Localization and Aggregation of Honokiol in the Lipid Membrane
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics Simulation
2.2. Molecular Dynamics Technical Specifications
2.3. Molecular Dynamics Analysis
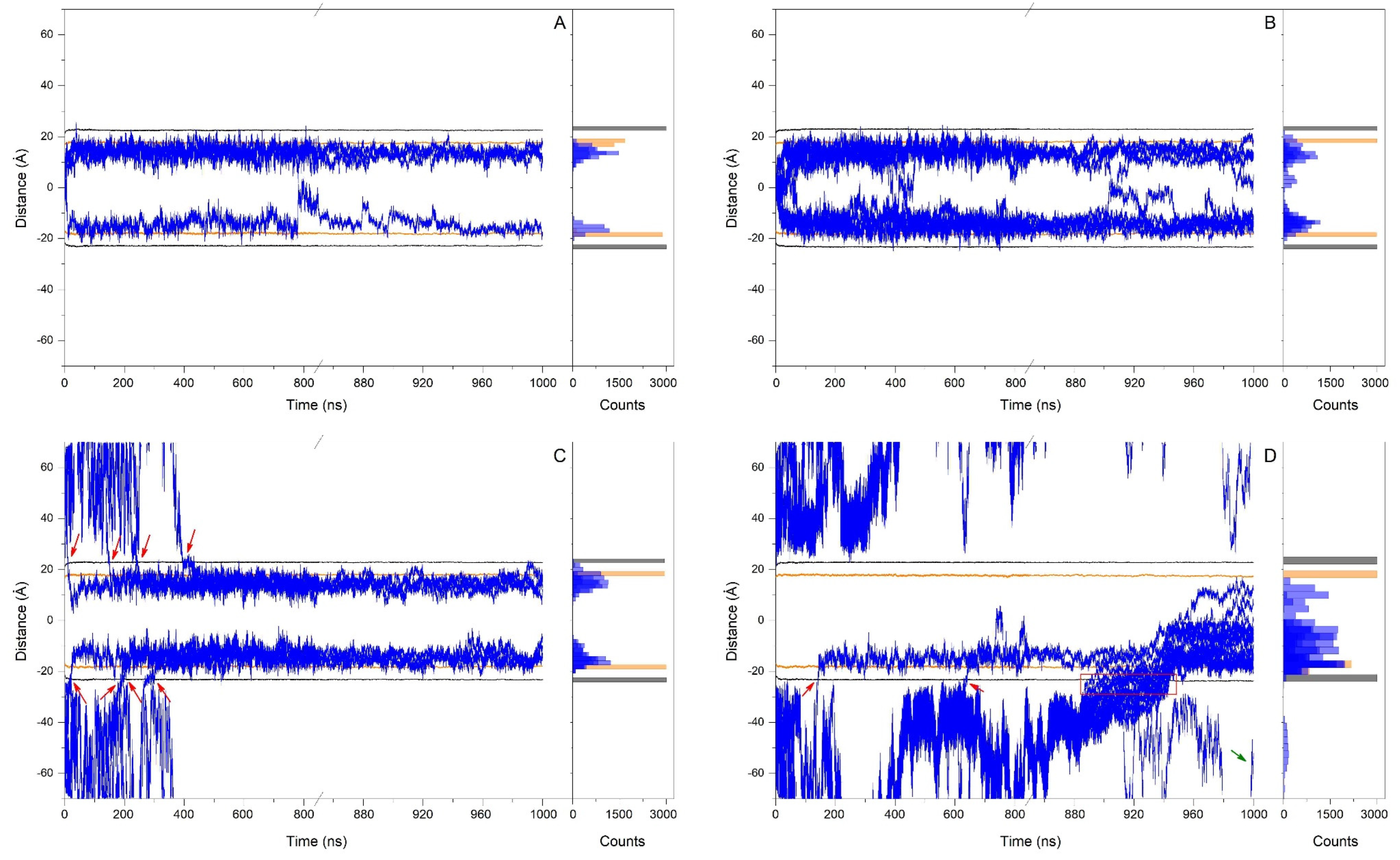
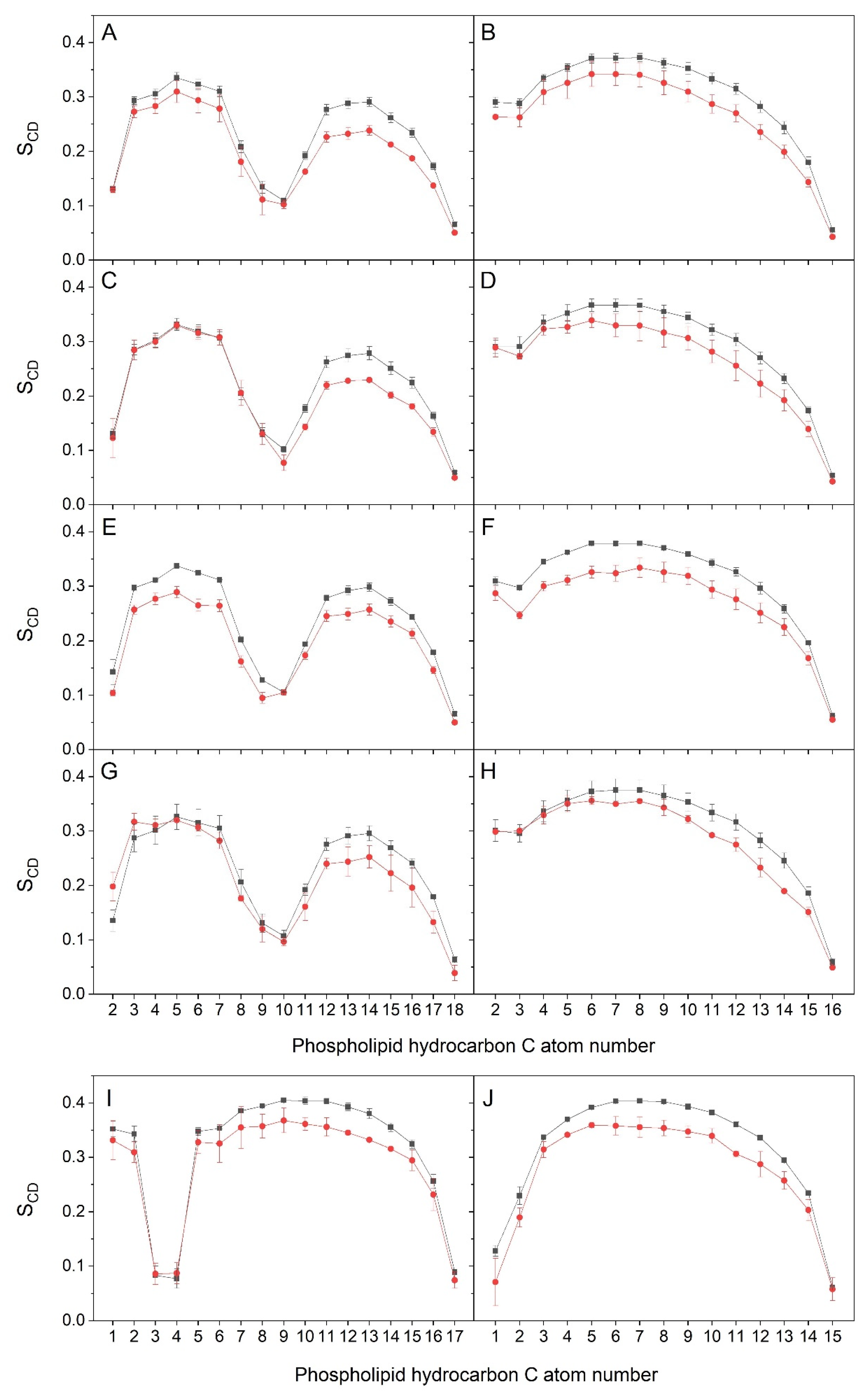
3. Results and Discussion
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CHOL | Cholesterol |
| HNK | Honokiol (2-(4-hydroxy-3-prop-2-enylphenyl)-4-prop-2-enylphenol) |
| MD | Molecular Dynamics |
| NAMD | Nanoscale Molecular Dynamics |
| PI-3P | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol-3-phosphorous |
| POPC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| POPE | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine |
| POPS | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine |
| PSM | N-Palmitoyl-D-erythro-sphingosylphosphorylcholine |
| SCD | Deuterium order parameter |
| VDW | Van der Waals |
| VMD | Visual Molecular Dynamics |
| z-COM | z-axis Centre-of-Mass (z direction normal to the bilayer plane) |
References
- Dzubak, P.; Hajduch, M.; Vydra, D.; Hustova, A.; Kvasnica, M.; Biedermann, D.; Markova, L.; Urban, M.; Sarek, J. Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep. 2006, 23, 394–411. [Google Scholar] [CrossRef] [PubMed]
- Itokawa, H.; Morris-Natschke, S.L.; Akiyama, T.; Lee, K.-H. Plant-derived natural product research aimed at new drug discovery. J. Nat. Med. 2008, 62, 263–280. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Rauf, A.; Olatunde, A.; Imran, M.; Alhumaydhi, F.A.; Aljohani, A.S.; Khan, S.A.; Uddin, S.; Mitra, S.; Bin Emran, T.; Khayrullin, M.; et al. Honokiol: A review of its pharmacological potential and therapeutic insights. Phytomedicine 2021, 90, 153647. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Wu, H.; Yu, X.; Zhang, X.; Lu, Y.; Fan, J.; Tang, L.; Wang, Z. A review of the phytochemistry and pharmacological activities of Magnoliae officinalis cortex. J. Ethnopharmacol. 2019, 236, 412–442. [Google Scholar] [CrossRef] [PubMed]
- Teponno, R.B.; Kusari, S.; Spiteller, M. Recent advances in research on lignans and neolignans. Nat. Prod. Rep. 2016, 33, 1044–1092. [Google Scholar] [CrossRef]
- Khatoon, F.; Ali, S.; Kumar, V.; Elasbali, A.M.; Alhassan, H.H.; Alharethi, S.H.; Islam, A.; Hassan, I. Pharmacological features, health benefits and clinical implications of honokiol. J. Biomol. Struct. Dyn. 2023, 41, 7511–7533. [Google Scholar] [CrossRef]
