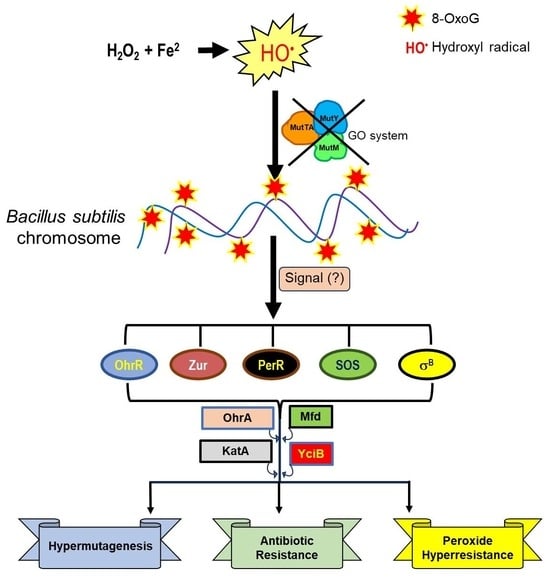
8-OxoG-Dependent Regulation of Global Protein Responses Leads to Mutagenesis and Stress Survival in Bacillus subtilis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Conditions, and Reagents
2.2. Cell Harvesting and Protein Extraction
2.3. Protein Identification and Quantitation with Mass Spectrometry Analysis LC-ESI-HDMSE
2.4. Treatment of Vegetative Cells with Oxidizing Agents
2.5. Determination of Mutation Frequencies
2.6. Bioinformatic Analysis
2.7. Statistical Analyses
3. Results
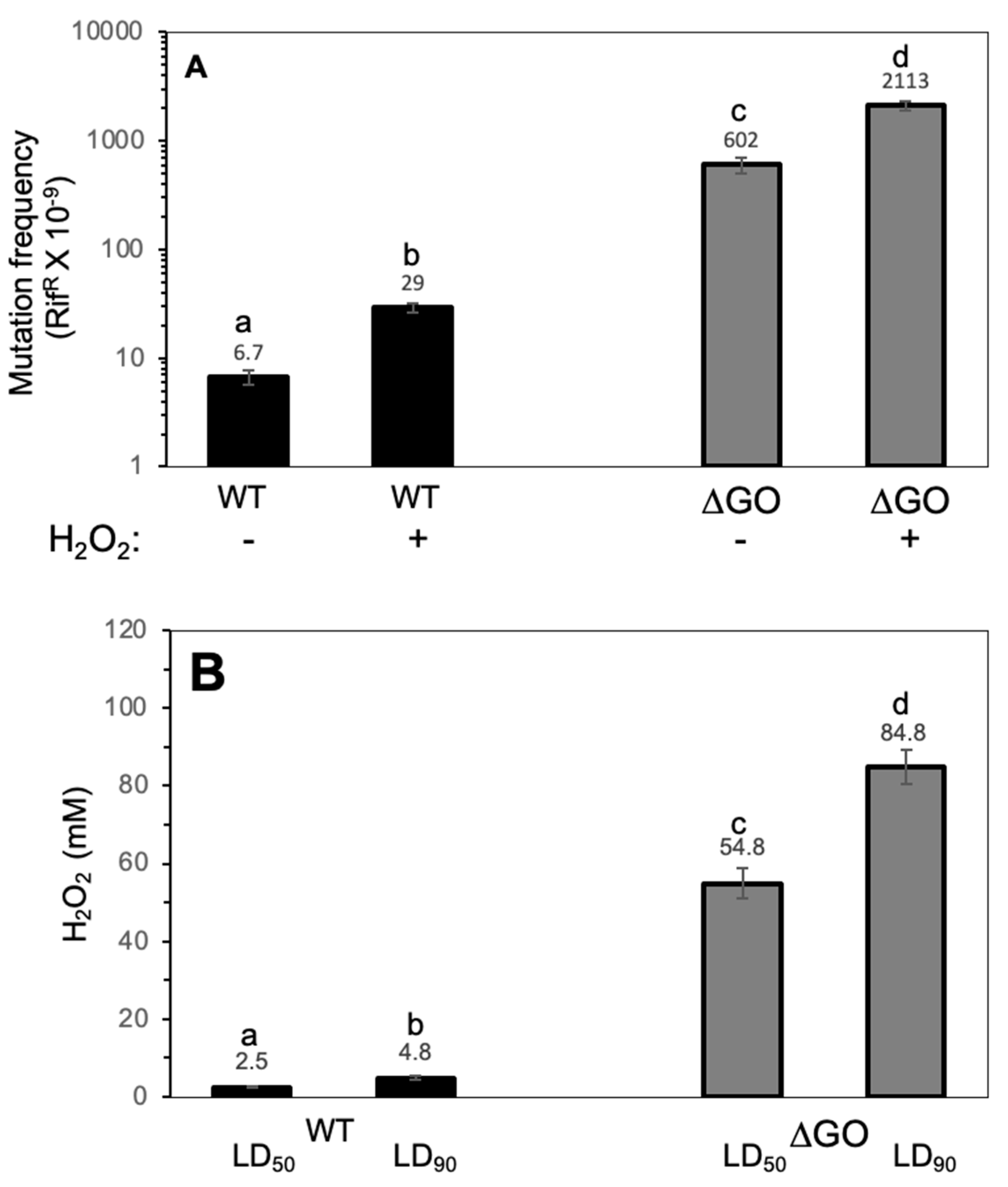
3.1. The Absence of the GO System Promotes Hypermutagenesis (HPM) and Hydrogen Peroxide Hyperresistance (HPHR) in Vegetative B. subtilis Cells
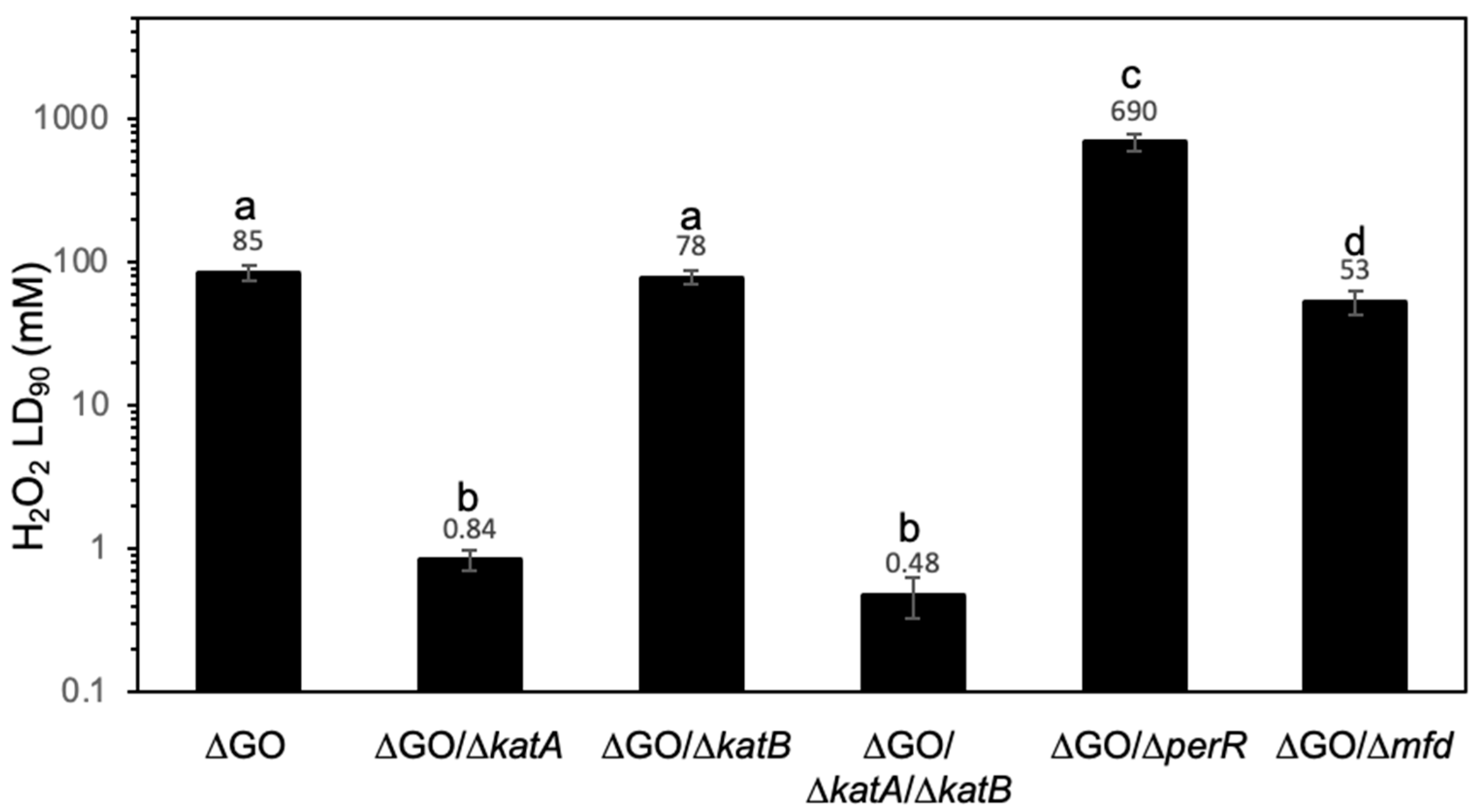
3.2. The HPHR of the ΔGO Strain Is katA Dependent, but perR Independent, and Influenced by Mfd
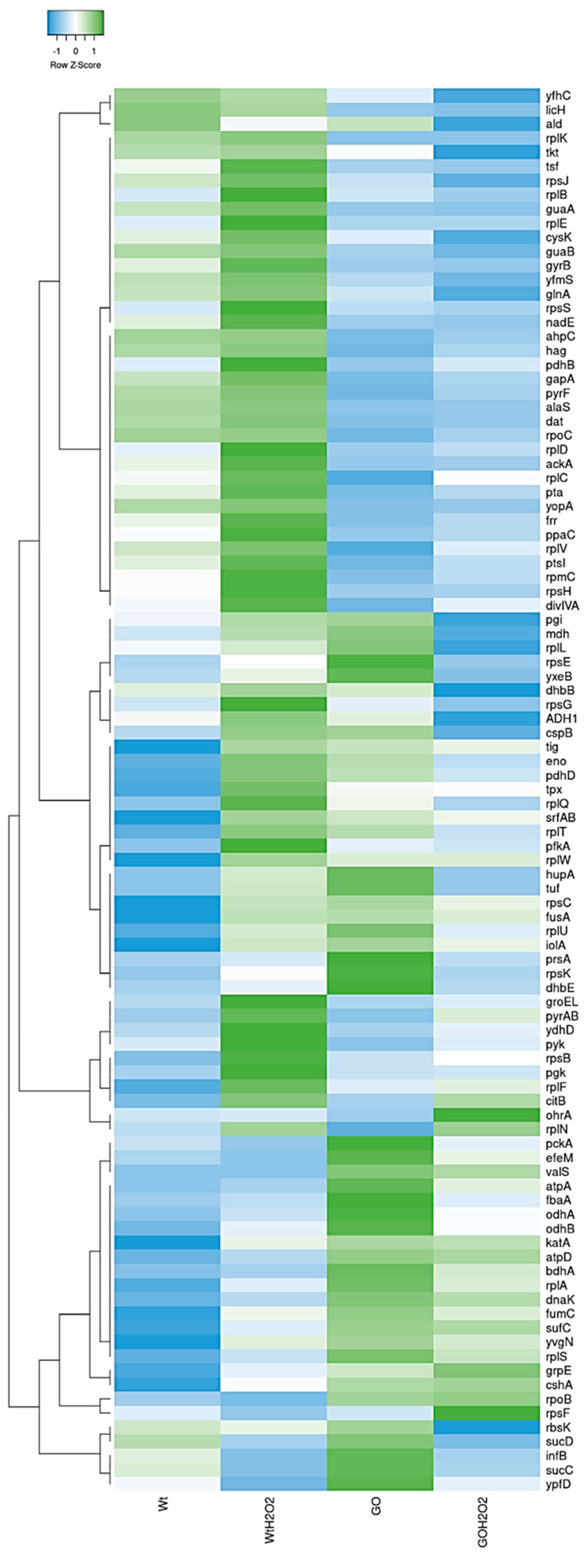
3.3. Global Protein Analysis Reveals Novel Factors Contributing to the HPM and HPHR of the Strain B. subtilis ∆GO
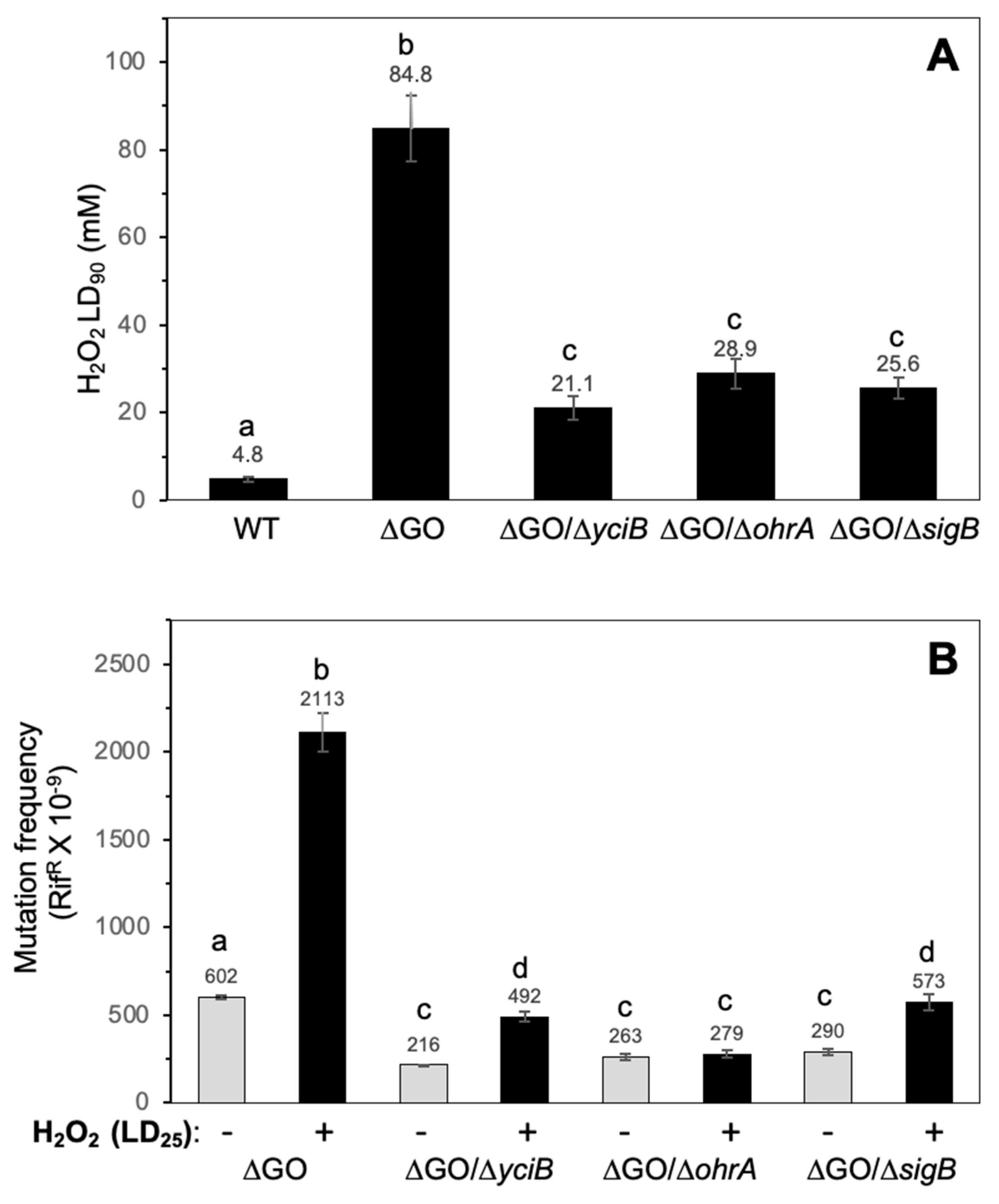
3.4. YciB, OhrA, and SigB Contribute to the HPM and HPHR of the Strain, B. subtilis ∆GO
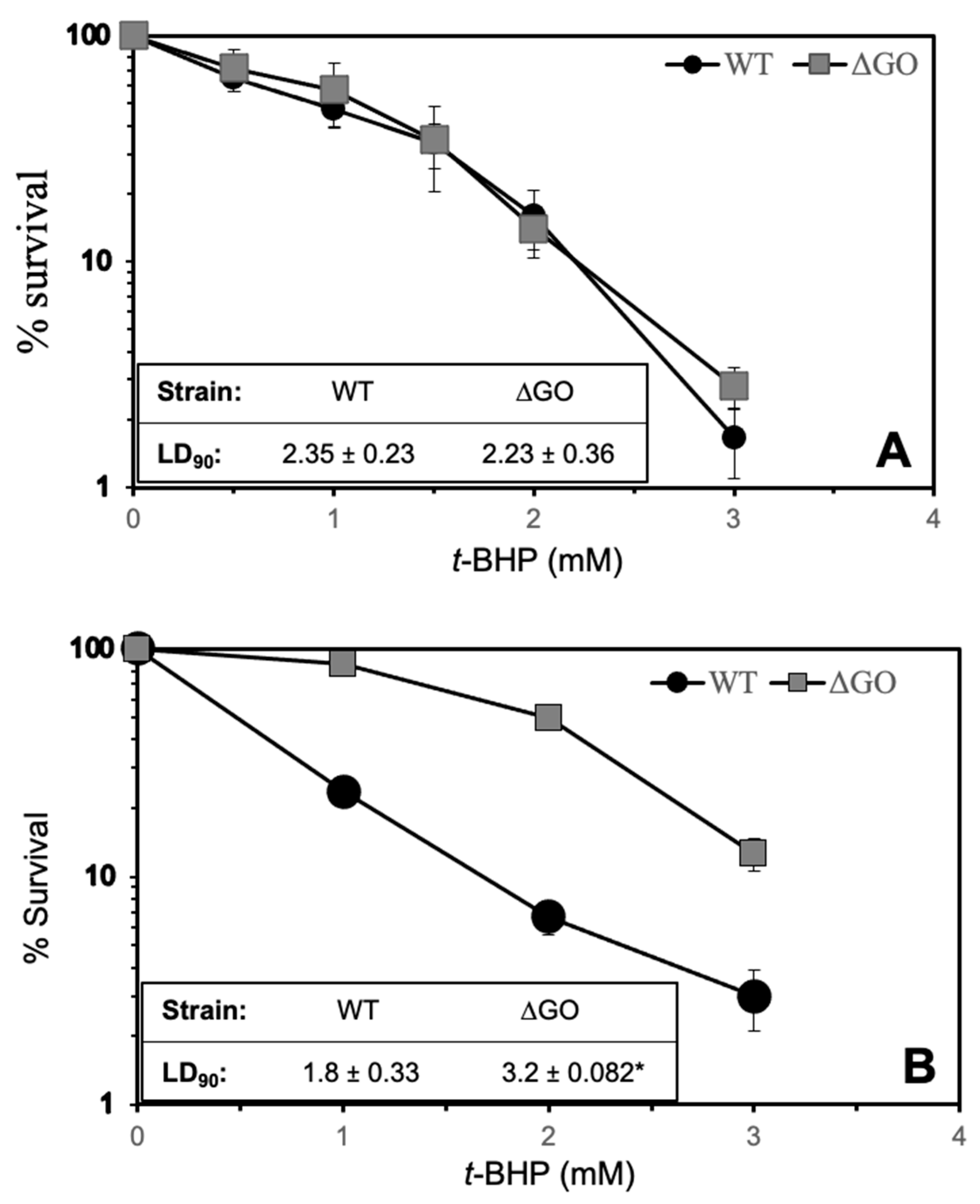
3.5. H2O2 Pretreatment Increases the Resistance of the GO-Deficient Strain to t-BHP
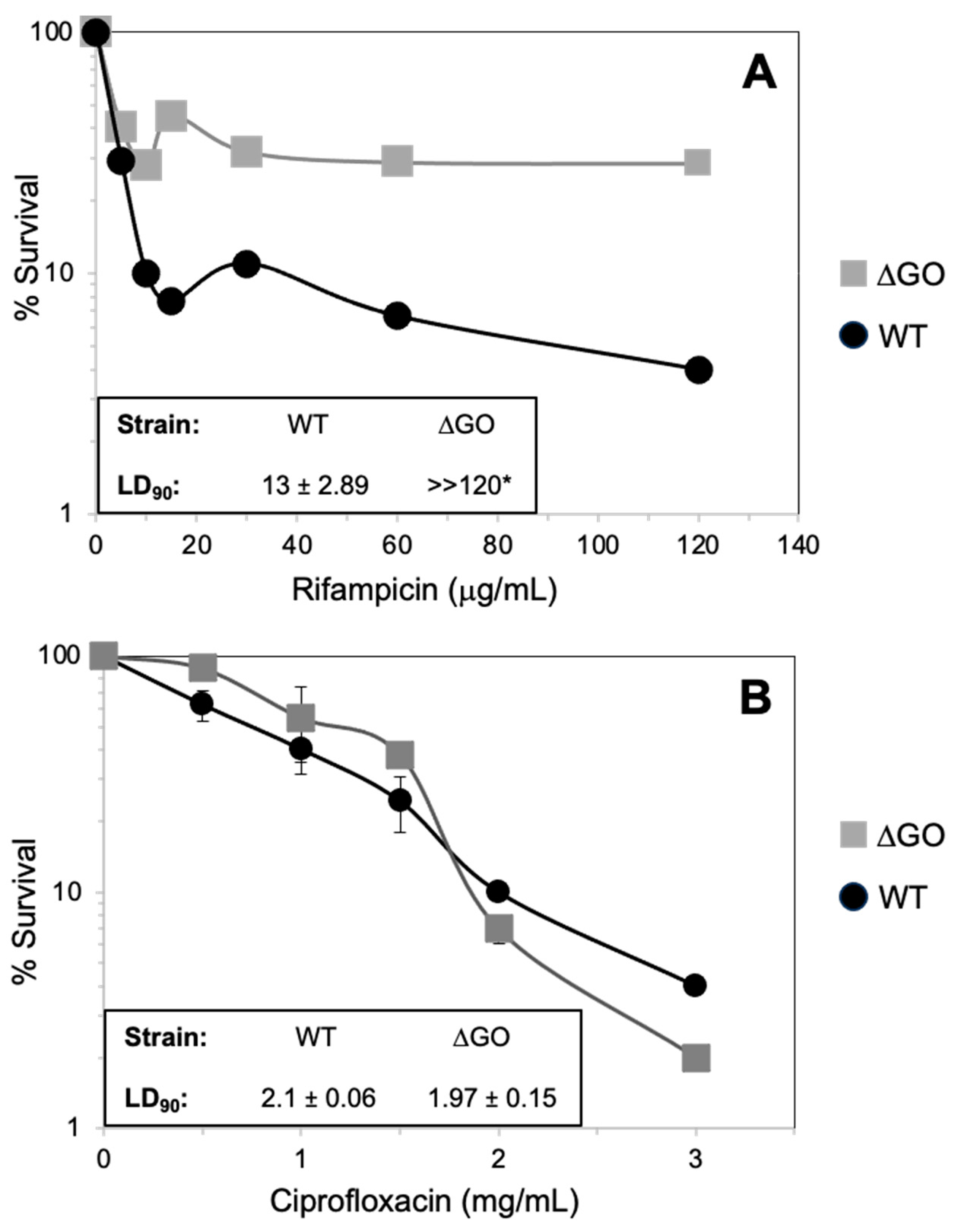
3.6. The Absence of a GO System Confers B. subtilis Resistance to Antibiotics That Target Transcription but Not DNA Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Imlay, J.A. Cellular defenses against superoxide and hydrogen peroxide. Ann. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Imlay, J.A. Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 2019, 21, 521–530. [Google Scholar] [CrossRef]
- Bonilla, C.Y. Generally Stressed Out Bacteria: Environmental Stress Response Mechanisms in Gram-Positive Bacteria. Integr. Comp. Biol. 2020, 60, 126–133. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Fasnacht, M.; Polacek, N. Oxidative stress in bacteria and the central dogma of molecular biology. Front. Mol. Biosci. 2021, 8, 671037. [Google Scholar] [CrossRef]
- Faulkner, M.J.; Helmann, J.D. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid. Redox. Signal 2011, 15, 175–189. [Google Scholar] [CrossRef]
- Henle, E.S.; Linn, S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 1997, 272, 19095–19098. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef]
- Keyer, K.; Imlay, J.A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 1996, 93, 13635–13640. [Google Scholar] [CrossRef]
