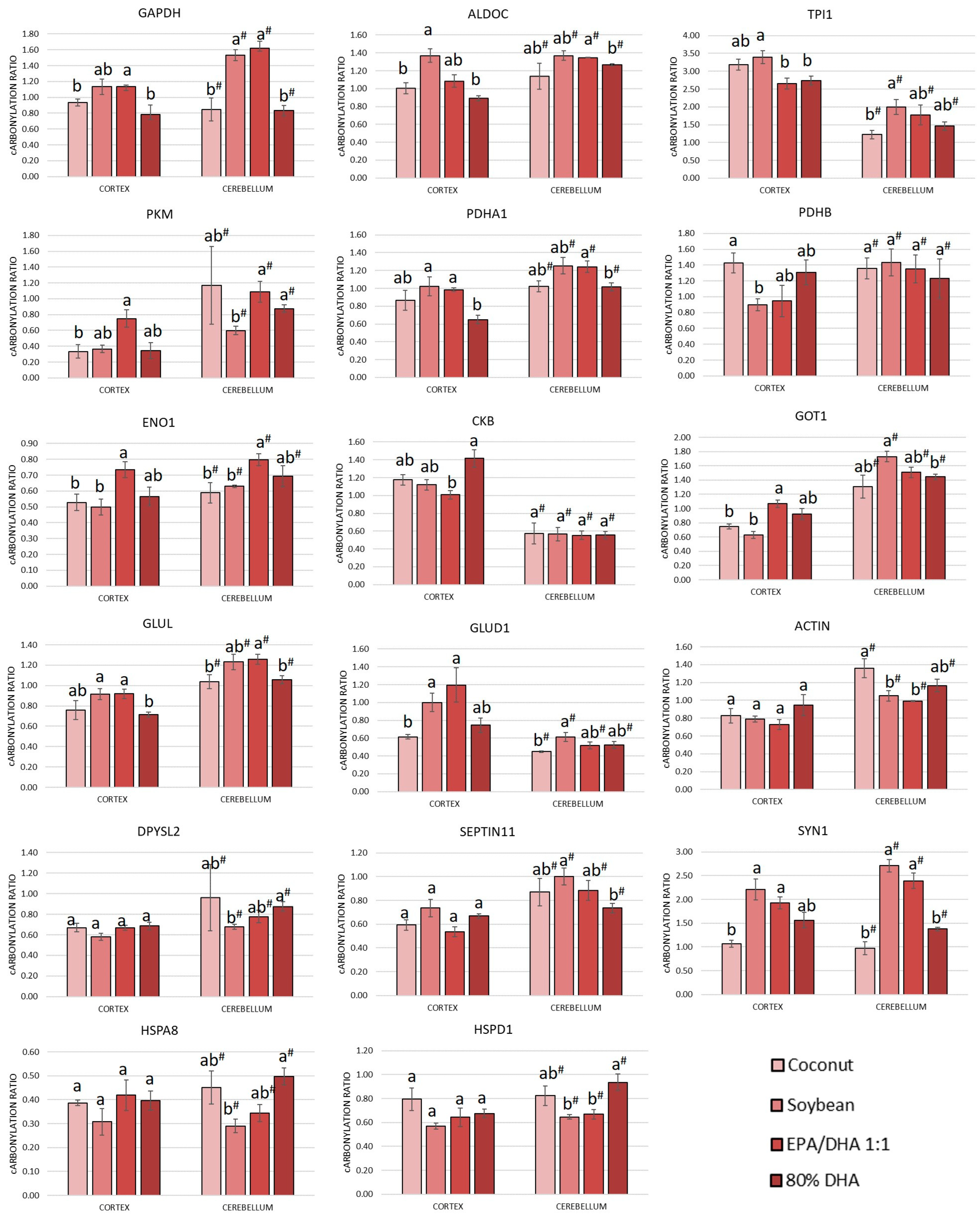
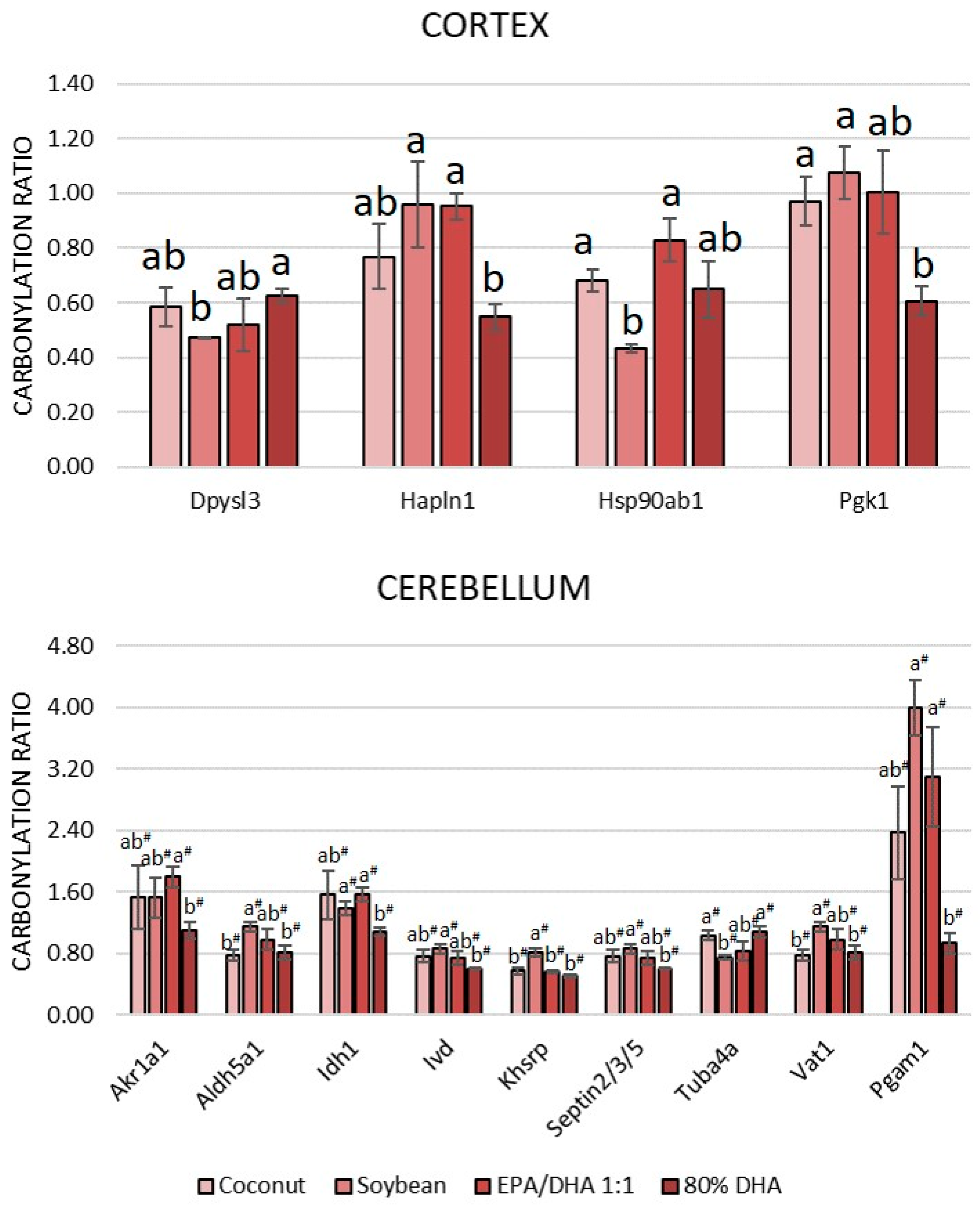
3.3. Comparison Between the Carbonylated Proteome of the Cortex and Cerebellum and Response to the Degree of Unsaturation of Fatty Acids in Oils Administered to the Rats
3.3.1. Comparison Between the Cortical and Cerebellar Proteome and Carbonylated Proteome
3.3.2. Influence of the Degree of Unsaturation of Fatty Acids in Oils Administered to the Rats on the Carbonylated Proteome
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Djuricic, I.; Calder, P.C. Beneficial Outcomes of Omega-6 and Omega-3 Polyunsaturated Fatty Acids on Human Health: An Update for 2021. Nutrients 2021, 13, 2421. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Ren, H.; Yao, X.; Shi, Z.; Liang, F.; Kang, J.X.; Wan, J.-B.; Pei, Z.; Su, K.P.; Su, H. Enriched Brain Omega-3 Polyunsaturated Fatty Acids Confer Neuroprotection against Microinfarction. EBioMedicine 2018, 32, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Kerdiles, O.; Layé, S.; Calon, F. Omega-3 Polyunsaturated Fatty Acids and Brain Health: Preclinical Evidence for the Prevention of Neurodegenerative Diseases. Trends Food Sci. Technol. 2017, 69, 203–213. [Google Scholar] [CrossRef]
- Bazinet, R.P.; Layé, S. Polyunsaturated Fatty Acids and Their Metabolites in Brain Function and Disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- von Schacky, C. Importance of EPA and DHA Blood Levels in Brain Structure and Function. Nutrients 2021, 13, 1074. [Google Scholar] [CrossRef]
- O’ Donovan, F.; Carney, S.; Kennedy, J.; Hayes, H.; Pender, N.; Boland, F.; Stanton, A.; O’Donovan, F.; Carney, S.; Kennedy, J.; et al. Associations and Effects of Omega-3 Polyunsaturated Fatty Acids on Cognitive Function and Mood in Healthy Adults: A Protocol for a Systematic Review of Observational and Interventional Studies. BMJ Open 2019, 9, 1–5. [Google Scholar] [CrossRef]
- Cobley, J.N.; Fiorello, M.L.; Bailey, D.M. 13 Reasons Why the Brain Is Susceptible to Oxidative Stress. Redox Biol. 2018, 15, 490–503. [Google Scholar] [CrossRef]
- Shichiri, M. The Role of Lipid Peroxidation in Neurological Disorders. J. Clin. Biochem. Nutr. 2014, 54, 151–160. [Google Scholar] [CrossRef]
- Fu, Y.; He, Y.; Phan, K.; Bhatia, S.; Pickford, R.; Wu, P.; Dzamko, N.; Halliday, G.M.; Kim, W.S. Increased Unsaturated Lipids Underlie Lipid Peroxidation in Synucleinopathy Brain. Acta Neuropathol. Commun. 2022, 10, 165. [Google Scholar] [CrossRef]
- Jové, M.; Pradas, I.; Dominguez-Gonzalez, M.; Ferrer, I.; Pamplona, R. Lipids and Lipoxidation in Human Brain Aging. Mitochondrial ATP-Synthase as a Key Lipoxidation Target. Redox Biol. 2019, 23, 101082. [Google Scholar] [CrossRef]
- Perrone, A.; Giovino, A.; Benny, J.; Martinelli, F. Advanced Glycation End Products (AGEs): Biochemistry, Signaling, Analytical Methods, and Epigenetic Effects. Oxid. Med. Cell. Longev. 2020, 2020, 3818196. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein Carbonylation in Human Diseases. Trends Mol. Med. 2003, 9, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, S.; Arsic, A.; Ristic-Medic, D.; Cvetkovic, Z.; Vucic, V. Lipid Peroxidation and Antioxidant Supplementation in Neurodegenerative Diseases: A Review of Human Studies. Antioxidants 2020, 9, 1128. [Google Scholar] [CrossRef] [PubMed]
