Ginsenosides Rg1 and Rg2 Activate Autophagy and Attenuate Oxidative Stress in Neuroblastoma Cells Overexpressing Aβ(1-42)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Regents and Chemicals
2.2. Cell Treatment and Cell Viability Assay
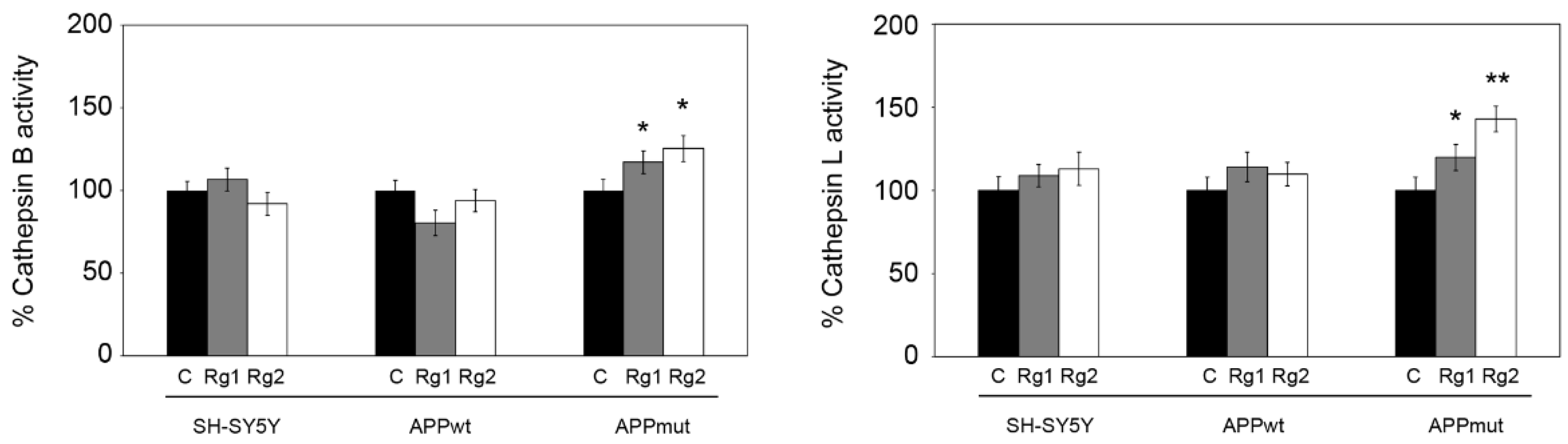
2.3. Cathepsin Activities
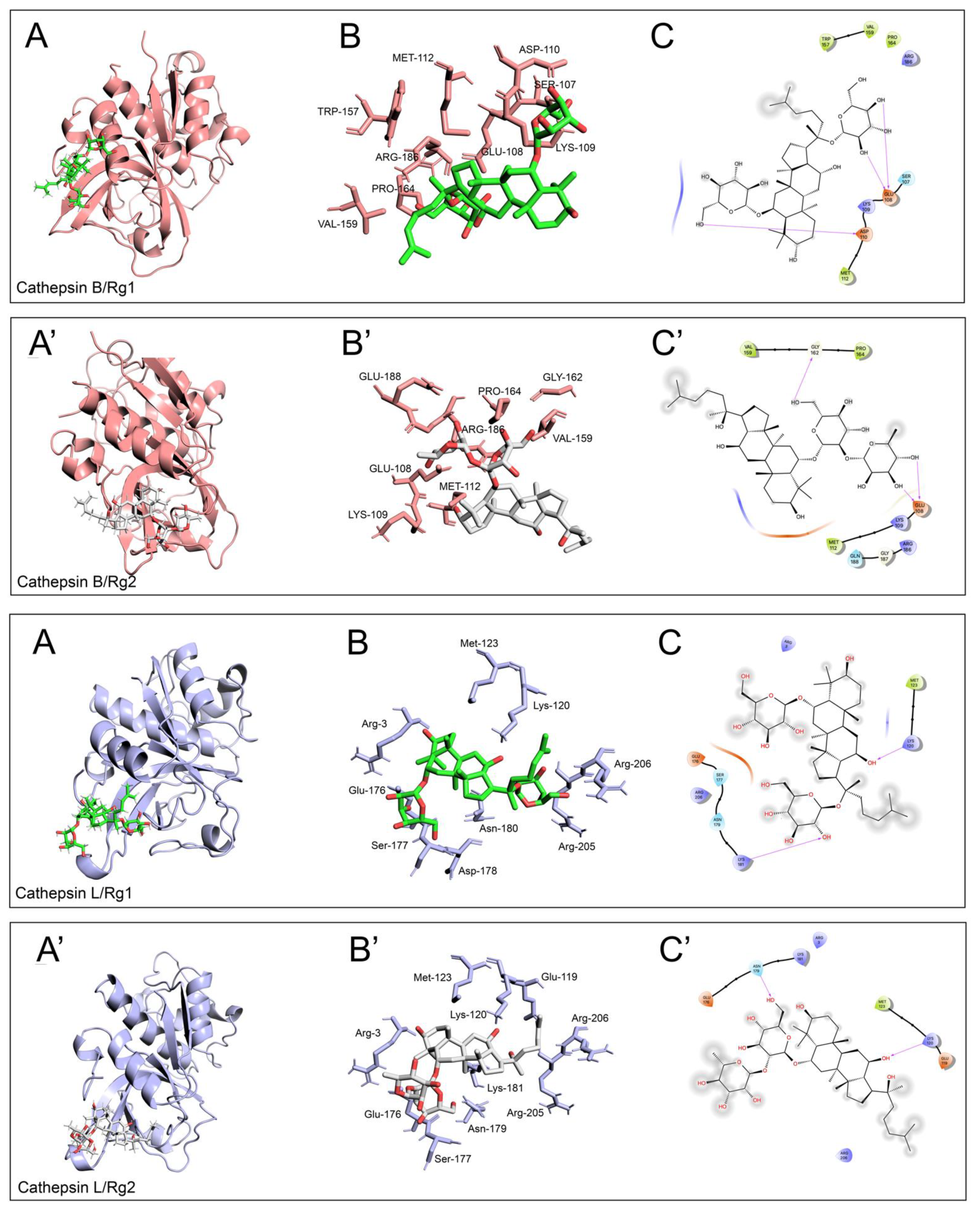
2.4. Molecular Docking
2.5. Western Blotting
2.6. Oxyblot Analysis
2.7. ROS Production and Mitochondrial Membrane Potential
2.8. Quantification of Aβ(1-42) in Cell Medium and Cell Lysates
2.9. Statistical Analysis
3. Results
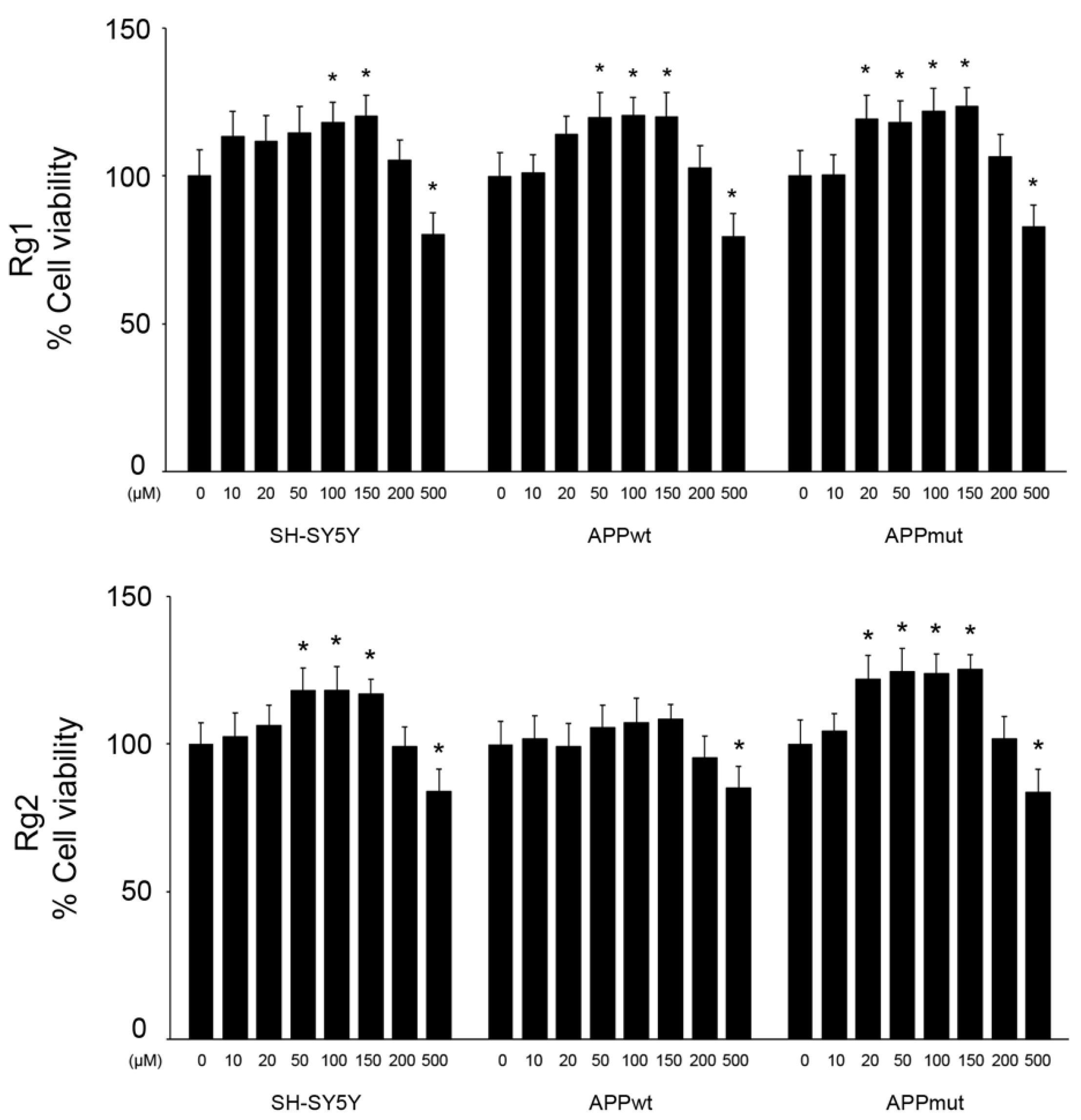
3.1. Effect of Rg1 and Rg2 on Cell Viability
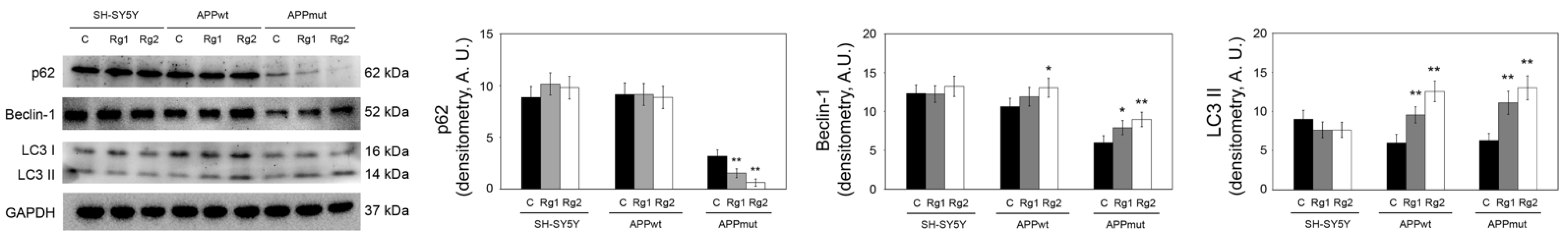
3.2. Effect of Rg1 and Rg2 on Autophagy-Related Proteins
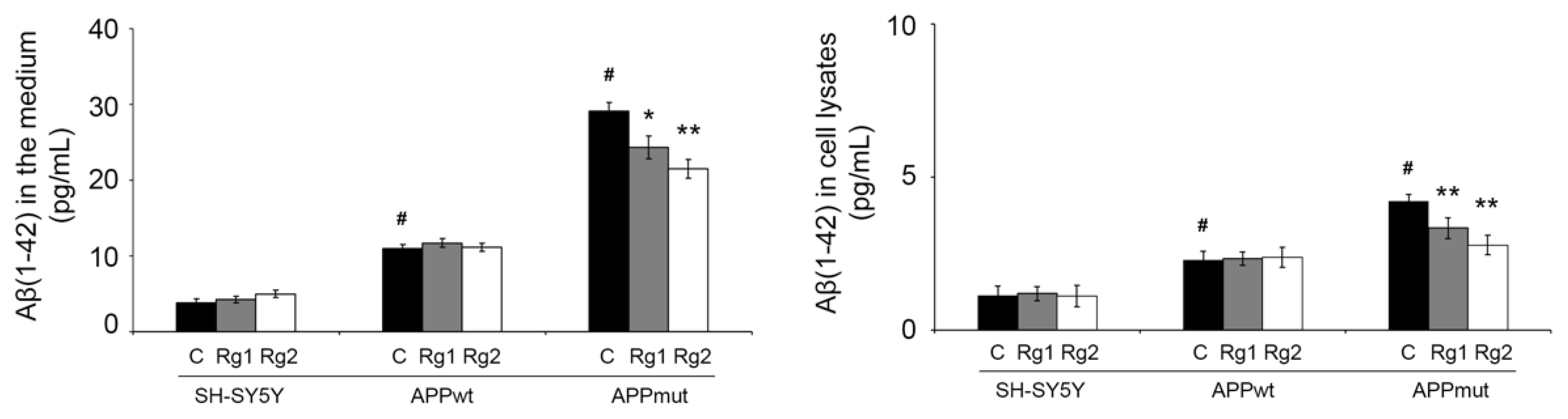
3.3. Determination of Aβ(1-42) in Cell Lysates and Cell Medium
3.4. ROS Production and Mitochondrial Membrane Potential
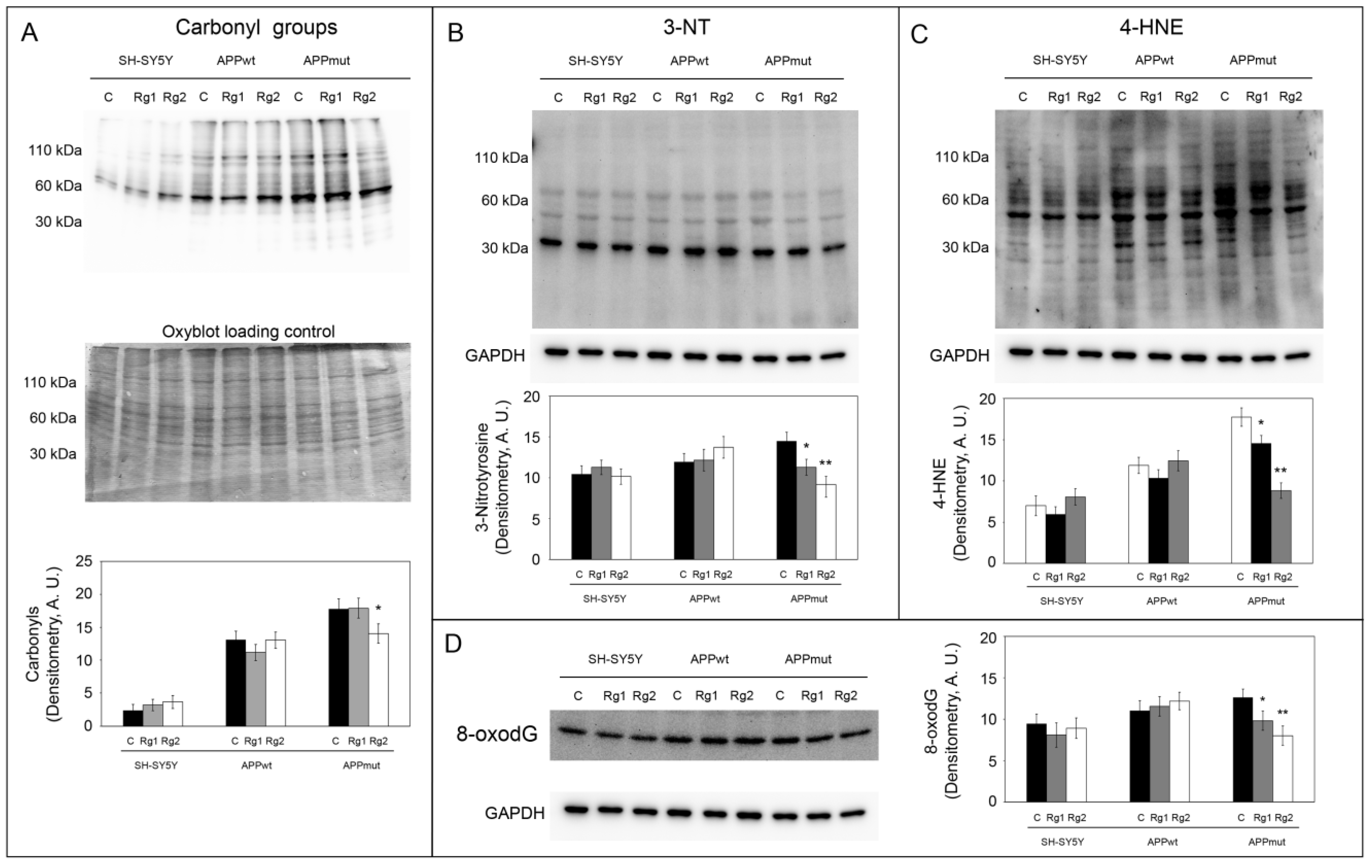
3.5. Oxidation of Cellular Macromolecules
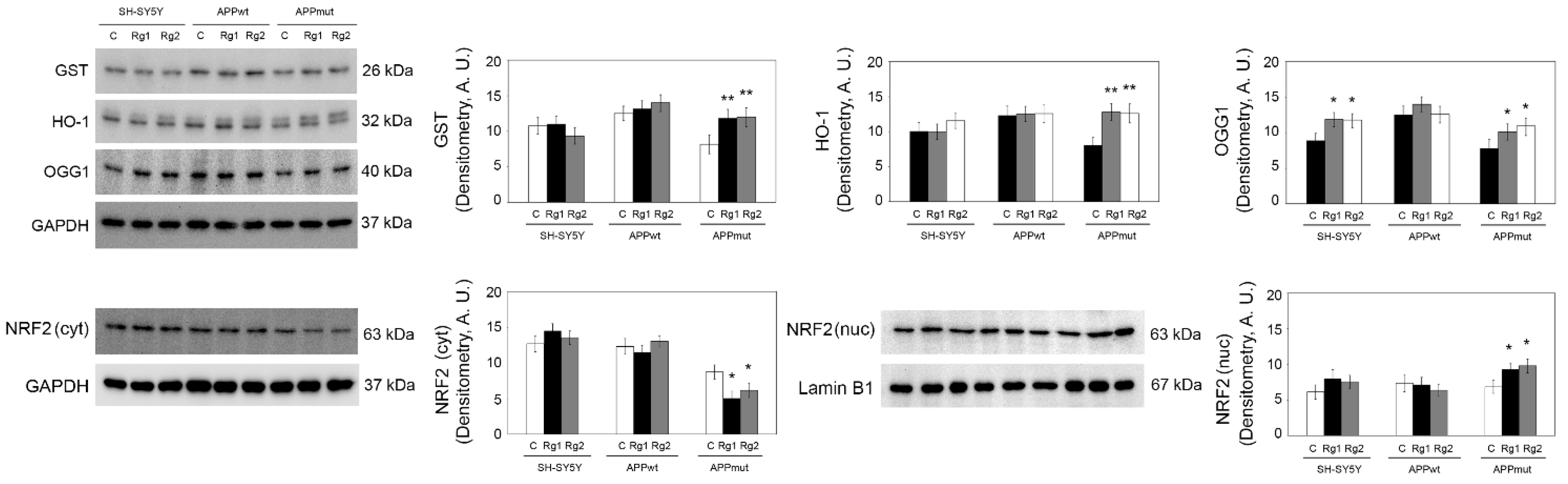
3.6. Upregulation of Antioxidant Enzyme Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2023, 19, 1598–1695. [Google Scholar] [CrossRef]
- Dröge, W. Free Radicals in the Physiological Control of Cell Function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Rottkamp, C.A.; Nunomura, A.; Raina, A.K.; Perry, G. Oxidative stress in Alzheimer’s disease. Biochim. Biophys. Acta 2000, 1502, 139–144. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Nixon, R.A.; Yang, D.S. Autophagy failure in Alzheimer’s disease—Locating the primary defect. Neurobiol. Dis. 2011, 43, 38–45. [Google Scholar] [CrossRef]