- Henle, E.S.; Han, Z.; Tang, N.; Rai, P.; Luo, Y.; Linn, S. Sequence-specific DNA cleavage by Fe2+-mediated fenton reactions has possible biological implications. J. Biol. Chem. 1999, 274, 962–971. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Helmann, J.D.; Wu, M.F.W.; Gaballa, A.; Kobel, P.A.; Morshedi, M.M.; Fawcett, P.; Paddon, C. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 2003, 185, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Price, C.W. General Stress Response. In Bacillus Subtilis and Its Closest Relatives: From Genes to Cells; American Society for Microbiology Press: Washington, DC, USA, 2014; pp. 369–384. [Google Scholar] [CrossRef]
- Nannapaneni, P.; Hertwig, F.; Depke, M.; Hecker, M.; Mäder, U.; Völker, U.; Steil, L.; van Hijum, S.A.F.T. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology 2012, 158, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Pinochet-Barros, A.; Helmann, J.D. Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid. Redox Signal. 2018, 29, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Fuangthong, M.; Atichartpongkul, S.; Mongkolsuk, S.; Helmann, J.D. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 2001, 183, 4134–4141. [Google Scholar] [CrossRef] [PubMed]
- Leichert, L.I.O.; Scharf, C.; Hecker, M. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 2003, 185, 1967–1975. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Kino, K.; Hirao-Suzuki, M.; Morikawa, M.; Sakaga, A.; Miyazawa, H. Generation, repair and replication of guanine oxidation products. Genes Environ. 2017, 39, 21. [Google Scholar] [CrossRef]
- Saito, I.; Nakamura, T.; Nakatani, K.; Yoshioka, Y.; Yamaguchi, K.; Sugiyama, H. Mapping of the hot spots for DNA Damage by one-electron oxidation: Efficacy of GG doublets and GGG triplets as a trap in long-range hole migration. J. Am. Chem. Soc. 1998, 120, 12686–12687. [Google Scholar] [CrossRef]
- Wood, M.L.; Dizdaroglu, M.; Gajewski, E.; Essigmann, J.M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry 1990, 29, 7024–7032. [Google Scholar] [CrossRef]
- Shibutani, S.; Takeshita, M.; Grollman, A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991, 349, 431–434. [Google Scholar] [CrossRef]
- De Rosa, M.; Johnson, S.A.; Opresko, P.L. Roles for the 8-Oxoguanine DNA repair system in protecting telomeres from oxidative stress. Front. Cell Dev. Biol. 2021, 9, 758402. [Google Scholar] [CrossRef]
- Michaels, M.L.; Miller, J.H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 1992, 174, 6321–6325. [Google Scholar] [CrossRef] [PubMed]
- Michaels, M.L.; Cruz, C.; Grollman, A.P.; Miller, J.H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 1992, 89, 7022–7025. [Google Scholar] [CrossRef] [PubMed]