- Mottaghi, S.; Abbaszadeh, H. Natural Lignans Honokiol and Magnolol as Potential Anticarcinogenic and Anticancer Agents. A Comprehensive Mechanistic Review. Nutr. Cancer 2022, 74, 761–778. [Google Scholar] [CrossRef]
- Pan, C.; Li, Q.; Xiong, S.; Yang, Y.; Yang, Y.; Huang, C.; Wang, Z. Delivery Strategies, Structural Modification, and Pharmacological Mechanisms of Honokiol: A Comprehensive Review. Chem. Biodivers. 2024, 21, e202302032. [Google Scholar] [CrossRef]
- Prasher, P.; Fatima, R.; Sharma, M.; Tynybekov, B.; Alshahrani, A.M.; Ateşşahin, D.A.; Sharifi-Rad, J.; Calina, D. Honokiol and its analogues as anticancer compounds: Current mechanistic insights and structure-activity relationship. Chem. Interact. 2023, 386, 110747. [Google Scholar] [CrossRef]
- Guo, M.M.M.; Wu, X.; Feng, Y.; Hu, Z. Research Progress on the Structural Modification of Magnolol and Honokiol and the Biological Activities of Their Derivatives. Chem. Biodivers. 2023, 20, e202300754. [Google Scholar] [CrossRef]
- Luo, L.; Wu, T.; Ji, M.; Xiang, J.; Zou, Y.; Liao, Y. Honokiol suppress the PD-L1 expression to improve anti-tumor immunity in lung cancer. Int. Immunopharmacol. 2024, 133, 112098. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, Y.; Xia, X.; Han, J.; Cao, L. Honokiol induces ferroptosis in ovarian cancer cells through the regulation of YAP by OTUB2. J. Obstet. Gynaecol. Res. 2024, 50, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Gui, S.; Zheng, Y.; Zhao, J.; Zhao, Y.; Li, Y.; Chen, X. The natural compounds, Magnolol or Honokiol, promote adipose tissue browning and resist obesity through modulating PPARalpha/gamma activity. Eur. J. Pharmacol. 2024, 969, 176438. [Google Scholar] [CrossRef]
- Szałabska-Rąpała, K.; Zych, M.; Borymska, W.; Londzin, P.; Dudek, S.; Kaczmarczyk-Żebrowska, I. Beneficial effect of honokiol and magnolol on polyol pathway and oxidative stress parameters in the testes of diabetic rats. Biomed. Pharmacother. 2024, 172, 116265. [Google Scholar] [CrossRef]
- Yang, F.; Yang, B.; Song, K.; Jin, Y.; Wang, G.; Li, P.; Yu, Q.; Ling, F. Natural product honokiol exhibits antiviral effects against Micropterus salmoides rhabdovirus (MSRV) both in vitro and in vivo. J. Fish Dis. 2024, 47, e13915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, T.; Yin, X.; Wang, W.; Li, W.; Chen, X.; Ma, J.; Long, Y. Honokiol inhibits Botryosphaeria dothidea, the causal pathogen of kiwifruit soft rot, by targeting membrane lipid biosynthesis. Pest Manag. Sci. 2024, 80, 1779–1794. [Google Scholar] [CrossRef]