- Sultana, R.; Perluigi, M.; Butterfield, D.A. Lipid Peroxidation Triggers Neurodegeneration: A Redox Proteomics View into the Alzheimer Disease Brain. Free Radic. Biol. Med. 2013, 62, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Xu, W.; Ou, Y.N.; Cao, X.P.; Tan, M.S.; Tan, L.; Yu, J.T. Diabetes Mellitus and Risks of Cognitive Impairment and Dementia: A Systematic Review and Meta-Analysis of 144 Prospective Studies. Ageing Res. Rev. 2019, 55, 100944. [Google Scholar] [CrossRef]
- Selman, A.; Burns, S.; Reddy, A.P.; Culberson, J.; Reddy, P.H. The Role of Obesity and Diabetes in Dementia. Int. J. Mol. Sci. 2022, 23, 9267. [Google Scholar] [CrossRef]
- Curtis, J.M.; Hahn, W.S.; Long, E.K.; Burrill, J.S.; Arriaga, E.A.; Bernlohr, D.A. Protein Carbonylation and Metabolic Control Systems. Trends Endocrinol. Metab. 2012, 23, 399–406. [Google Scholar] [CrossRef]
- Mora, I.; Arola, L.; Caimari, A.; Escoté, X.; Puiggròs, F. Structured Long-Chain Omega-3 Fatty Acids for Improvement of Cognitive Function during Aging. Int. J. Mol. Sci. 2022, 23, 3472. [Google Scholar] [CrossRef]
- Avramovic, N.; Dragutinovic, V.; Krstic, D.; Colovic, M.B.; Trbovic, A.; de Luka, S.; Milovanovic, I.; Popovic, T. The Effecects of Omega 3 Fatty Acid Supplementation on Brain Tissue Oxidative Status in Aged Wistar Rats. Hippokratia 2012, 16, 241–245. [Google Scholar]
- Park, Y.H.; Shin, S.J.; Kim, H.s.; Hong, S.B.; Kim, S.; Nam, Y.; Kim, J.-J.; Lim, K.; Kim, J.-S.; Kim, J.; et al. Omega-3 Fatty Acid-Type Docosahexaenoic Acid Protects against Aβ-Mediated Mitochondrial Deficits and Pathomechanisms in Alzheimer’s Disease-Related Animal Model. Int. J. Mol. Sci. 2020, 21, 3879. [Google Scholar] [CrossRef]
- Zugno, A.I.; Chipindo, H.L.; Volpato, A.M.; Budni, J.; Steckert, A.V.; de Oliveira, M.B.; Heylmann, A.S.; da Rosa Silveira, F.; Mastella, G.A.; Maravai, S.G.; et al. Omega-3 Prevents Behavior Response and Brain Oxidative Damage in the Ketamine Model of Schizophrenia. Neuroscience 2014, 259, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.; Méndez, L.; Raner, A.; Miralles-Pérez, B.; Romeu, M.; Ramos-Romero, S.; Torres, J.L.; Medina, I. Fish Oil Supplementation Counteracts the Effect of High-Fat and High-Sucrose Diets on the Carbonylated Proteome in the Rat Cerebral Cortex. Biomed. Pharmacother. 2023, 168, 115708. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.; Méndez, L.; Raner, A.; Miralles-Pérez, B.; Romeu, M.; Ramos-Romero, S.; Torres, J.L.; Medina, I. Dietary Marine Oils Selectively Decrease Obesogenic Diet-Derived Carbonylation in Proteins Involved in ATP Homeostasis and Glutamate Metabolism in the Rat Cerebellum. Antioxidants 2024, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Miralles-Pérez, B.; Méndez, L.; Nogués, M.R.; Sánchez-Martos, V.; Fortuño-Mar, À.; Ramos-Romero, S.; Hereu, M.; Medina, I.; Romeu, M. Effects of a Fish Oil Rich in Docosahexaenoic Acid on Cardiometabolic Risk Factors and Oxidative Stress in Healthy Rats. Mar. Drugs 2021, 19, 555. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Ceccarelli, V.; Codini, M.; Fettucciari, K.; Calvitti, M.; Cataldi, S.; Albi, E.; Vecchini, A.; Beccari, T. The Polyunsaturated Fatty Acid EPA, but Not DHA, Enhances Neurotrophic Factor Expression through Epigenetic Mechanisms and Protects against Parkinsonian Neuronal Cell Death. Int. J. Mol. Sci. 2022, 23, 6176. [Google Scholar] [CrossRef]
- Opinion, S. Scientific Opinion on the Tolerable Upper Intake Level of Eicosapentaenoic Acid (EPA), Docosahexaenoic Acid (DHA) and Docosapentaenoic Acid (DPA). EFSA J. 2012, 10, 2815. [Google Scholar] [CrossRef]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose Translation from Animal to Human Studies Revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct Transesterification of All Classes of Lipids in a One-Step Reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar] [CrossRef]
