SARS-CoV-2 Spike Protein Stimulates Macropinocytosis in Murine and Human Macrophages via PKC-NADPH Oxidase Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Primary Human Macrophages
2.4. Animals
2.5. Isolation of Murine Macrophages
2.5.1. Thioglycollate-Elicited Peritoneal Macrophages
2.5.2. Bone-Marrow-Derived Macrophages
2.6. 2′,7′-Dichlorodihydrofluorescein Diacetate (H2DCFDA) Assay
2.7. Flow Cytometry
2.8. Scanning Electron Microscopy
2.9. Statistical Analysis
3. Results
3.1. Recombinant SARS-CoV-2 Spike Proteins Stimulate Macropinocytosis in Murine Macrophages
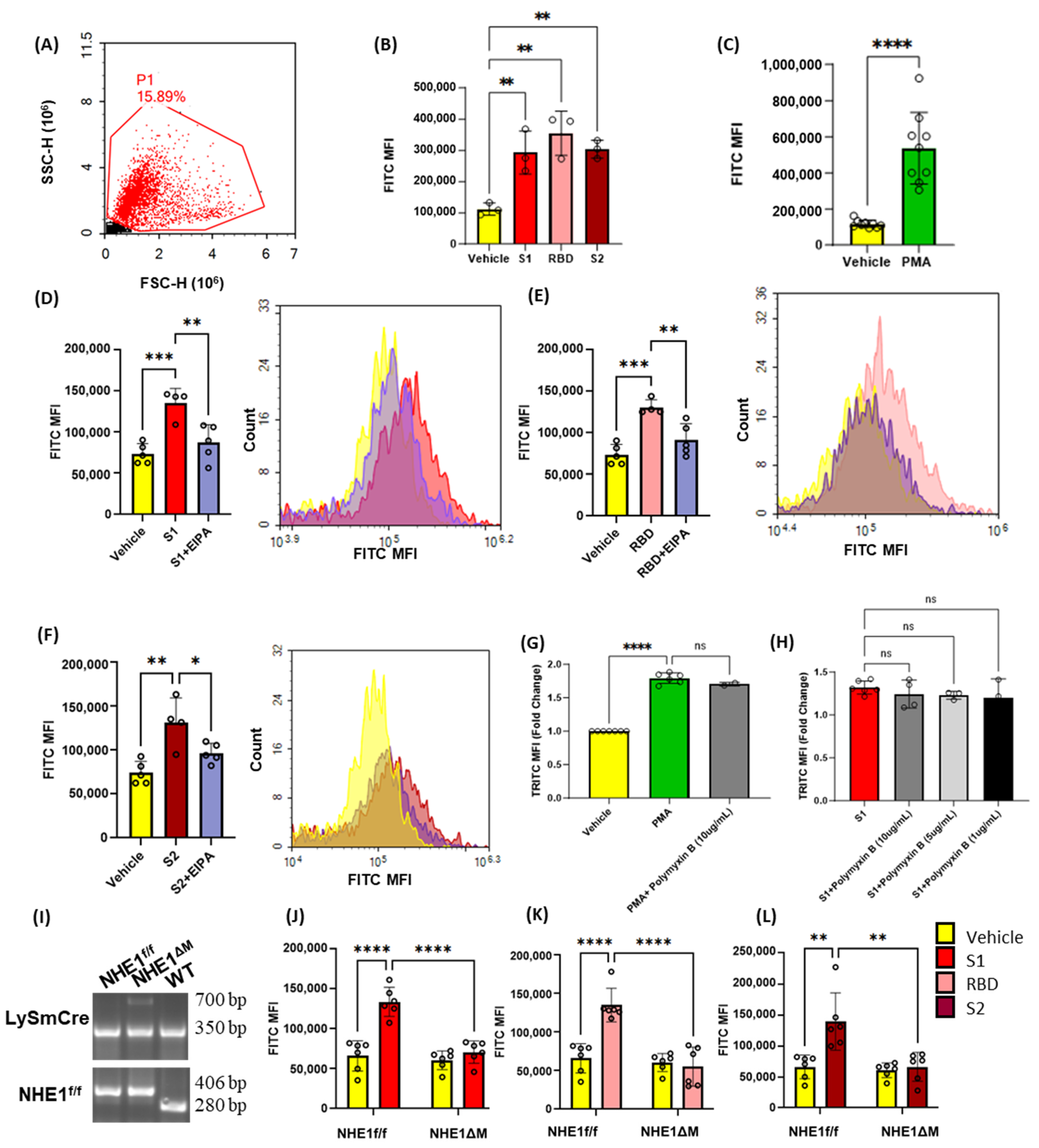
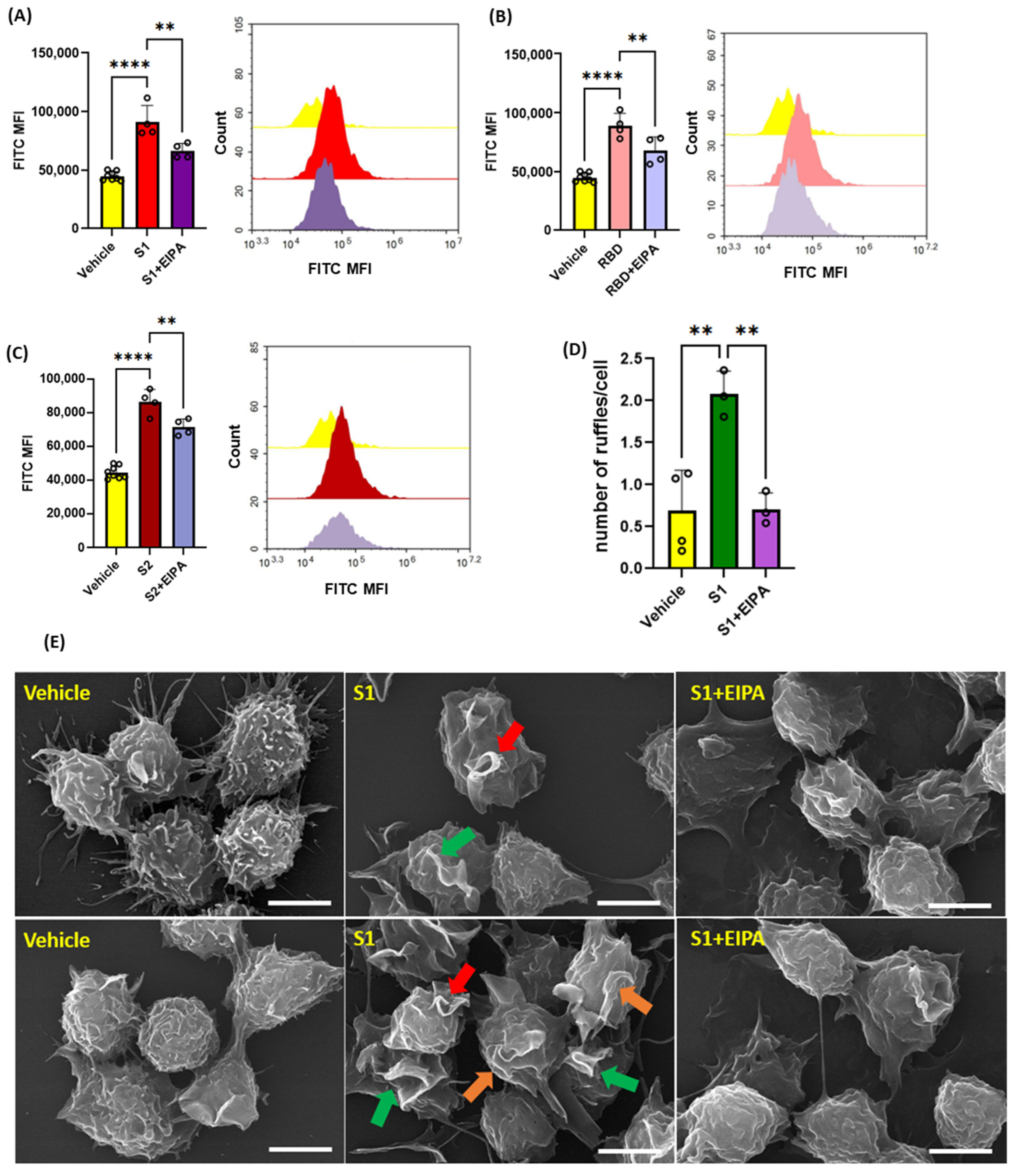
3.2. SARS-CoV2 Spike Protein Subunits Stimulate Macropinocytosis in Primary Human Macrophages
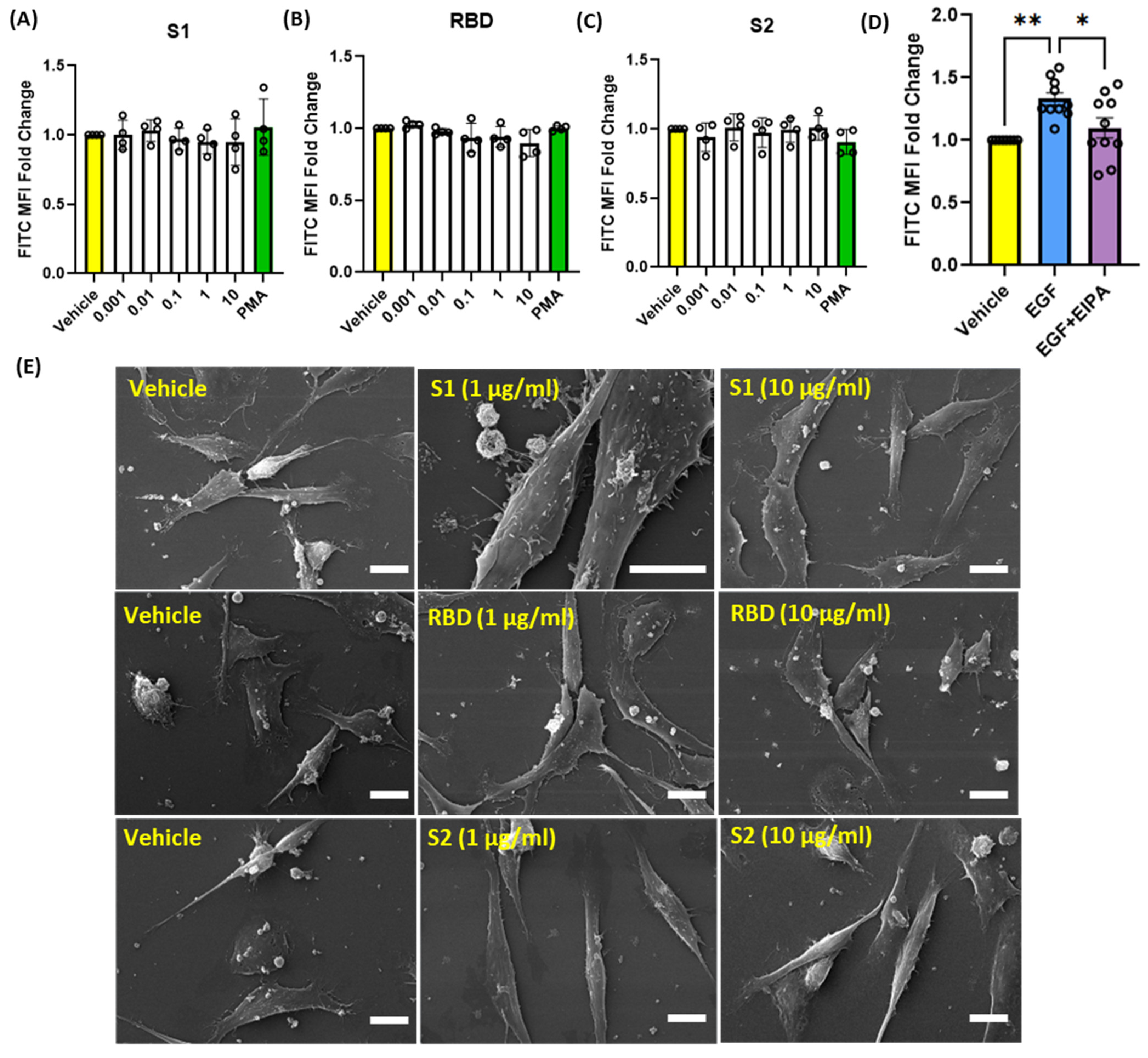
3.3. SARS-CoV-2 Spike Proteins Do Not Stimulate Macropinocytosis in Human Alveolar Epithelial Cells
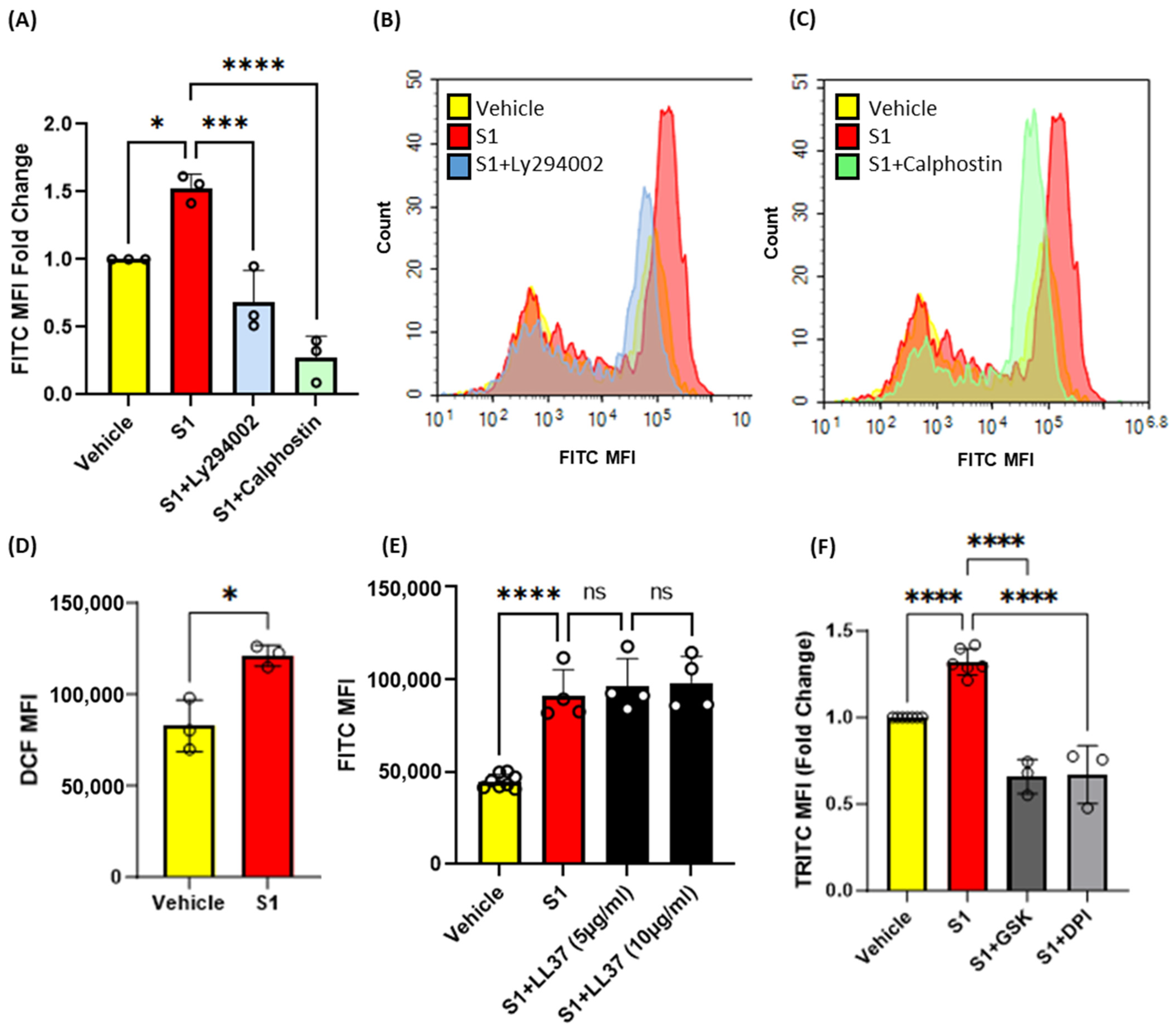
3.4. Inhibition of Protein Kinase C, Phosphoinositide 3-Kinase and Nox2 Blocks SARS-CoV-2 Spike-Protein-Induced Macropinocytosis in Macrophages
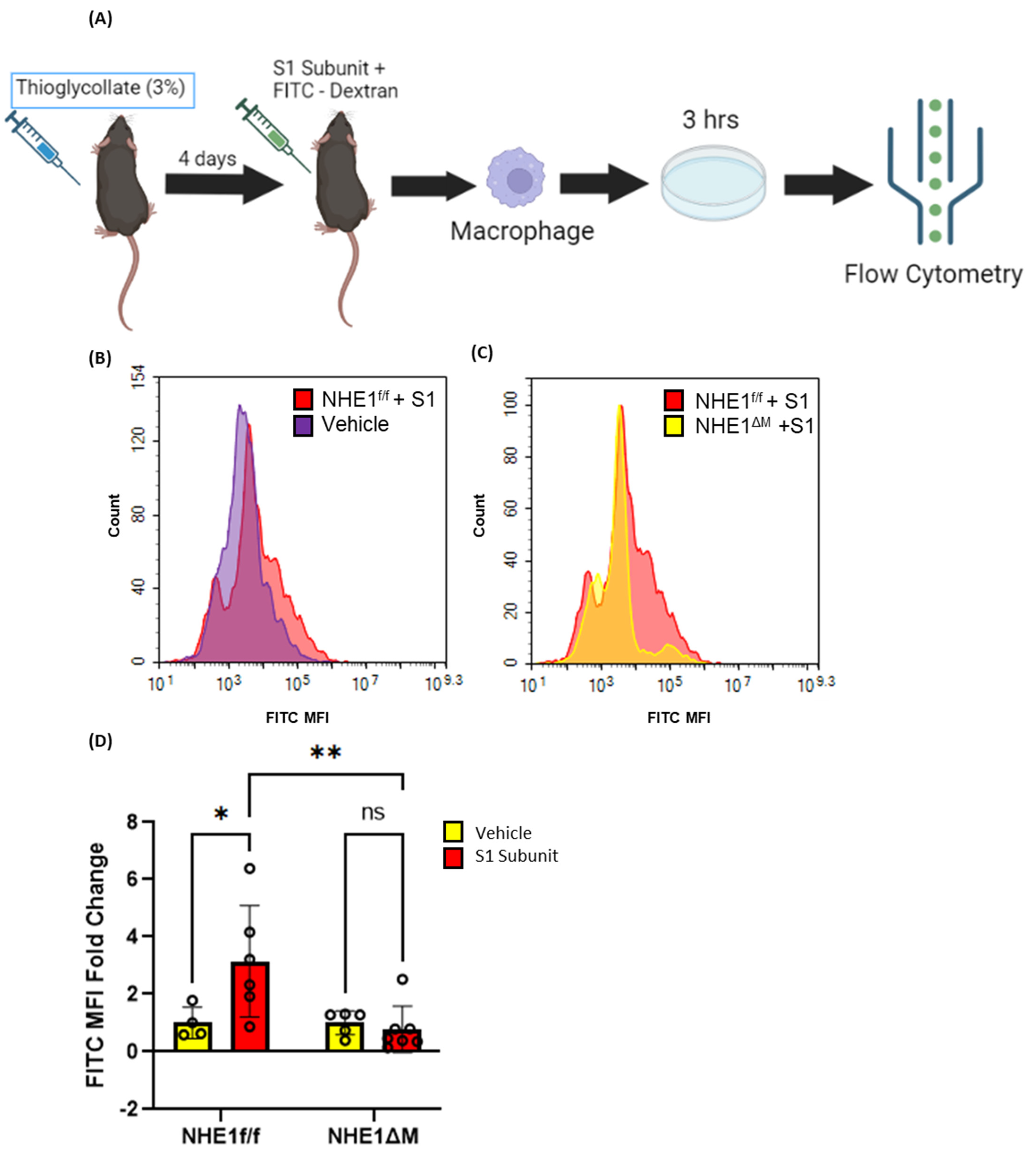
3.5. SARS-CoV-2 Spike Proteins Stimulate Macrophage Macropinocytosis In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collaborators, C.-E.M. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020–2021. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef]
- Hulswit, R.J.; de Haan, C.A.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Bridges, J.P.; Vladar, E.K.; Huang, H.; Mason, R.J. Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax 2022, 77, 203–209. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef]