- Liu, J.; Li, L. Targeting Autophagy for the Treatment of Alzheimer’s Disease: Challenges and Opportunities. Front. Mol. Neurosci. 2019, 12, 203. [Google Scholar] [CrossRef] [PubMed]
- Talebi, M.; Mohammadi Vadoud, S.A.; Haratian, A.; Talebi, M.; Farkhondeh, T.; Pourbagher-Shahri, A.M.; Samarghandian, S. The interplay between oxidative stress and autophagy: Focus on the development of neurological diseases. Behav. Brain Funct. 2022, 18, 3. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Interactions between reactive oxygen species and autophagy: Special issue: Death mechanisms in cellular homeostasis. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119041. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Pajares, M.; Jimenez-Moreno, N.; Garcia-Yague, A.J.; Escoll, M.; de Ceballos, M.L.; Van Leuven, F.; Rabano, A.; Yamamoto, M.; Rojo, A.I.; Cuadrado, A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 2016, 12, 1902–1916. [Google Scholar] [CrossRef]
- He, B.; Chen, D.; Zhang, X.; Yang, R.; Yang, Y.; Chen, P.; Shen, Z. Oxidative Stress and Ginsenosides: An Update on the Molecular Mechanisms. Oxidative Med. Cell. Longev. 2022, 2022, 9299574. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Cha, S.; Kim, M.Y.; Cho, J.Y. Ginsenosides are active ingredients in Panax ginseng with immunomodulatory properties from cellular to organismal levels. J. Ginseng Res. 2022, 46, 711–721. [Google Scholar] [CrossRef]