- American Oil Chemists’ Society Official and Tentative Methods of the American Oil Chemists’ Society. Analyst 1947, 72, 157. [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to ‘The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets’. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef] [PubMed]
- Misra, H.P.; Fridovich, I. The Role of Superoxide Anion in the Autoxidation of Epinephrine and a Simple Assay for Superoxide Dismutase. J. Biol. Chem. 1972, 247, 3170–3175. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.; Dembiec, D.; Marcus, J. Measurement of Catalase Activity in Tissue Extracts. Anal. Biochem. 1970, 34, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, C.R.; Salzman, J.A.; Elsayed, N.M.; Omaye, S.T.; Korte, D.W. Automated Assays for Superoxide Dismutase, Catalase, Glutathione Peroxidase, and Glutathione Reductase Activity. Anal. Biochem. 1990, 184, 193–199. [Google Scholar] [CrossRef]
- Hissin, P.J.; Hilf, R. A Fluorometric Method for Determination of Oxidized and Reduced Glutathione in Tissues. Anal. Biochem. 1976, 74, 214–226. [Google Scholar] [CrossRef]
- Chavko, M.; Nemoto, E.M.; Melick, J.A. Regional Lipid Composition in the Rat Brain. Mol. Chem. Neuropathol. 1993, 18, 123–131. [Google Scholar] [CrossRef]
- Firląg, M.; Kamaszewski, M.; Ostaszewski, P.; Balasinska, B. Fatty Acid Composition in Cerebral Cortex, Hippocampus and Cerebellum in Adult Rats Receiving Salmon Oil for 6 Months. J. Pre-Clin. Clin. Res. 2014, 8, 30–33. [Google Scholar] [CrossRef]
- Rule, D.C.; Melson, E.A.; Alexander, B.M.; Brown, T.E. Dietary Fatty Acid Composition Impacts the Fatty Acid Profiles of Different Regions of the Bovine Brain. Animals 2022, 12, 2696. [Google Scholar] [CrossRef]
- Bascoul-Colombo, C.; Guschina, I.A.; Maskrey, B.H.; Good, M.; O’Donnell, V.B.; Harwood, J.L. Dietary DHA Supplementation Causes Selective Changes in Phospholipids from Different Brain Regions in Both Wild Type Mice and the Tg2576 Mouse Model of Alzheimer’s Disease. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2016, 1861, 524–537. [Google Scholar] [CrossRef]
- Aoyama, K. Glutathione in the Brain. Int. J. Mol. Sci. 2021, 22, 5010. [Google Scholar] [CrossRef] [PubMed]
- Haseena, P.A.; Diwakar, L.; Ravindranath, V. Protein Glutathionylation and Glutaredoxin: Role in Neurodegenerative Diseases. Antioxidants 2022, 11, 2334. [Google Scholar] [CrossRef] [PubMed]
- Pantano, P.; Baron, J.C.; Lebrun-Grandié, P.; Duquesnoy, N.; Bousser, M.G.; Comar, D. Regional Cerebral Blood Flow and Oxygen Consumption in Human Aging. Stroke 1984, 15, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Michaelis, E.K. Selective Neuronal Vulnerability to Oxidative Stress in the Brain. Front. Aging Neurosci. 2010, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Aschner, M.; Syversen, T. Role of Glutathione in Determining the Differential Sensitivity between the Cortical and Cerebellar Regions towards Mercury-Induced Oxidative Stress. Toxicology 2007, 230, 164–177. [Google Scholar] [CrossRef]
- Andres, R.H.; Ducray, A.D.; Schlattner, U.; Wallimann, T.; Widmer, H.R. Functions and Effects of Creatine in the Central Nervous System. Brain Res. Bull. 2008, 76, 329–343. [Google Scholar] [CrossRef]