- Konopka, K.E.; Nguyen, T.; Jentzen, J.M.; Rayes, O.; Schmidt, C.J.; Wilson, A.M.; Farver, C.F.; Myers, J.L. Diffuse alveolar damage (DAD) resulting from coronavirus disease 2019 Infection is Morphologically Indistinguishable from Other Causes of DAD. Histopathology 2020, 77, 570–578. [Google Scholar] [CrossRef]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and Macrophages in COVID-19. Front. Immunol. 2021, 12, 720109. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Grant, R.A.; Morales-Nebreda, L.; Markov, N.S.; Swaminathan, S.; Querrey, M.; Guzman, E.R.; Abbott, D.A.; Donnelly, H.K.; Donayre, A.; Goldberg, I.A.; et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021, 590, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Glebov, O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020, 287, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Liang, R.; Wang, S.J.; Ye, Z.W.; Wang, T.Y.; Chen, M.; Liu, J.; Na, L.; Yang, Y.L.; Yang, Y.B.; et al. SARS-CoV-2 hijacks macropinocytosis to facilitate its entry and promote viral spike-mediated cell-to-cell fusion. J. Biol. Chem. 2022, 298, 102511. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.P.; Mintern, J.D.; Gleeson, P.A. Macropinocytosis in Different Cell Types: Similarities and Differences. Membranes 2020, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.; Singla, B.; Marshall, B.; Csanyi, G. Visualizing Membrane Ruffle Formation using Scanning Electron Microscopy. J. Vis. Exp. 2021, 171, e62658. [Google Scholar] [CrossRef]
- Marechal, V.; Prevost, M.C.; Petit, C.; Perret, E.; Heard, J.M.; Schwartz, O. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J. Virol. 2001, 75, 11166–11177. [Google Scholar] [CrossRef]
- Aleksandrowicz, P.; Marzi, A.; Biedenkopf, N.; Beimforde, N.; Becker, S.; Hoenen, T.; Feldmann, H.; Schnittler, H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011, 204 (Suppl. S3), S957–S967. [Google Scholar] [CrossRef]
- Nicola, A.V.; Hou, J.; Major, E.O.; Straus, S.E. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 2005, 79, 7609–7616. [Google Scholar] [CrossRef]