- Cecarini, V.; Cuccioloni, M.; Gong, C.; Liu, Z.; Bonfili, L.; Angeletti, M.; Angeloni, S.; Alessandroni, L.; Sagratini, G.; Liu, H.; et al. Role of Panax ginseng and ginsenosides in regulating cholesterol homeostasis. Food Biosci. 2023, 56, 103256. [Google Scholar] [CrossRef]
- Angeloni, S.; Marinelli, O.; Aguzzi, C.; Nabissi, M.; Bordoni, L.; Petracci, I.; Gabbianelli, R.; Cecarini, V.; Angeletti, M.; Eleuteri, A.M.; et al. Anti-inflammatory and antioxidant activities of ginsenosides and ginseng root extract to counteract obesity. Eur. Food Res. Technol. 2021, 250, 119–133. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Zhang, C.; Guo, Y.; Li, J.; Wu, C.; Jiao, J.; Zheng, H. Ginsenoside Rg1 improves Alzheimer’s disease by regulating oxidative stress, apoptosis, and neuroinflammation through Wnt/GSK-3β/β-catenin signaling pathway. Chem. Biol. Drug Des. 2022, 99, 884–896. [Google Scholar] [CrossRef]
- Wang, N.; Yang, J.; Chen, R.; Liu, Y.; Liu, S.; Pan, Y.; Lei, Q.; Wang, Y.; He, L.; Song, Y.; et al. Ginsenoside Rg1 ameliorates Alzheimer’s disease pathology via restoring mitophagy. J. Ginseng Res. 2023, 47, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Shan, R.; Cao, Y.; Zhou, Y.; Liu, C.; Fan, Y. Protective effects of ginsenoside Rg2 against memory impairment and neuronal death induced by Aβ25-35 in rats. J. Ethnopharmacol. 2021, 266, 113466. [Google Scholar] [CrossRef]
- Ye, X.; Shao, S.; Wang, Y.; Su, W. Ginsenoside Rg2 alleviates neurovascular damage in 3xTg-AD mice with Alzheimer’s disease through the MAPK-ERK pathway. J. Chem. Neuroanat. 2023, 133, 102346. [Google Scholar] [CrossRef]