- Valvona, C.J.; Fillmore, H.L.; Nunn, P.B.; Pilkington, G.J. The Regulation and Function of Lactate Dehydrogenase A: Therapeutic Potential in Brain Tumor. Brain Pathol. 2016, 26, 3–17. [Google Scholar] [CrossRef]
- Horvat, S.; Kos, J.; Pišlar, A. Multifunctional Roles of γ-Enolase in the Central Nervous System: More than a Neuronal Marker. Cell Biosci. 2024, 14, 61. [Google Scholar] [CrossRef]
- Pal, M.M. Glutamate: The Master Neurotransmitter and Its Implications in Chronic Stress and Mood Disorders. Front. Hum. Neurosci. 2021, 15, 722323. [Google Scholar] [CrossRef]
- Lau, A.; Tymianski, M. Glutamate Receptors, Neurotoxicity and Neurodegeneration. Pflügers Arch.—Eur. J. Physiol. 2010, 460, 525–542. [Google Scholar] [CrossRef]
- Gasiorowska, A.; Wydrych, M.; Drapich, P.; Zadrozny, M.; Steczkowska, M.; Niewiadomski, W.; Niewiadomska, G. The Biology and Pathobiology of Glutamatergic, Cholinergic, and Dopaminergic Signaling in the Aging Brain. Front. Aging Neurosci. 2021, 13, 654931. [Google Scholar] [CrossRef] [PubMed]
- Botvinick, M.; Braver, T. Motivation and Cognitive Control: From Behavior to Neural Mechanism. Annu. Rev. Psychol. 2015, 66, 83–113. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. GABA and Synaptic Transmission in the Cerebellum BT—Handbook of the Cerebellum and Cerebellar Disorders; Manto, M., Gruol, D., Schmahmann, J., Koibuchi, N., Sillitoe, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–14. ISBN 978-3-319-97911-3. [Google Scholar]
- Bishop, P.; Rocca, D.; Henley, J.M. Ubiquitin C-Terminal Hydrolase L1 (UCH-L1): Structure, Distribution and Roles in Brain Function and Dysfunction. Biochem. J. 2016, 473, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Watkins, P.A. Fatty Acid Activation. Prog. Lipid Res. 1997, 36, 55–83. [Google Scholar] [CrossRef] [PubMed]
- Mashek, D.G.; Li, L.O.; Coleman, R.A. Long-Chain Acyl-CoA Synthetases and Fatty Acid Channeling. Future Lipidol. 2007, 2, 465–476. [Google Scholar] [CrossRef]
- Lawrence, G.D. Perspective: The Saturated Fat-Unsaturated Oil Dilemma: Relations of Dietary Fatty Acids and Serum Cholesterol, Atherosclerosis, Inflammation, Cancer, and All-Cause Mortality. Adv. Nutr. 2021, 12, 647–656. [Google Scholar] [CrossRef]
- Sauer, S.W.; Kölker, S.; Hoffmann, G.F.; ten Brink, H.J.; Jakobs, C.; Gibson, K.M.; Okun, J.G. Enzymatic and Metabolic Evidence for a Region Specific Mitochondrial Dysfunction in Brains of Murine Succinic Semialdehyde Dehydrogenase Deficiency (Aldh5a1-/- Mice). Neurochem. Int. 2007, 50, 653–659. [Google Scholar] [CrossRef]
- Pham, X.; Song, G.; Lao, S.; Goff, L.; Zhu, H.; Valle, D.; Avramopoulos, D. The DPYSL2 Gene Connects MTOR and Schizophrenia. Transl. Psychiatry 2016, 6, 4–11. [Google Scholar] [CrossRef]
- Matsunuma, R.; Chan, D.W.; Kim, B.J.; Singh, P.; Han, A.; Saltzman, A.B.; Cheng, C.; Lei, J.T.; Wang, J.; Roberto da Silva, L.; et al. DPYSL3 Modulates Mitosis, Migration, and Epithelial-to-Mesenchymal Transition in Claudin-Low Breast Cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E11978–E11987. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Carini, M.; Vistoli, G.; Gamberoni, L.; Giustarini, D.; Colombo, R.; Maffei Facino, R.; Rossi, R.; Milzani, A.; Aldini, G. Actin Cys374 as a Nucleophilic Target of α,β-Unsaturated Aldehydes. Free Radic. Biol. Med. 2007, 42, 583–598. [Google Scholar] [CrossRef]