- Ghoshal, P.; Singla, B.; Lin, H.; Feck, D.M.; Cantu-Medellin, N.; Kelley, E.E.; Haigh, S.; Fulton, D.; Csanyi, G. Nox2-Mediated PI3K and Cofilin Activation Confers Alternate Redox Control of Macrophage Pinocytosis. Antioxid. Redox Signal 2017, 26, 902–916. [Google Scholar] [CrossRef] [PubMed]
- Bedard, K.; Krause, K.H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Singla, B.; Lin, H.P.; Ghoshal, P.; Cherian-Shaw, M.; Csanyi, G. PKCdelta stimulates macropinocytosis via activation of SSH1-cofilin pathway. Cell Signal 2019, 53, 111–121. [Google Scholar] [CrossRef]
- Yoshida, S.; Gaeta, I.; Pacitto, R.; Krienke, L.; Alge, O.; Gregorka, B.; Swanson, J.A. Differential signaling during macropinocytosis in response to M-CSF and PMA in macrophages. Front. Physiol. 2015, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Kerr, M.C.; Hammond, L.A.; Joseph, S.R.; Mostov, K.E.; Teasdale, R.D.; Stow, J.L. EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. J. Cell Sci. 2007, 120, 1818–1828. [Google Scholar] [CrossRef]
- Yoshida, S.; Pacitto, R.; Yao, Y.; Inoki, K.; Swanson, J.A. Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J. Cell Biol. 2015, 211, 159–172. [Google Scholar] [CrossRef]
- Zhang, X.; Goncalves, R.; Mosser, D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008, 14, 14.1.1–14.1.14. [Google Scholar] [CrossRef]
- Tardelli, M.; Zeyda, K.; Moreno-Viedma, V.; Wanko, B.; Grun, N.G.; Staffler, G.; Zeyda, M.; Stulnig, T.M. Osteopontin is a key player for local adipose tissue macrophage proliferation in obesity. Mol. Metab. 2016, 5, 1131–1137. [Google Scholar] [CrossRef]
- Kruth, H.S.; Jones, N.L.; Huang, W.; Zhao, B.; Ishii, I.; Chang, J.; Combs, C.A.; Malide, D.; Zhang, W.Y. Macropinocytosis is the endocytic pathway that mediates macrophage foam cell formation with native low density lipoprotein. J. Biol. Chem. 2005, 280, 2352–2360. [Google Scholar] [CrossRef]