- Cecarini, V.; Bonfili, L.; Cuccioloni, M.; Mozzicafreddo, M.; Rossi, G.; Buizza, L.; Uberti, D.; Angeletti, M.; Eleuteri, A.M. Crosstalk between the ubiquitin-proteasome system and autophagy in a human cellular model of Alzheimer’s disease. Biochim. Biophys. Acta 2012, 1822, 1741–1751. [Google Scholar] [CrossRef]
- Zampagni, M.; Evangelisti, E.; Cascella, R.; Liguri, G.; Becatti, M.; Pensalfini, A.; Uberti, D.; Cenini, G.; Memo, M.; Bagnoli, S.; et al. Lipid rafts are primary mediators of amyloid oxidative attack on plasma membrane. J. Mol. Med. 2010, 88, 597–608. [Google Scholar] [CrossRef]
- Cecarini, V.; Cuccioloni, M.; Zheng, Y.; Bonfili, L.; Gong, C.; Angeletti, M.; Mena, P.; Del Rio, D.; Eleuteri, A.M. Flavan-3-ol Microbial Metabolites Modulate Proteolysis in Neuronal Cells Reducing Amyloid-beta (1–42) Levels. Mol. Nutr. Food Res. 2021, 65, e2100380. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Bonfili, L.; Zheng, Y.; Cecarini, V.; Cuccioloni, M.; Angeletti, M.; Dematteis, G.; Tapella, L.; Genazzani, A.A.; Lim, D.; et al. Immortalized Alzheimer’s Disease Astrocytes: Characterization of Their Proteolytic Systems. Mol. Neurobiol. 2023, 60, 2787–2800. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Turk, D.; Podobnik, M.; Popovic, T.; Katunuma, N.; Bode, W.; Huber, R.; Turk, V. Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: A basis for the design of specific epoxysuccinyl inhibitors. Biochemistry 1995, 34, 4791–4797. [Google Scholar] [CrossRef] [PubMed]
- Asaad, N.; Bethel, P.A.; Coulson, M.D.; Dawson, J.E.; Ford, S.J.; Gerhardt, S.; Grist, M.; Hamlin, G.A.; James, M.J.; Jones, E.V.; et al. Dipeptidyl nitrile inhibitors of Cathepsin L. Bioorg. Med. Chem. Lett. 2009, 19, 4280–4283. [Google Scholar] [CrossRef]