- Pan, N.; Shi, J.; Du, S.; Qiu, Z.; Ran, Q.; Guo, Y.; Ma, A.; Zhang, Q.; Sang, A.; Yang, X. Honokiol Attenuates Choroidal Neovascularization by Inhibiting the Hypoxia-Inducible Factor-alpha/Vascular Endothelial Growth Factor Axis via Nuclear Transcription Factor-Kappa B Activation. Curr. Eye Res. 2024, 49, 88–96. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.-F.; Gao, L.; Li, Y.-J.; Feng, D.-X. Honokiol Prevents Intestinal Barrier Dysfunction in Mice with Severe Acute Pancreatitis and Inhibits JAK/STAT1 Pathway and Acetylation of HMGB1. Chin. J. Integr. Med. 2023, 30, 534–542. [Google Scholar] [CrossRef]
- Mei, M.; Tang, L.; Zhou, H.; Xue, N.; Li, M. Honokiol prevents lung metastasis of triple-negative breast cancer by regulating polarization and recruitment of macrophages. Eur. J. Pharmacol. 2023, 959, 176076. [Google Scholar] [CrossRef]
- Borgonetti, V.; Galeotti, N. Honokiol-Rich Magnolia officinalis Bark Extract Attenuates Trauma-Induced Neuropathic Pain. Antioxidants 2023, 12, 1518. [Google Scholar] [CrossRef]
- Wei, X.; Wang, Y.; Lao, Y.; Weng, J.; Deng, R.; Li, S.; Lu, J.; Yang, S.; Liu, X. Effects of honokiol protects against chronic kidney disease via BNIP3/NIX and FUNDC1-mediated mitophagy and AMPK pathways. Mol. Biol. Rep. 2023, 50, 6557–6568. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Yang, Y.; Xia, M.; Deng, Y.; Zuo, Y.; Lei, L.; Hu, T. A Chinese herb preparation, honokiol, inhibits Streptococcus mutans biofilm formation. Arch. Oral Biol. 2023, 147, 105610. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Z.; Bai, J.; Wang, X.; Yuan, Q.; Mi, Y.; Zhang, C. Bioactive Lignan Honokiol Alleviates Ovarian Oxidative Stress in Aging Laying Chickens by Regulating SIRT3/AMPK Pathway. Antioxidants 2024, 13, 377. [Google Scholar] [CrossRef] [PubMed]
- Jayakumari, N.R.; Rajendran, R.S.; Sivasailam, A.; Parambil, S.T.; Reghuvaran, A.C.; Sreelatha, H.V.; Gopala, S. Honokiol regulates mitochondrial substrate utilization and cellular fatty acid metabolism in diabetic mice heart. Eur. J. Pharmacol. 2021, 896, 173918. [Google Scholar] [CrossRef]
- Caballero, E.P.; Mariz-Ponte, N.; Rigazio, C.S.; Santamaria, M.H.; Corral, R.S. Honokiol attenuates oxidative stress-dependent heart dysfunction in chronic Chagas disease by targeting AMPK/NFE2L2/SIRT3 signaling pathway. Free Radic. Biol. Med. 2020, 156, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, Q.-W.; Ye, Y.; Lin, L.-G. Honokiol: A naturally occurring lignan with pleiotropic bioactivities. Chin. J. Nat. Med. 2021, 19, 481–490. [Google Scholar] [CrossRef]