- Jayashankar, V.; Edinger, A.L. Macropinocytosis confers resistance to therapies targeting cancer anabolism. Nat. Commun. 2020, 11, 1121. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Singla, B.; Ahn, W.; Ghoshal, P.; Blahove, M.; Cherian-Shaw, M.; Chen, A.; Haller, A.; Hui, D.Y.; Dong, K.; et al. Receptor-independent fluid-phase macropinocytosis promotes arterial foam cell formation and atherosclerosis. Sci. Transl. Med. 2022, 14, eadd2376. [Google Scholar] [CrossRef] [PubMed]
- West, M.A.; Bretscher, M.S.; Watts, C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 1989, 109, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Singla, B.; Ghoshal, P.; Lin, H.; Wei, Q.; Dong, Z.; Csanyi, G. PKCdelta-Mediated Nox2 Activation Promotes Fluid-Phase Pinocytosis of Antigens by Immature Dendritic Cells. Front. Immunol. 2018, 9, 537. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef]
- Freeman, M.C.; Peek, C.T.; Becker, M.M.; Smith, E.C.; Denison, M.R. Coronaviruses induce entry-independent, continuous macropinocytosis. mBio 2014, 5, e01340-14. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Miura, H.S.; Nakagaki, K.; Taguchi, F. N-terminal domain of the murine coronavirus receptor CEACAM1 is responsible for fusogenic activation and conformational changes of the spike protein. J. Virol. 2004, 78, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zeng, R.; von Brunn, A.; Lei, J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol. Biomed. 2020, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhong, K.; Wang, G.; Lu, Q.; Li, H.; Wu, Z.; Zhang, Z.; Yang, N.; Zheng, M.; Wang, Y.; et al. Loss of furin site enhances SARS-CoV-2 spike protein pseudovirus infection. Gene 2023, 856, 147144. [Google Scholar] [CrossRef]