- Maki, H.; Sekiguchi, M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 1992, 355, 273–275. [Google Scholar] [CrossRef]
- Ramírez, M.I.; Castellanos-Juárez, F.X.; Yasbin, R.E.; Pedraza-Reyes, M. The ytkD(mutTA) gene of Bacillus subtilis encodes a functional antimutator 8-Oxo-(dGTP/GTP)ase and is under dual control of Sigma A and Sigma F RNA polymerases. J. Bacteriol. 2004, 186, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Juárez, F.X.; Álvarez-Álvarez, C.; Yasbin, R.E.; Setlow, B.; Setlow, P.; Pedraza-Reyes, M. YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress. J. Bacteriol. 2006, 188, 2285–2289. [Google Scholar] [CrossRef] [PubMed]
- Tajiri, T.; Maki, H.; Sekiguchi, M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 1995, 336, 257–267. [Google Scholar] [CrossRef]
- Gómez-Marroquín, M.; Vidales, L.E.; Debora, B.N.; Santos-Escobar, F.; Obregón-Herrera, A.; Robleto, E.A.; Pedraza-Reyes, M. Role of Bacillus subtilis DNA glycosylase MutM in counteracting oxidatively induced DNA damage and in stationary-phase-associated mutagenesis. J. Bacteriol. 2015, 197, 1963–1971. [Google Scholar] [CrossRef]
- Debora, B.N.; Vidales, L.E.; Ramírez, R.; Ramírez, M.; Robleto, E.A.; Yasbin, R.E.; Pedraza-Reyes, M. Mismatch repair modulation of MutY activity drives Bacillus subtilis stationary-phase mutagenesis. J. Bacteriol. 2011, 193, 236–245. [Google Scholar] [CrossRef]
- Vidales, L.E.; Cárdenas, L.C.; Robleto, E.; Yasbin, R.E.; Pedraza-Reyes, M. Defects in the error prevention oxidized guanine system potentiate stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 2009, 191, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Sánchez, H.C.; Villegas-Negrete, N.; Abundiz-Yañez, K.; Yasbin, R.E.; Robleto, E.A.; Pedraza-Reyes, M. Role of Mfd and GreA in Bacillus subtilis base excision repair-dependent stationary-phase mutagenesis. J. Bacteriol. 2020, 202, e00807-19. [Google Scholar] [CrossRef]
- Suárez, V.P.; Martínez, L.E.; Leyva-Sánchez, H.C.; Valenzuela-García, L.I.; Lara-Martínez, R.; Jiménez-García, L.F.; Ramírez-Ramírez, N.; Obregon-Herrera, A.; Cuéllar-Cruz, M.; Robleto, E.A.; et al. Transcriptional coupling and repair of 8-OxoG activate a RecA-dependent checkpoint that controls the onset of sporulation in Bacillus subtilis. Sci. Rep. 2021, 11, 2513. [Google Scholar] [CrossRef]
- Bagyan, I.; Noback, M.; Bron, S.; Paidhungat, M.; Setlow, P. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 1998, 212, 179–188. [Google Scholar] [CrossRef]
- Spizizen, J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 1958, 44, 1072–1078. [Google Scholar] [CrossRef]