- Cardullo, N.; Monti, F.; Muccilli, V.; Amorati, R.; Baschieri, A. Reaction with ROO* and HOO* Radicals of Honokiol-Related Neolignan Antioxidants. Molecules 2023, 28, 735. [Google Scholar] [CrossRef]
- Bentinger, M.; Brismar, K.; Dallner, G. The antioxidant role of coenzyme Q. Mitochondrion 2007, 7, S41–S50. [Google Scholar] [CrossRef]
- Sood, B.; Patel, P.; Keenaghan, M. Coenzyme Q10; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Cores, A.; Carmona-Zafra, N.; Clerigué, J.; Villacampa, M.; Menéndez, J.C. Quinones as Neuroprotective Agents. Antioxidants 2023, 12, 1464. [Google Scholar] [CrossRef]
- Amorati, R.; Zotova, J.; Baschieri, A.; Valgimigli, L. Antioxidant Activity of Magnolol and Honokiol: Kinetic and Mechanistic Investigations of Their Reaction with Peroxyl Radicals. J. Org. Chem. 2015, 80, 10651–10659. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Araiza, C.; Alvarez-Mejia, A.L.; Sanchez-Torres, S.; Farfan-Garcia, E.; Mondragon-Lozano, R.; Pin-to-Almazan, R.; Salgado-Ceballos, H. Effect of natural exogenous antioxidants on aging and on neurodegenerative dis-eases. Free Radic. Res. 2013, 47, 451–462. [Google Scholar] [CrossRef]
- Dikalov, S.; Losik, T.; Arbiser, J.L. Honokiol is a potent scavenger of superoxide and peroxyl radicals. Biochem. Pharmacol. 2008, 76, 589–596. [Google Scholar] [CrossRef]
- Wang, J.; Nisar, M.; Huang, C.; Pan, X.; Lin, D.; Zheng, G.; Jin, H.; Chen, D.; Tian, N.; Huang, Q.; et al. Small molecule natural compound agonist of SIRT3 as a therapeutic target for the treatment of intervertebral disc degeneration. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ingólfsson, H.I.; Melo, M.N.; van Eerden, F.J.; Arnarez, C.; Lopez, C.A.; Wassenaar, T.A.; Periole, X.; de Vries, A.H.; Tieleman, D.P.; Marrink, S.J. Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 2014, 136, 14554–14559. [Google Scholar] [CrossRef] [PubMed]
- Perricone, U.; Gulotta, M.R.; Lombino, J.; Parrino, B.; Cascioferro, S.; Diana, P.; Cirrincione, G.; Padova, A. An overview of recent molecular dynamics applications as medicinal chemistry tools for the undruggable site challenge. MedChemComm 2018, 9, 920–936. [Google Scholar] [CrossRef]
- Villalaín, J. Bergamottin: Location, aggregation and interaction with the plasma membrane. J. Biomol. Struct. Dyn. 2023, 41, 12026–12037. [Google Scholar] [CrossRef]