- Mulay, A.; Konda, B.; Garcia, G., Jr.; Yao, C.; Beil, S.; Villalba, J.M.; Koziol, C.; Sen, C.; Purkayastha, A.; Kolls, J.K.; et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021, 35, 109055. [Google Scholar] [CrossRef]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Zheng, J.; Miao, J.; Guo, R.; Guo, J.; Fan, Z.; Kong, X.; Gao, R.; Yang, L. Mechanism of COVID-19 Causing ARDS: Exploring the Possibility of Preventing and Treating SARS-CoV-2. Front. Cell Infect. Microbiol. 2022, 12, 931061. [Google Scholar] [CrossRef]
- Abdullaev, A.; Odilov, A.; Ershler, M.; Volkov, A.; Lipina, T.; Gasanova, T.; Lebedin, Y.; Babichenko, I.; Sudarikov, A. Viral Load and Patterns of SARS-CoV-2 Dissemination to the Lungs, Mediastinal Lymph Nodes, and Spleen of Patients with COVID-19 Associated Lymphopenia. Viruses 2021, 13, 1410. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, O.; Nechipurenko, Y.; Lagutkin, D.; Yegorov, Y.E.; Kzhyshkowska, J. SARS-CoV-2 infection of phagocytic immune cells and COVID-19 pathology: Antibody-dependent as well as independent cell entry. Front. Immunol. 2022, 13, 1050478. [Google Scholar] [CrossRef] [PubMed]
- Jum’ah, H.; Kundrapu, S.; Jabri, A.; Kondapaneni, M.; Tomashefski, J.F.; Loeffler, A.G. Cardiac macrophage density in Covid-19 infection: Relationship to myocyte necrosis and acute lung injury. Cardiovasc. Pathol. 2022, 60, 107447. [Google Scholar] [CrossRef] [PubMed]
- Labzin, L.I.; Chew, K.Y.; Eschke, K.; Wang, X.; Esposito, T.; Stocks, C.J.; Rae, J.; Patrick, R.; Mostafavi, H.; Hill, B.; et al. Macrophage ACE2 is necessary for SARS-CoV-2 replication and subsequent cytokine responses that restrict continued virion release. Sci. Signal 2023, 16, eabq1366. [Google Scholar] [CrossRef]
- Bost, P.; Giladi, A.; Liu, Y.; Bendjelal, Y.; Xu, G.; David, E.; Blecher-Gonen, R.; Cohen, M.; Medaglia, C.; Li, H.; et al. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell 2020, 181, 1475–1488.e1412. [Google Scholar] [CrossRef] [PubMed]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, A.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Knyazev, E.; Nersisyan, S.; Tonevitsky, A. Endocytosis and Transcytosis of SARS-CoV-2 Across the Intestinal Epithelium and Other Tissue Barriers. Front. Immunol. 2021, 12, 636966. [Google Scholar] [CrossRef]