- Novinec, M.; Rebernik, M.; Lenarcic, B. An allosteric site enables fine-tuning of cathepsin K by diverse effectors. FEBS Lett. 2016, 590, 4507–4518. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Uddin, M.S.; Stachowiak, A.; Mamun, A.A.; Tzvetkov, N.T.; Takeda, S.; Atanasov, A.G.; Bergantin, L.B.; Abdel-Daim, M.M.; Stankiewicz, A.M. Autophagy and Alzheimer’s Disease: From Molecular Mechanisms to Therapeutic Implications. Front. Aging Neurosci. 2018, 10, 4. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.-Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.-X.; Zhang, D.-F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 2019, 16, 52–69. [Google Scholar] [CrossRef]
- Luo, F.; Sandhu, A.F.; Rungratanawanich, W.; Williams, G.E.; Akbar, M.; Zhou, S.; Song, B.-J.; Wang, X. Melatonin and Autophagy in Aging-Related Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 7174. [Google Scholar] [CrossRef]
- Drobny, A.; Prieto Huarcaya, S.; Dobert, J.; Kluge, A.; Bunk, J.; Schlothauer, T.; Zunke, F. The role of lysosomal cathepsins in neurodegeneration: Mechanistic insights, diagnostic potential and therapeutic approaches. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119243. [Google Scholar] [CrossRef]
- Ata, S.; Imran Din, M.; Rasool, A.; Qasim, I.; Ul Mohsin, I. Equilibrium, thermodynamics, and kinetic sorption studies for the removal of coomassie brilliant blue on wheat bran as a low-cost adsorbent. J. Anal. Methods Chem. 2012, 2012, 405980. [Google Scholar] [CrossRef]