- Kurahashi, T.; Kwon, M.; Homma, T.; Saito, Y.; Lee, J.; Takahashi, M.; Yamada, K.I.; Miyata, S.; Fujii, J. Reductive Detoxification of Acrolein as a Potential Role for Aldehyde Reductase (AKR1A) in Mammals. Biochem. Biophys. Res. Commun. 2014, 452, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Olguin, S.L.; Patel, P.; Buchanan, C.N.; Dell’Orco, M.; Gardiner, A.S.; Cole, R.; Vaughn, L.S.; Sundararajan, A.; Mudge, J.; Allan, A.M.; et al. KHSRP Loss Increases Neuronal Growth and Synaptic Transmission and Alters Memory Consolidation through RNA Stabilization. Commun. Biol. 2022, 5, 672. [Google Scholar] [CrossRef] [PubMed]
- Lal, J.; Kumar, S.; Madambath, I. Coconut Palm. In Encyclopedia of Food Sciences and Nutrition; Academic Press: London, UK, 2003; pp. 1464–1475. [Google Scholar] [CrossRef]

| Region | Total 3 | Coconut | Soybean | EPA/DHA 1:1 | 80% DHA | |
|---|---|---|---|---|---|---|
| Brain weight (g) | - | 1.93 (0.05) | 2.0 (0.1) | 1.9 (0.1) | 1.9 (0.1) | 1.9 (0.1) |
| Brain-to-body weight index (%) 2 | - | 0.45 (0.01) | 0.46 (0.02) | 0.43 (0.02) | 0.45 (0.03) | 0.44 (0.02) |
| FAT (%) * | Cortex | 6.60 (0.19) | 6.69 (0.31) | 6.35 (0.13) | 6.56 (0.43) | 6.80 (0.02) |
| Cerebellum | 7.64 * (0.18) | 7.61 (0.32) | 7.82 (0.08) | 7.72 (0.27) | 7.40 (0.42) | |
| Total SFAs (%) * | Cortex | 47.29 (0.37) | 47.24 (0.27) | 47.37 (0.64) | 47.42 (0.32) | 47.14 (0.30) |
| Cerebellum | 43.41 * (0.27) | 43.57 (0.32) | 43.30 (0.07) | 43.40 (0.13) | 43.38 (0.46) | |
| Total MUFAs (%) * | Cortex | 23.38 (0.73) | 23.60 (0.79) | 23.16 (1.12) | 23.22 (0.75) | 23.53 (0.55) |
| Cerebellum | 30.92 * (0.80) | 30.73 (1.16) | 30.87 (0.10) | 31.18 (0.62) | 30.89 (1.28) | |
| Total PUFAs (%) * | Cortex | 29.12 (0.20) | 28.90 (0.54) | 29.39 (0.75) | 29.09 (1.06) | 29.09 (0.35) |
| Cerebellum | 25.27 * (0.17) | 25.30 (0.93) | 25.43 (0.07) | 25.02 (0.54) | 25.31 (0.88) | |
| ω-6 (%) *$ | Cortex | 15.28 (0.91) | 15.87 a (0.46) | 16.21 a (0.43) | 14.72 ab (0.57) | 14.30 b (0.53) |
| Cerebellum | 10.88 * (0.77) | 11.46 c (0.46) | 11.61 c (0.16) | 10.35 cd (0.28) | 10.09 d (0.16) | |
| ω-3 (%) *$ | Cortex | 13.84 (0.87) | 13.02 a (0.23) | 13.18 a (0.36) | 14.37 ab (0.50) | 14.79 b (0.38) |
| Cerebellum | 14.39 * (0.68) | 13.84 ab (0.48) | 13.82 ab (0.09) | 14.67 b (0.62) | 15.21 b (0.94) | |
| ω-3/ω-6 *$ | Cortex | 0.91 (0.12) | 0.82 a (0.03) | 0.81 a (0.01) | 0.98 ab (0.01) | 1.04 b (0.06) |
| Cerebellum | 1.33 * (0.16) | 1.21 c (0.01) | 1.19 bc (0.02) | 1.42 d (0.09) | 1.51 d (0.11) | |
| ARA (%) *$ | Cortex | 10.16 (0.50) | 10.46 a (0.39) | 10.65 a (0.34) | 9.97 ab (0.44) | 9.55 b (0.42) |
| Cerebellum | 6.69 * (0.48) | 7.04 c (0.33) | 7.15 cd (0.09) | 6.41 cd (0.18) | 6.16 d (0.15) | |
| DHA (%) *$ | Cortex | 13.60 (0.81) | 12.84 a (0.23) | 12.98 a (0.37) | 14.11 ab (0.53) | 14.47 b (0.37) |
| Cerebellum | 14.06 * (0.57) | 13.60 ab (0.50) | 13.60 ab (0.10) | 14.26 ab (0.61) | 14.78 b (0.90) |
| Total 2 | Coconut | Soybean | EPA/DHA 1:1 | 80% DHA | |
|---|---|---|---|---|---|
| Brain | |||||
| SOD (U/g brain) | 663.75 (43.78) | 671 (127) | 702 (190) | 601 (111) | 681 (118) |
| CAT (mmol H2O2 decomposed/g brain) | 0.060 (0.01) | 0.059 (0.014) | 0.052 (0.016) | 0.049 (0.027) | 0.061 (0.024) |
| GPx (U/g brain) | 0.70 (0.05) | 0.71 (0.09) | 0.68 (0.13) | 0.65 (0.12) | 0.76 (0.07) |
| SOD/CAT | 11,062.50 | 11,372.88 | 13,500.00 | 12,265.31 | 11,163.93 |
| SOD/GPx | 948.21 | 945.07 | 1032.35 | 924.62 | 896.05 |
| SOD/GPx + CAT | 873.36 | 872.56 | 959.02 | 859.80 | 829.48 |
| GR (U/g brain) $ | 0.78 (0.15) | 0.66 a (0.14) | 0.96 b (0.38) | 0.66 a (0.17) | 0.84 ab (0.06) |
| GSH (µmol/g brain) $ | 0.28 (0.03) | 0.26 ab (0.03) | 0.24 a (0.04) | 0.30 bc (0.04) | 0.30 c (0.04) |