- Neuhoff, V.; Arold, N.; Taube, D.; Ehrhardt, W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using coomassie brilliant blue G-250 and R-250. Electrophoresis 1988, 9, 255–262. [Google Scholar] [CrossRef]
- Zuccoli, G.S.; Martins-de-Souza, D.; Guest, P.C.; Rehen, S.K.; Nascimento, J.M. Combining patient-reprogrammed neural cells and proteomics as a model to study psychiatric disorders. Proteomic Methods Neuropsychiatr. Res. 2017, 974, 279–287. [Google Scholar] [CrossRef]
- Ramírez-Flores, C.J.; Cruz-Mirón, R.; Mondragón-Castelán, M.E.; González-Pozos, S.; Ríos-Castro, E.; Mondragón-Flores, R. Proteomic and structural characterization of self-assembled vesicles from excretion/secretion products of Toxoplasma gondii. J. Proteom. 2019, 208, 103490. [Google Scholar] [CrossRef]
- Geromanos, S.J.; Hughes, C.; Golick, D.; Ciavarini, S.; Gorenstein, M.V.; Richardson, K.; Hoyes, J.B.; Vissers, J.P.; Langridge, J.I. Simulating and validating proteomics data and search results. Proteomics 2011, 11, 1189–1211. [Google Scholar] [CrossRef]
- Valentine, S.J.; Ewing, M.A.; Dilger, J.M.; Glover, M.S.; Geromanos, S.; Hughes, C.; Clemmer, D.E. Using ion mobility data to improve peptide identification: Intrinsic Amino acid size parameters. J. Proteome Res. 2011, 10, 2318–2329. [Google Scholar] [CrossRef]
- Skipp, P.J.S.; Hughes, C.; McKenna, T.; Edwards, R.; Langridge, J.; Thomson, N.R.; Clarke, I.N. Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PLoS ONE 2016, 11, e0149011. [Google Scholar] [CrossRef]
- Engelmann, S.; Hecker, M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 1996, 145, 63–69. [Google Scholar] [CrossRef]
- Pybus, C.; Pedraza-Reyes, M.; Ross, C.A.; Martin, H.; Ona, K.; Yasbin, R.E.; Robleto, E. Transcription-associated mutation in Bacillus subtilis cells under stress. J. Bacteriol. 2010, 192, 3321–3328. [Google Scholar] [CrossRef]
- Ross, C.; Pybus, C.; Pedraza-Reyes, M.; Sung, H.-M.; Yasbin, R.E.; Robleto, E. Novel role of mfd: Effects on stationary-phase mutagenesis in Bacillus subtilis. J. Bacteriol. 2006, 188, 7512–7520. [Google Scholar] [CrossRef]
- Martin, H.A.; Porter, K.E.; Vallin, C.; Ermi, T.; Contreras, N.; Pedraza-Reyes, M.; Robleto, E.A. Mfd protects against oxidative stress in Bacillus subtilis independently of its canonical function in DNA repair. BMC Microbiol. 2019, 19, 26. [Google Scholar] [CrossRef]
- Moszer, I.; Jones, L.M.; Moreira, S.; Fabry, C.; Danchin, A. SubtiList: The reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002, 30, 62–65. Available online: http://genolist.pasteur.fr/ (accessed on 1 May 2022). [CrossRef] [PubMed]
- Pedreira, T.; Elfmann, C.; Stülke, J. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res. 2022, 50, 875–882. [Google Scholar] [CrossRef]