- Villalaín, J. Envelope E protein of dengue virus and phospholipid binding to the late endosomal membrane. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183889. [Google Scholar] [CrossRef]
- Galiano, V.; Villalaín, J. Aggregation of 25-hydroxycholesterol in a complex biomembrane. Differences with cholesterol. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183413. [Google Scholar] [CrossRef]
- Villalaín, J. Labyrinthopeptin A2 disrupts raft domains. Chem. Phys. Lipids 2023, 253, 105303. [Google Scholar] [CrossRef]
- Villalaín, J. Procyanidin C1 Location, Interaction, and Aggregation in Two Complex Biomembranes. Membranes 2022, 12, 692. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Espósito, D.L.A.; Nguyen, J.B.; DeWitt, D.C.; Rhoades, E.; Modis, Y. Physicochemical requirements and kinetics of membrane fusion of flavivirus-like particles. J. Gen. Virol. 2015, 96, 1702–1711. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Murzyn, K.; Róg, T.; Jezierski, G.; Takaoka, Y.; Pasenkiewicz-Gierula, M. Effects of Phospholipid Unsaturation on the Membrane/Water Interface: A Molecular Simulation Study. Biophys. J. 2001, 81, 170–183. [Google Scholar] [CrossRef]
- Košinová, P.; Berka, K.; Wykes, M.; Otyepka, M.; Trouillas, P. Positioning of Antioxidant Quercetin and Its Metabolites in Lipid Bilayer Membranes: Implication for Their Lipid-Peroxidation Inhibition. J. Phys. Chem. B 2012, 116, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Galiano, V.; Villalaín, J. The Location of the Protonated and Unprotonated Forms of Arbidol in the Membrane: A Molecular Dynamics Study. J. Membr. Biol. 2016, 249, 381–391. [Google Scholar] [CrossRef]
- Galiano, V.; Villalaín, J. Oleuropein aglycone in lipid bilayer membranes. A molecular dynamics study. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2849–2858. [Google Scholar] [CrossRef]
- Gotzias, A.; Tocci, E.; Sapalidis, A. Solvent-Assisted Graphene Exfoliation from Graphite Using Umbrella Sampling Simulations. Langmuir 2023, 39, 18437–18446. [Google Scholar] [CrossRef]
- Gotzias, A. Umbrella Sampling Simulations of Carbon Nanoparticles Crossing Immiscible Solvents. Molecules 2022, 27, 956. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Guixà-González, R.; Rodriguez-Espigares, I.; Ramírez-Anguita, J.M.; Carrió-Gaspar, P.; Martinez-Seara, H.; Giorgino, T.; Selent, J. MEMBPLUGIN: Studying membrane complexity in VMD. Bioinformatics 2014, 30, 1478–1480. [Google Scholar] [CrossRef]