| GSSG (µmol/g brain) | 0.11 (0.01) | 0.11 (0.03) | 0.12 (0.03) | 0.11 (0.02) | 0.09 (0.01) |
| GSSG/GSH ratio $ | 0.41 (0.08) | 0.44 ab (0.12) | 0.50 a (0.15) | 0.36 bc (0.08) | 0.32 c (0.08) |
| Region | Total 2 | Coconut | Soybean | EPA/DHA 1:1 | 80% DHA | |
|---|---|---|---|---|---|---|
| CD (mmoles hydroperoxide/kg lipid) | Cortex | 5.72 (0.38) | 5.19 (0.78) | 5.88 (0.55) | 5.74 (0.92) | 6.08 (0.54) |
| Cerebellum | 5.97 (0.84) | 5.52 (0.27) | 5.09 (0.26) | 6.25 (1.41) | 7.00 (1.59) | |
| MDA (a.u./µg lipid) * | Cortex | 138.33 (2.89) | 137.37 (9.98) | 135.15 (5.19) | 142.05 (10.33) | 138.74 (19.58) |
| Cerebellum | 107.72 * (3.78) | 106.10 (4.32) | 112.04 (3.60) | 105.02 (5.39) | 102.78 (5.01) | |
| HNE (a.u./µg lipid) * | Cortex | 136.61 (3.35) | 135.24 (6.42) | 132.76 (4.52) | 140.51 (7.00) | 137.93 (24.63) |
| Cerebellum | 115.00 * (3.35) | 119.97 (10.23) | 113.93 (3.77) | 113.40 (4.77) | 112.69 (5.70) | |
| HHE (a.u./µg lipid) * | Cortex | 263.61 (7.62) | 266.06 (16.69) | 252.42 (19.80) | 266.42 (16.72) | 269.52 (64.00) |
| Cerebellum | 210.06 * (4.98) | 217.38 (5.22) | 208.99 (0.79) | 206.63 (6.42) | 207.23 (9.90) | |
| oxPC (a.u./µg lipid) * | Cortex | 617.99 (64.62) | 626.12 (59.09) | 590.50 (44.01) | 625.32 (42.97) | 630.04 (112.42) |
| Cerebellum | 544.60 * (29.55) | 524.79 (29.51) | 535.05 (25.62) | 544.41 (41.10) | 574.15 (21.95) | |
| oxPC (a.u./µg PC) * | Cortex | 456.37 (62.97) | 483.05 (71.97) | 498.31 (50.01) | 438.73 (35.63) | 405.39 (94.26) |
| Cerebellum | 445.24 * (30.89) | 454.25 (45.91) | 506.01 (31.71) | 422.02 (25.84) | 398.69 (20.11) | |
| MDA-protein adducts (a.u./µg protein) * | Cortex | 1.20 (0.31) | 1.19 (0.30) | 1.23 (0.39) | 1.32 (0.31) | 1.05 (0.24) |
| Cerebellum | 1.03 * (0.21) | 1.01 (0.20) | 1.06 (0.18) | 0.97 (0.16) | 1.10 (0.32) | |
| HNE-protein adducts (a.u./µg protein) * | Cortex | 1.91 (0.65) | 1.75 (0.64) | 1.54 (0.44) | 2.01 (0.80) | 2.32 (0.51) |
| Cerebellum | 0.80 * (0.16) | 0.79 (0.11) | 0.75 (0.14) | 0.68 (0.15) | 0.72 (0.15) | |
| HHE-protein adducts (a.u./µg protein) * | Cortex | 1.10 (0.25) | 1.13 (0.22) | 0.98 (0.15) | 1.10 (0.28) | 1.20 (0.33) |
| Cerebellum | 1.12 (0.41) | 1.17 (0.29) | 0.98 (0.20) | 1.30 (0.70) | 1.03 (0.32) | |
| oxPC-protein adducts (a.u./µg protein) * | Cortex | 1.05 (0.18) | 1.07 (0.23) | 1.11 (0.20) | 1.05 (0.14) | 0.97 (0.13) |
| Cerebellum | 0.95 * (0.16) | 0.85 (0.02) | 0.92 (0.15) | 1.12 (0.14) | 0.91 (0.12) | |
| Total protein carbonylation (a.u./µg protein) *$ | Cortex | 2.94 (0.44) | 3.18 a (0.39) | 3.05 a (0.05) | 3.10 a (0.41) | 2.41 b& (0.40) |
| Cerebellum | 1.30 * (0.17) | 1.23 c (0.16) | 1.34 c (0.07) | 1.28 c (0.30) | 1.37 c (0.16) | |
| Cytosolic protein carbonylation (a.u./µg protein) * | Cortex | 0.43 (0.07) | 0.47 (0.07) | 0.39 (0.08) | 0.39 (0.01) | 0.46 (0.07) |
| Cerebellum | 0.22 * (0.02) | 0.23 (0.02) | 0.21 (0.03) | 0.21 (0.01) | 0.21 (0.02) | |
| Myofibrillar protein carbonylation (a.u./µg protein) *$ | Cortex | 2.51 (0.47) | 2.71 a (0.46) | 2.66 a (0.05) | 2.71 a (0.41) | 1.95 b& (0.47) |
| Cerebellum | 1.09 * (0.18) | 1.00 c (0.16) | 1.13 c (0.10) | 1.07 c (0.29) | 1.16 c (0.17) | |
| Protein-SSG (a.u./µg protein) * | Cortex | 0.50 (0.02) | 0.52 (0.08) | 0.49 (0.08) | 0.49 (0.11) | 0.49 (0.09) |
| Cerebellum | 0.16 * (0.02) | 0.17 (0.05) | 0.17 (0.03) | 0.14 (0.03) | 0.14 (0.04) |
| Protein Name | Gene Name | Uniprot Code | Region | Relative Protein Amount (a.u.) 2 | Protein Carbonylation Index |