- Fuangthong, M.; Herbig, A.F.; Bsat, N.; Helmann, J.D. Regulation of the Bacillus subtilis fur and perR genes by PerR: Not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 2002, 184, 3276–3286. [Google Scholar] [CrossRef]
- Cheo, D.L.; Bayles, K.W.; E Yasbin, R. Elucidation of regulatory elements that control damage induction and competence induction of the Bacillus subtilis SOS system. J. Bacteriol. 1993, 175, 5907–5915. [Google Scholar] [CrossRef] [PubMed]
- Au, N.; Kuester-Schoeck, E.; Mandava, V.; Bothwell, L.E.; Canny, S.P.; Chachu, K.; Colavito, S.A.; Fuller, S.N.; Groban, E.S.; Hensley, L.A.; et al. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 2005, 187, 7655–7666. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.T.; Helmann, J.D. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 2007, 153, 4123–4128. [Google Scholar] [CrossRef]
- Antelmann, H.; Engelmann, S.; Schmid, R.; Hecker, M. General and oxidative stress responses in Bacillus subtilis: Cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 1996, 178, 6571–6578. [Google Scholar] [CrossRef]
- Bsat, N.; Chen, L.; Helmann, J.D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 1996, 178, 6579–6586. [Google Scholar] [CrossRef]
- Even, S.; Burguière, P.; Auger, S.; Soutourina, O.; Danchin, A.; Martin-Verstraete, I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 2006, 188, 2184–2197. [Google Scholar] [CrossRef]
- Esquilin-Lebron, K.; Dubrac, S.; Barras, F.; Boyd, J.M. Bacterial approaches for assembling iron-sulfur proteins. mBio 2021, 12, e0242521. [Google Scholar] [CrossRef]
- Gaballa, A.; Wang, T.; Ye, R.W.; Helmann, J.D. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 2002, 184, 6508–6514. [Google Scholar] [CrossRef]
- Bramkamp, M. The putative Bacillus subtilis L,D-transpeptidase YciB is a lipoprotein that localizes to the cell poles in a divisome-dependent manner. Arch. Microbiol. 2010, 192, 57–68. [Google Scholar] [CrossRef]
- Micka, B.; Groch, N.; Heinemann, U.; A Marahiel, M. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 1991, 173, 3191–3198. [Google Scholar] [CrossRef]
- Karaboja, X.; Wang, X. HBsu Is Required for the Initiation of DNA Replication in Bacillus subtilis. J. Bacteriol. 2022, 204, e00119-22. [Google Scholar] [CrossRef]
- Price, C.W.; Fawcett, P.; Cérémonie, H.; Su, N.; Murphy, C.K.; Youngman, P. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 2001, 41, 757–774. [Google Scholar] [CrossRef]
- Baichoo, N.; Wang, T.; Ye, R.; Helmann, J.D. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 2002, 45, 1613–1629. [Google Scholar] [CrossRef]