- Song, J.; Xu, P.; Xiang, H.; Su, Z.; Storer, A.C.; Ni, F. The active-site residue Cys-29 is responsible for the neutral-pH inactivation and the refolding barrier of human cathepsin B. FEBS Lett. 2000, 475, 157–162. [Google Scholar] [CrossRef]
- Wu, Y.H.; Hsieh, H.L. Roles of Heme Oxygenase-1 in Neuroinflammation and Brain Disorders. Antioxidants 2022, 11, 923. [Google Scholar] [CrossRef] [PubMed]
- Spitz, D.R.; Sullivan, S.J.; Malcolm, R.R.; Roberts, R.J. Glutathione dependent metabolism and detoxification of 4-hydroxy-2-nonenal. Free Radic. Biol. Med. 1991, 11, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Anton, N.; Fernandez-Gines, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxidative Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef]
- Briggs, R.; Kennelly, S.P.; O’Neill, D. Drug treatments in Alzheimer’s disease. Clin. Med. 2016, 16, 247–253. [Google Scholar] [CrossRef]
- Kim, H.J.; Jung, S.W.; Kim, S.Y.; Cho, I.H.; Kim, H.C.; Rhim, H.; Kim, M.; Nah, S.Y. Panax ginseng as an adjuvant treatment for Alzheimer’s disease. J. Ginseng Res. 2018, 42, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.H.; Lee, S.T.; Chu, K.; Oh, M.J.; Park, H.J.; Shim, J.Y.; Kim, M. An open-label trial of Korean red ginseng as an adjuvant treatment for cognitive impairment in patients with Alzheimer’s disease. Eur. J. Neurol. 2008, 15, 865–868. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, H.; Zhou, X.; Yang, W.; Lin, Y. Pharmacological effects of natural medicine ginsenosides against Alzheimer’s disease. Front. Pharmacol. 2022, 13, 952332. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Li, P.; Wei, N.; Zhao, Z.; Liang, H.; Ji, X.; Chen, W.; Xue, M.; Wei, J. The Ambiguous Relationship of Oxidative Stress, Tau Hyperphosphorylation, and Autophagy Dysfunction in Alzheimer’s Disease. Oxidative Med. Cell. Longev. 2015, 2015, 352723. [Google Scholar] [CrossRef]
- Yun, H.R.; Jo, Y.H.; Kim, J.; Shin, Y.; Kim, S.S.; Choi, T.G. Roles of Autophagy in Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 3289. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, N.; Rocchi, A.; Zhang, W.; Vassar, R.; Zhou, Y.; He, C. Identification of natural products with neuronal and metabolic benefits through autophagy induction. Autophagy 2017, 13, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.M.; Nixon, R.A. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc. Natl. Acad. Sci. USA 1990, 87, 3861–3865. [Google Scholar] [CrossRef]
- Mueller-Steiner, S.; Zhou, Y.; Arai, H.; Roberson, E.D.; Sun, B.; Chen, J.; Wang, X.; Yu, G.; Esposito, L.; Mucke, L.; et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: Implications for Alzheimer’s disease. Neuron 2006, 51, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sun, B.; Zhou, Y.; Grubb, A.; Gan, L. Cathepsin B degrades amyloid-beta in mice expressing wild-type human amyloid precursor protein. J. Biol. Chem. 2012, 287, 39834–39841. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.M.; Felsenstein, K.M.; Brenneman, D.E. Cathepsins B and L differentially regulate amyloid precursor protein processing. J. Pharmacol. Exp. Ther. 2009, 328, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Hettiarachchi, N.; Dallas, M.; Al-Owais, M.; Griffiths, H.; Hooper, N.; Scragg, J.; Boyle, J.; Peers, C. Heme oxygenase-1 protects against Alzheimer’s amyloid-beta(1–42)-induced toxicity via carbon monoxide production. Cell Death Dis. 2014, 5, e1569. [Google Scholar] [CrossRef] [PubMed]
- Yachie, A. Heme Oxygenase-1 Deficiency and Oxidative Stress: A Review of 9 Independent Human Cases and Animal Models. Int. J. Mol. Sci. 2021, 22, 1514. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.K.; Chen, S.E.; Chang, L.C. The Role of HO-1 and Its Crosstalk with Oxidative Stress in Cancer Cell Survival. Cells 2021, 10, 2401. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Cecarini, V.; Cuccioloni, M.; Bonfili, L.; Gong, C.; Angeletti, M.; Eleuteri, A.M. Ginsenosides Rg1 and Rg2 Activate Autophagy and Attenuate Oxidative Stress in Neuroblastoma Cells Overexpressing Aβ(1-42). Antioxidants 2024, 13, 310. https://doi.org/10.3390/antiox13030310
Liu Z, Cecarini V, Cuccioloni M, Bonfili L, Gong C, Angeletti M, Eleuteri AM. Ginsenosides Rg1 and Rg2 Activate Autophagy and Attenuate Oxidative Stress in Neuroblastoma Cells Overexpressing Aβ(1-42). Antioxidants. 2024; 13(3):310. https://doi.org/10.3390/antiox13030310
Chicago/Turabian StyleLiu, Ziqi, Valentina Cecarini, Massimiliano Cuccioloni, Laura Bonfili, Chunmei Gong, Mauro Angeletti, and Anna Maria Eleuteri. 2024. "Ginsenosides Rg1 and Rg2 Activate Autophagy and Attenuate Oxidative Stress in Neuroblastoma Cells Overexpressing Aβ(1-42)" Antioxidants 13, no. 3: 310. https://doi.org/10.3390/antiox13030310