|---|---|---|---|---|---|
| Creatine kinase B-type OS = Rattus norvegicus OX = 10116 GN = Ckb PE = 1 SV = 2 | Ckb | P07335 | Cortex | 11.17 (1.18) | 1.18 (0.20) |
| Cerebellum | 6.40 * (0.90) | 0.46 * (0.13) | |||
| ATP synthase subunit beta mitochondrial OS = Rattus norvegicus OX = 10116 GN = Atp5f1b PE = 1 SV = 2 | Atp5f1b | P10719 | Cortex | 0.60 (0.12) | 0.65 (0.14) |
| Cerebellum | 3.97 * (0.55) | 1.08 * (0.19) | |||
| Pyruvate kinase PKM OS = Rattus norvegicus OX = 10116 GN = Pkm PE = 1 SV = 3 | Pkm | P11980 | Cortex | 1.35 (0.59) | 0.45 (0.24) |
| Cerebellum | 1.42 * (0.32) | 0.94 * (0.51) | |||
| Pyruvate dehydrogenase E1 component subunit alpha somatic form mitochondrial OS = Rattus norvegicus OX = 10116 GN = Pdha1 PE = 1 SV = 2 | Pdha1 | P26284 | Cortex | 0.76 (0.13) | 0.93 (0.19) |
| Cerebellum | 1.97 * (0.32) | 1.11 (0.18) | |||
| Pyruvate dehydrogenase E1 component subunit beta mitochondrial OS = Rattus norvegicus OX = 10116 GN = Pdhb PE = 1 SV = 2 | Pdhb | P49432 | Cortex | 0.42 (0.07) | 1.15 (0.34) |
| Cerebellum | 0.59 * (0.08) | 1.34 (0.33) | |||
| Aconitate hydratase mitochondrial OS = Rattus norvegicus OX = 10116 GN = Aco2 PE = 1 SV = 2 | Aco2 | Q9ER34 | Cortex | 0.52 (0.13) | 0.67 (0.15) |
| Cerebellum | 1.33 * (0.29) | 1.03 * (0.27) | |||
| L-lactate dehydrogenase B chain OS = Rattus norvegicus OX = 10116 GN = Ldhb PE = 1 SV = 2 | Ldhb | P42123 | Cortex | 0.72 (0.13) | 0.84 (0.21) |
| Cerebellum | 2.58 * (0.29) | 0.57 * (0.15) | |||
| Fructose-bisphosphate aldolase A OS = Rattus norvegicus OX = 10116 GN = Aldoa PE = 1 SV = 2 | Aldoa | P05065 | Cortex | 1.18 (0.26) | 1.30 (0.31) |
| Cerebellum | 0.57 * (0.14) | 1.53 (0.78) | |||
| Fructose-bisphosphate aldolase C OS = Rattus norvegicus OX = 10116 GN = Aldoc PE = 1 SV = 3 | Aldoc | P09117 | Cortex | 2.96 (0.35) | 1.08 (0.22) |
| Cerebellum | 3.32 (0.41) | 1.22 (0.16) | |||
| Triosephosphate isomerase OS = Rattus norvegicus OX = 10116 GN = Tpi1 PE = 1 SV = 2 | Tpi1 | P48500 | Cortex | 1.38 (0.15) | 2.78 (0.41) |
| Cerebellum | 1.20 (0.25) | 1.61 * (0.45) | |||
| Glyceraldehyde-3-phosphate dehydrogenase OS = Rattus norvegicus OX = 10116 GN = Gapdh PE = 1 SV = 3 | Gapdh | P04797 | Cortex | 2.38 (0.48) | 0.96 (0.17) |
| Cerebellum | 2.94 (0.78) | 1.21 (0.41) | |||
| Alpha-enolase OS = Rattus norvegicus OX = 10116 GN = Eno1 PE = 1 SV = 4 | Eno1 | P04764 | Cortex | 1.77 (0.48) | 0.58 (0.13) |
| Cerebellum | 2.39 * (0.44) | 0.64 (0.14) | |||
| Gamma-enolase OS = Rattus norvegicus OX = 10116 GN = Eno2 PE = 1 SV = 2 | Eno2 | P07323 | Cortex | 1.39 (0.34) | 0.45 (0.10) |
| Cerebellum | 1.37 (0.24) | 0.53 (0.19) | |||
| Aspartate aminotransferase cytoplasmic OS = Rattus norvegicus OX = 10116 GN = Got1 PE = 1 SV = 3 | Got1 | P13221 | Cortex | 0.39 (0.07) | 0.84 (0.20) |
| Cerebellum | 0.90 * (0.21) | 1.43 * (0.26) | |||
| Glutamine synthetase OS = Rattus norvegicus OX = 10116 GN = Glul PE = 1 SV = 3 | Glul | P09606 | Cortex | 2.66 (0.36) | 0.83 (0.13) |
| Cerebellum | 2.63 (0.33) | 1.12 * (0.14) | |||
| Glutamate dehydrogenase 1 mitochondrial OS = Rattus norvegicus OX = 10116 GN = Glud1 PE = 1 SV = 2 | Glud1 | P10860 | Cortex | 0.54 (0.08) | 0.89 (0.31) |
| Cerebellum | 3.76 * (0.62) | 0.58 * (0.10) | |||
| Actin cytoplasmic 1 OS = Rattus norvegicus OX = 10116 GN = Actb PE = 1 SV = 1/Actin cytoplasmic 2 OS = Rattus norvegicus OX = 10116 GN = Actg1 PE = 1 SV = 1 | Actb/Actg1 | P60711/P63259 | Cortex | 3.83 (0.85) | 0.86 (0.25) |
| Cerebellum | 8.89 * (1.33) | 1.14 * (0.19) | |||