- Santos-Escobar, F.; Gutiérrez-Corona, J.F.; Pedraza-Reyes, M. Role of Bacillus subtilis error prevention oxidized guanine system in counteracting hexavalent chromium-promoted oxidative DNA damage. Appl. Environ. Microbiol. 2014, 80, 5493–5502. [Google Scholar] [CrossRef]
- Faulkner, M.J.; Ma, Z.; Fuangthong, M.; Helmann, J.D. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J. Bacteriol. 2012, 194, 1226–1235. [Google Scholar] [CrossRef]
- Aliashkevich, A.; Cava, F. LD-transpeptidases: The great unknown among the peptidoglycan cross-linkers. FEBS J. 2022, 289, 4718–4730. [Google Scholar] [CrossRef]
- Bernal-Cabas, M.; Ayala, J.A.; Raivio, T.L. The Cpx envelope stress response modifies peptidoglycan cross-linking via the L,D-transpeptidase LdtD and the novel protein YgaU. J. Bacteriol. 2015, 197, 603–614. [Google Scholar] [CrossRef]
- Delhaye, A.; Collet, J.-F.; Laloux, G. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 2016, 7, e00047-16. [Google Scholar] [CrossRef]
- Tanous, C.; Soutourina, O.; Raynal, B.; Hullo, M.-F.; Mervelet, P.; Gilles, A.-M.; Noirot, P.; Danchin, A.; England, P.; Martin-Verstraete, I. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J. Biol. Chem. 2008, 283, 35551–35560. [Google Scholar] [CrossRef] [PubMed]
- Hullo, M.-F.; Martin-Verstraete, I.; Soutourina, O. Complex phenotypes of a mutant inactivated for CymR, the global regulator of cysteine metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 2010, 309, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Cha, M.-K.; Bae, Y.-J.; Kim, K.-J.; Park, B.-J.; Kim, I.-H. Characterization of two alkyl hydroperoxide reductase C homologs alkyl hydroperoxide reductase C_H1 and alkyl hydroperoxide reductase C_H2 in Bacillus subtilis. World J. Biol. Chem. 2015, 6, 249–264. [Google Scholar] [CrossRef]
- Gout, I. Coenzyme A: A protective thiol in bacterial antioxidant defence. Biochem. Soc. Trans. 2019, 47, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; Soonsanga, S.; Helmann, J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA 2007, 104, 8743–8748. [Google Scholar] [CrossRef]
- Höper, D.; Völker, U.; Hecker, M. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 2005, 187, 2810–2826. [Google Scholar] [CrossRef]
- Yeak, K.Y.C.; Boekhorst, J.; Wels, M.; Abee, T.; Wells-Bennik, M.H.J. Prediction and validation of novel SigB regulon members in Bacillus subtilis and regulon structure comparison to Bacillales members. BMC Microbiol. 2023, 23, 17. [Google Scholar] [CrossRef]