- Garcia-Nicolas, O.; Godel, A.; Zimmer, G.; Summerfield, A. Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation. Cell Mol. Immunol. 2023, 20, 835–849. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef]
- Bain, C.C.; Lucas, C.D.; Rossi, A.G. Pulmonary macrophages and SARS-Cov2 infection. Int. Rev. Cell Mol. Biol. 2022, 367, 1–28. [Google Scholar] [CrossRef]
- Quinn, S.E.; Huang, L.; Kerkvliet, J.G.; Swanson, J.A.; Smith, S.; Hoppe, A.D.; Anderson, R.B.; Thiex, N.W.; Scott, B.L. The structural dynamics of macropinosome formation and PI3-kinase-mediated sealing revealed by lattice light sheet microscopy. Nat. Commun. 2021, 12, 4838. [Google Scholar] [CrossRef]
- Redka, D.S.; Gutschow, M.; Grinstein, S.; Canton, J. Differential ability of proinflammatory and anti-inflammatory macrophages to perform macropinocytosis. Mol. Biol. Cell 2018, 29, 53–65. [Google Scholar] [CrossRef]
- Condon, N.D.; Heddleston, J.M.; Chew, T.L.; Luo, L.; McPherson, P.S.; Ioannou, M.S.; Hodgson, L.; Stow, J.L.; Wall, A.A. Macropinosome formation by tent pole ruffling in macrophages. J. Cell Biol. 2018, 217, 3873–3885. [Google Scholar] [CrossRef]
- Li, D.; Chen, P.; Shi, T.; Mehmood, A.; Qiu, J. HD5 and LL-37 Inhibit SARS-CoV and SARS-CoV-2 Binding to Human ACE2 by Molecular Simulation. Interdiscip. Sci. 2021, 13, 766–777. [Google Scholar] [CrossRef]
- Liu, C.L.; Zhang, X.; Liu, J.; Wang, Y.; Sukhova, G.K.; Wojtkiewicz, G.R.; Liu, T.; Tang, R.; Achilefu, S.; Nahrendorf, M.; et al. Na(+)-H(+) exchanger 1 determines atherosclerotic lesion acidification and promotes atherogenesis. Nat. Commun. 2019, 10, 3978. [Google Scholar] [CrossRef] [PubMed]