| Tubulin beta-2A chain OS = Rattus norvegicus OX = 10116 GN = Tubb2a PE = 1 SV = 1 | Tubb2a | P85108 | Cortex | 3.72 (1.07) | 1.40 (0.27) |
| Cerebellum | 1.66 * (0.55) | 0.61 * (0.17) | |||
| Septin-11 OS = Rattus norvegicus OX = 10116 GN = Septin11 PE = 1 SV = 1 | Septin11 | B3GNI6 | Cortex | 0.51 (0.07) | 0.63 (0.11) |
| Cerebellum | 0.45 (0.06) | 0.87 * (0.17) | |||
| Syntaxin-binding protein 1 OS = Rattus norvegicus OX = 10116 GN = Stxbp1 PE = 1 SV = 1 | Stxbp1 | P61765 | Cortex | 1.09 (0.20) | 0.44 (0.09) |
| Cerebellum | 0.56 * (0.11) | 0.89 * (0.26) | |||
| Synapsin-1 OS = Rattus norvegicus OX = 10116 GN = Syn1 PE = 1 SV = 3 | Syn1 | P09951 | Cortex | 1.79 (0.54) | 1.91 (0.65) |
| Cerebellum | 0.70 * (0.19) | 1.86 (0.77) | |||
| Dihydropyrimidinase-related protein 2 OS = Rattus norvegicus OX = 10116 GN = Dpysl2 PE = 1 SV = 1 | Dpysl2 | P47942 | Cortex | 2.93 (0.35) | 0.66 (0.07) |
| Cerebellum | 1.60 * (0.16) | 0.82 (0.30) | |||
| Guanine nucleotide-binding protein G(o) subunit alpha OS = Rattus norvegicus OX = 10116 GN = Gnao1 PE = 1 SV = 2 | Gnao1 | P59215 | Cortex | 0.53 (0.11) | 0.76 (0.21) |
| Cerebellum | 0.44 (0.10) | 0.84 (0.25) | |||
| Heat shock cognate 71 kDa protein OS = Rattus norvegicus OX = 10116 GN = Hspa8 PE = 1 SV = 1 | Hspa8 | P63018 | Cortex | 1.92 (0.44) | 0.38 (0.09) |
| Cerebellum | 3.34 * (0.94) | 0.40 (0.12) | |||
| 60 kDa heat shock protein mitochondrial OS = Rattus norvegicus OX = 10116 GN = Hspd1 PE = 1 SV = 1 | Hspd1 | P63039 | Cortex | 0.50 (0.07) | 0.67 (0.14) |
| Cerebellum | 0.97 * (0.13) | 0.73 (0.16) | |||
| Albumin OS = Rattus norvegicus OX = 10116 GN = Alb PE = 1 SV = 2 | Alb | P02770 | Cortex | 1.14 (0.13) | 0.89 (0.13) |
| Cerebellum | 0.46 * (0.09) | 1.18 (0.39) | |||
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 OS = Rattus norvegicus OX = 10116 GN = Uchl1 PE = 1 SV = 2 | Uchl1 | Q00981 | Cortex | 0.80 (0.08) | 1.19 (0.24) |
| Cerebellum | <0.001 * | <0.001 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno, F.; Méndez, L.; Fernández, I.; Miralles-Pérez, B.; Giralt, M.; Romeu, M.; Ramos-Romero, S.; Torres, J.L.; Medina, I. Influence of the Degree of Unsaturation in Fish Oil Supplements on Oxidative Stress and Protein Carbonylation in the Cerebral Cortex and Cerebellum of Healthy Rats. Antioxidants 2024, 13, 1408. https://doi.org/10.3390/antiox13111408
Moreno F, Méndez L, Fernández I, Miralles-Pérez B, Giralt M, Romeu M, Ramos-Romero S, Torres JL, Medina I. Influence of the Degree of Unsaturation in Fish Oil Supplements on Oxidative Stress and Protein Carbonylation in the Cerebral Cortex and Cerebellum of Healthy Rats. Antioxidants. 2024; 13(11):1408. https://doi.org/10.3390/antiox13111408
Chicago/Turabian StyleMoreno, Francisco, Lucía Méndez, Ingrid Fernández, Bernat Miralles-Pérez, Montserrat Giralt, Marta Romeu, Sara Ramos-Romero, Josep Lluís Torres, and Isabel Medina. 2024. "Influence of the Degree of Unsaturation in Fish Oil Supplements on Oxidative Stress and Protein Carbonylation in the Cerebral Cortex and Cerebellum of Healthy Rats" Antioxidants 13, no. 11: 1408. https://doi.org/10.3390/antiox13111408
APA StyleMoreno, F., Méndez, L., Fernández, I., Miralles-Pérez, B., Giralt, M., Romeu, M., Ramos-Romero, S., Torres, J. L., & Medina, I. (2024). Influence of the Degree of Unsaturation in Fish Oil Supplements on Oxidative Stress and Protein Carbonylation in the Cerebral Cortex and Cerebellum of Healthy Rats. Antioxidants, 13(11), 1408. https://doi.org/10.3390/antiox13111408