- Smits, W.K.; Dubois, J.-Y.F.; Bron, S.; van Dijl, J.M.; Kuipers, O.P. Tricksy business: Transcriptome analysis reveals the involvement of thioredoxin A in redox homeostasis, oxidative stress, sulfur metabolism, and cellular differentiation in Bacillus subtilis. J. Bacteriol. 2005, 187, 3921–3930. [Google Scholar] [CrossRef] [PubMed]
- Garibyan, L.; Huang, T.; Kim, M.; Wolff, E.; Nguyen, A.; Nguyen, T.; Diep, A.; Hu, K.; Iverson, A.; Yang, H.; et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2003, 2, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Price, L.B.; Vogler, A.; Pearson, T.; Busch, J.D.; Schupp, J.M.; Keim, P. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob. Agents Chemother. 2003, 47, 2362–2365. [Google Scholar] [CrossRef] [PubMed]
- Leehan, J.D.; Nicholson, W.L. The spectrum of spontaneous rifampin resistance mutations in the Bacillus subtilis rpoB gene depends on the growth environment. Appl. Environ. Microbiol. 2021, 87, e01237-21. [Google Scholar] [CrossRef] [PubMed]
- Leehan, J.D.; Nicholson, W.L. Environmental dependence of competitive fitness in rifampin-resistant rpoB mutants of Bacillus subtilis. Appl. Environ. Microbiol. 2022, 88, e0242221. [Google Scholar] [CrossRef]
- Patel, Y.; Soni, V.; Rhee, K.Y.; Helmann, J.D. Mutations in rpoB that confer rifampicin resistance can alter levels of peptidoglycan precursors and affect β-lactam susceptibility. mBio 2023, 14, e0316822. [Google Scholar] [CrossRef] [PubMed]
- Selby, C.P.; Lindsey-Boltz, L.A.; Li, W.; Sancar, A. Molecular mechanisms of transcription-coupled repair. Annu. Rev. Biochem. 2023, 92, 115–144. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.A.; Sundararajan, A.; Ermi, T.S.; Heron, R.; Gonzales, J.; Lee, K.; Anguiano-Mendez, D.; Schilkey, F.; Pedraza-Reyes, M.; Robleto, E.A. Mfd affects global transcription and the physiology of stressed Bacillus subtilis cells. Front. Appl. Environ. Microbiol. 2021, 12, 625705. [Google Scholar] [CrossRef] [PubMed]
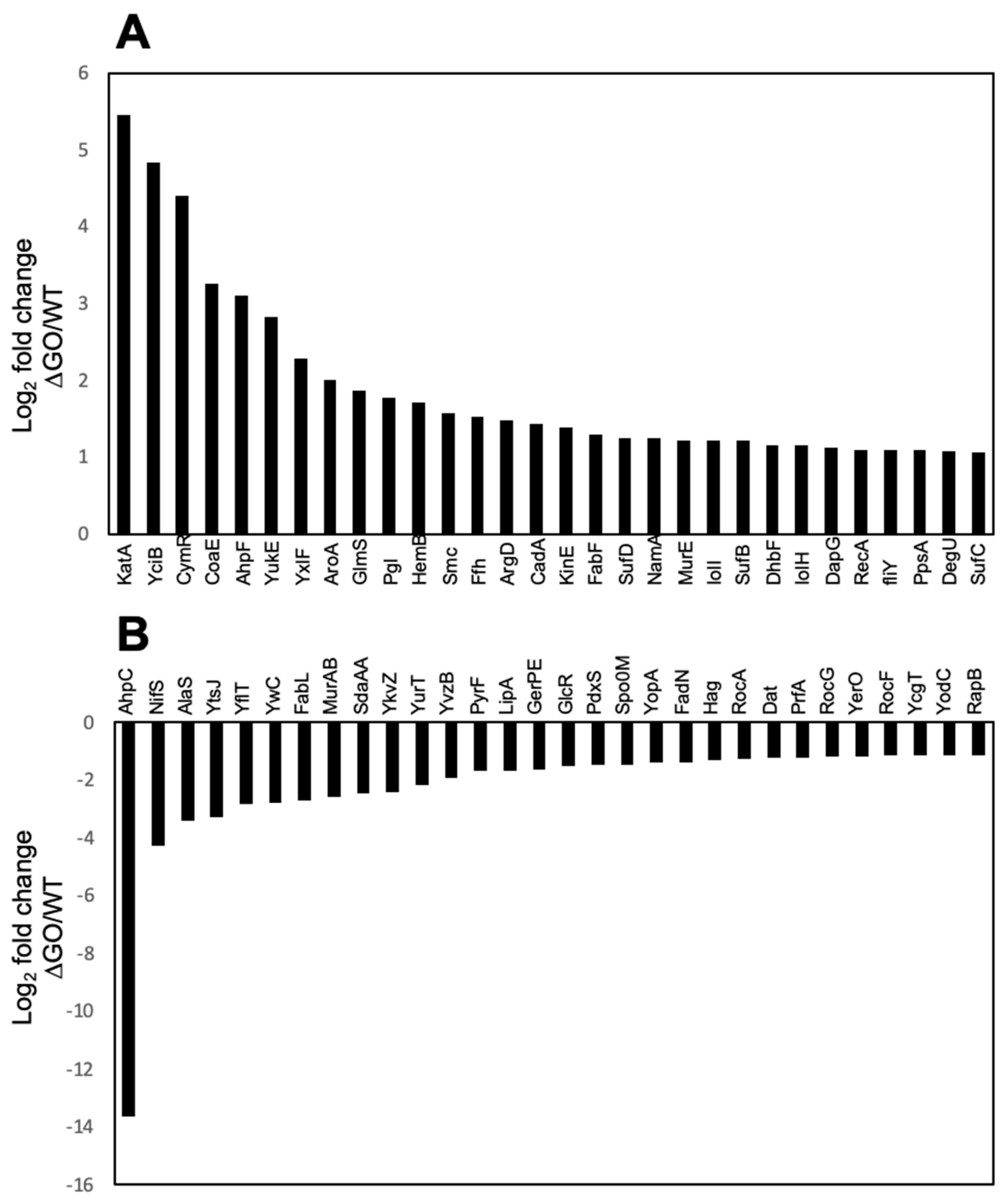
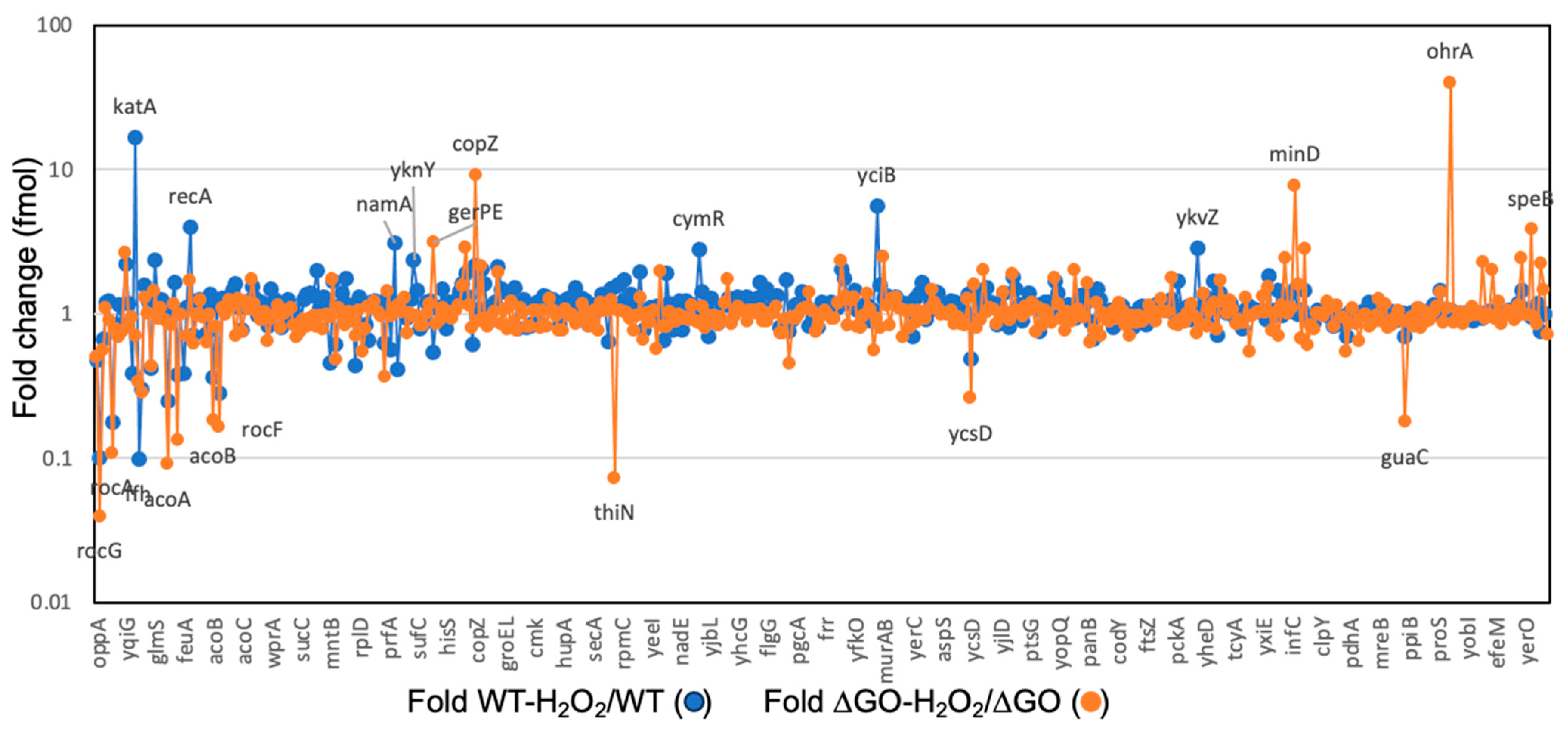
| Protein | Cell Function | Essential | Protein Abundance (fmol) | ∆GO/WT | ∆GO H2O2/GO | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | ∆GO | WT H2O2 | ∆GO H2O2 | Max Fold | Log2 Max Fold | Max Fold | Log2 Max Fold | |||
| KatA | Oxidative stress | No | 5.21 | 227.71 | 87.7 | 161 | 43.5 | 5.45 | 0.7 | −0.51 |
| YciB | Zur regulon | No | 0.54 | 15.6 | 3.02 | 14.37 | 28.9 | 4.84 | 0.92 | −0.12 |
| CoaE | Peroxide stress | Yes | 0.67 | 6.39 | 0.26 | 6.02 | 9.5 | 3.25 | 0.94 | −0.09 |
| AhpF | Oxidative stress | No | 19.55 | 169 | 39.2 | 137.36 | 8.64 | 3.1 | 0.81 | −0.3 |
| AhpC | Oxidative stress | No | 250.3 | 0.02 | 478.3 | 0.06 | 7.60 × 10−5 | −13.7 | 3 | 1.58 |
| OhrA | Oxidative stress | No | 0.42 | 0.23 | 0.45 | 9.3 | 0.53 | −0.76 | 40.43 | 5.3 |