- Saied, E.M.; Bediwy, A.S. Expression of epidermal growth factor receptor (EGFR) in the bronchial epithelium of patients with chronic obstructive pulmonary disease (COPD). Eur. Respir. J. 2011, 38, p3823. [Google Scholar]
- Kobayashi, E.; Nakano, H.; Morimoto, M.; Tamaoki, T. Calphostin C (UCN-1028C), a novel microbial compound, is a highly potent and specific inhibitor of protein kinase C. Biochem. Biophys. Res. Commun. 1989, 159, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, S.I.; Zvelebil, M.J.; Shuttleworth, S.J.; Hancox, T.; Saghir, N.; Timms, J.F.; Waterfield, M.D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007, 404, 15–21. [Google Scholar] [CrossRef]
- Piszczatowska, K.; Przybylska, D.; Sikora, E.; Mosieniak, G. Inhibition of NADPH Oxidases Activity by Diphenyleneiodonium Chloride as a Mechanism of Senescence Induction in Human Cancer Cells. Antioxidants 2020, 9, 1248. [Google Scholar] [CrossRef]
- Altenhofer, S.; Radermacher, K.A.; Kleikers, P.W.; Wingler, K.; Schmidt, H.H. Evolution of NADPH Oxidase Inhibitors: Selectivity and Mechanisms for Target Engagement. Antioxid. Redox Signal 2015, 23, 406–427. [Google Scholar] [CrossRef]
- Hirano, K.; Chen, W.S.; Chueng, A.L.; Dunne, A.A.; Seredenina, T.; Filippova, A.; Ramachandran, S.; Bridges, A.; Chaudry, L.; Pettman, G.; et al. Discovery of GSK2795039, a Novel Small Molecule NADPH Oxidase 2 Inhibitor. Antioxid. Redox Signal 2015, 23, 358–374. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.G.; Monica, F.Z.; Passos, G.R.; Victorio, J.A.; Davel, A.P.; Oliveira, A.L.L.; Parada, C.A.; D’Ancona, C.A.L.; Hill, W.G.; Antunes, E. Selective Pharmacological Inhibition of NOX2 by GSK2795039 Improves Bladder Dysfunction in Cyclophosphamide-Induced Cystitis in Mice. Antioxidants 2022, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Bozkurt, B. Shedding Light on Mechanisms of Myocarditis With COVID-19 mRNA Vaccines. Circulation 2023, 147, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Petrovszki, D.; Walter, F.R.; Vigh, J.P.; Kocsis, A.; Valkai, S.; Deli, M.A.; Der, A. Penetration of the SARS-CoV-2 Spike Protein across the Blood-Brain Barrier, as Revealed by a Combination of a Human Cell Culture Model System and Optical Biosensing. Biomedicines 2022, 10, 188. [Google Scholar] [CrossRef]
- Lim, S.; Zhang, M.; Chang, T.L. ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses 2022, 14, 2535. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, W.; Burnett, F.N.; Pandey, A.; Ghoshal, P.; Singla, B.; Simon, A.B.; Derella, C.C.; A. Addo, S.; Harris, R.A.; Lucas, R.; et al. SARS-CoV-2 Spike Protein Stimulates Macropinocytosis in Murine and Human Macrophages via PKC-NADPH Oxidase Signaling. Antioxidants 2024, 13, 175. https://doi.org/10.3390/antiox13020175
Ahn W, Burnett FN, Pandey A, Ghoshal P, Singla B, Simon AB, Derella CC, A. Addo S, Harris RA, Lucas R, et al. SARS-CoV-2 Spike Protein Stimulates Macropinocytosis in Murine and Human Macrophages via PKC-NADPH Oxidase Signaling. Antioxidants. 2024; 13(2):175. https://doi.org/10.3390/antiox13020175
Chicago/Turabian StyleAhn, WonMo, Faith N. Burnett, Ajay Pandey, Pushpankur Ghoshal, Bhupesh Singla, Abigayle B. Simon, Cassandra C. Derella, Stephen A. Addo, Ryan A. Harris, Rudolf Lucas, and et al. 2024. "SARS-CoV-2 Spike Protein Stimulates Macropinocytosis in Murine and Human Macrophages via PKC-NADPH Oxidase Signaling" Antioxidants 13, no. 2: 175. https://doi.org/10.3390/antiox13020175
APA StyleAhn, W., Burnett, F. N., Pandey, A., Ghoshal, P., Singla, B., Simon, A. B., Derella, C. C., A. Addo, S., Harris, R. A., Lucas, R., & Csányi, G. (2024). SARS-CoV-2 Spike Protein Stimulates Macropinocytosis in Murine and Human Macrophages via PKC-NADPH Oxidase Signaling. Antioxidants, 13(2), 175. https://doi.org/10.3390/antiox13020175