| CymR | Redox stress and stringent response | No | 1.32 | 27 | 3.7 | 24 | 20.4 | 3.2 | 0.88 | −0.02 |
| CysS | Redox stress and stringent response | Yes | 7.32 | 8.25 | 8.42 | 7.87 | 1.12 | 0.16 | 0.95 | −0.07 |
| CysK | Redox stress and stringent response | No | 94 | 91 | 97.8 | 89.5 | 0.96 | −0.06 | 0.98 | −0.03 |
| SufB | Redox stress | Yes | 26.7 | 62.1 | 31.4 | 56.1 | 2.32 | 1.21 | 0.9 | −0.12 |
| SufD | Redox stress | Yes | 25.5 | 61.2 | 30.9 | 58.86 | 2.4 | 1.26 | 0.96 | −0.07 |
| SufC | Redox stress | Yes | 34.6 | 77.1 | 53.1 | 73.8 | 2.2 | 1.13 | 0.95 | −0.07 |
| SufS | Redox stress | Yes | 10.8 | 16 | 17.6 | 17.2 | 1.5 | 0.58 | 1.07 | 0.1 |
| YutI(Nfu) | Redox stress | No | 15.8 | 14.3 | 16.6 | 13.9 | 0.9 | −0.15 | 0.97 | −0.08 |
| RecA | SOS response | No | 2.7 | 5.8 | 10.9 | 10 | 2.1 | 1.07 | 1.7 | 0.76 |
| Hupa | SOS response | 303.4 | 401.5 | 359.1 | 306.4 | 1.32 | 0.4 | 0.76 | −0.39 | |
| YceD | General stress | No | 21.12 | 34.9 | 18.9 | 32.1 | 1.65 | 0.72 | 0.91 | −13 |
| TrxA | General stress | Yes | 1.1 | 2.34 | 1.9 | 1.9 | 1.99 | 0.99 | 0.82 | −0.28 |
| YceE | General stress | No | 19.2 | 28.22 | 25.3 | 24.9 | 1.46 | 0.55 | 0.88 | −0.02 |
| YflT | General stress | No | 13.3 | 1.9 | 12.4 | 3.82 | 0.14 | −1.32 | 2 | 1 |
| SmC | Stringent response | 3 | 9.3 | 4.9 | 8.76 | 3.07 | 1.63 | 0.94 | −0.09 | |
| RpoB | Stringent response | 59.54 | 68.71 | 58.57 | 69.2 | 1.15 | 0.2 | 1 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, L.E.; Gómez, G.; Ramírez, N.; Franco, B.; Robleto, E.A.; Pedraza-Reyes, M. 8-OxoG-Dependent Regulation of Global Protein Responses Leads to Mutagenesis and Stress Survival in Bacillus subtilis. Antioxidants 2024, 13, 332. https://doi.org/10.3390/antiox13030332
Martínez LE, Gómez G, Ramírez N, Franco B, Robleto EA, Pedraza-Reyes M. 8-OxoG-Dependent Regulation of Global Protein Responses Leads to Mutagenesis and Stress Survival in Bacillus subtilis. Antioxidants. 2024; 13(3):332. https://doi.org/10.3390/antiox13030332
Chicago/Turabian StyleMartínez, Lissett E., Gerardo Gómez, Norma Ramírez, Bernardo Franco, Eduardo A. Robleto, and Mario Pedraza-Reyes. 2024. "8-OxoG-Dependent Regulation of Global Protein Responses Leads to Mutagenesis and Stress Survival in Bacillus subtilis" Antioxidants 13, no. 3: 332. https://doi.org/10.3390/antiox13030332
APA StyleMartínez, L. E., Gómez, G., Ramírez, N., Franco, B., Robleto, E. A., & Pedraza-Reyes, M. (2024). 8-OxoG-Dependent Regulation of Global Protein Responses Leads to Mutagenesis and Stress Survival in Bacillus subtilis. Antioxidants, 13(3), 332. https://doi.org/10.3390/antiox13030332