- Villalaín, J. Epigallocatechin-3-gallate location and interaction with late endosomal and plasma membrane model membranes by molecular dynamics. J. Biomol. Struct. Dyn. 2019, 37, 3122–3134. [Google Scholar] [CrossRef] [PubMed]
- Giorgino, T. Computing 1-D atomic densities in macromolecular simulations: The density profile tool for VMD. Comput. Phys. Commun. 2014, 185, 317–322. [Google Scholar] [CrossRef]
- Kandt, C.; Ash, W.L.; Tieleman, D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods 2007, 41, 475–488. [Google Scholar] [CrossRef]
- Anézo, C.; de Vries, A.H.; Höltje, H.-D.; Tieleman, D.P.; Marrink, S.-J. Methodological Issues in Lipid Bilayer Simulations. J. Phys. Chem. B 2003, 107, 9424–9433. [Google Scholar] [CrossRef]
- Bera, I.; Klauda, J.B. Molecular Simulations of Mixed Lipid Bilayers with Sphingomyelin, Glycerophospholipids, and Cholesterol. J. Phys. Chem. B 2017, 121, 5197–5208. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Monticelli, L.; Tieleman, D.P. Molecular dynamics simulation of a palmitoyl-oleoyl phosphati-dylserine bilayer with Na+ counterions and NaCl. Biophys. J. 2004, 86, 1601–1609. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef]
- Tsai, H.-H.G.; Lee, J.-B.; Li, H.-S.; Hou, T.-Y.; Chu, W.-Y.; Shen, P.-C.; Chen, Y.-Y.; Tan, C.-J.; Hu, J.-C.; Chiu, C.-C. Geometrical effects of phospholipid olefinic bonds on the structure and dynamics of membranes: A molecular dynamics study. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1234–1247. [Google Scholar] [CrossRef]
- Böckmann, R.A.; Hac, A.; Heimburg, T.; Grubmüller, H. Effect of Sodium Chloride on a Lipid Bilayer. Biophys. J. 2003, 85, 1647–1655. [Google Scholar] [CrossRef] [PubMed]



| SYSTEMS | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| HNK | 4 | 9 | 8 | 18 |
| POPC | 144 | 144 | 144 | 144 |
| POPE | 86 | 86 | 86 | 86 |
| PI-3P | 28 | 28 | 28 | 28 |
| POPS | 32 | 32 | 32 | 32 |
| PSM | 60 | 60 | 60 | 60 |
| CHOL | 150 | 150 | 150 | 150 |
| No. LIPIDS | 500 | 500 | 500 | 500 |
| ATOMS | 166,463 | 166,653 | 166,255 | 166,159 |
| H2O | 36,413 | 36,413 | 36,293 | 36,135 |
| H2O/LIPID | 72.5 | 72.8 | 72.6 | 72.3 |
| Na+ | 219 | 219 | 219 | 218 |
| Cl− | 103 | 103 | 103 | 102 |
| INITIAL DIMENSIONS x/y/z (Å) | 128/129/120 | 128/129/120 | 128/129/120 | 128/128/120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalaín, J. Localization and Aggregation of Honokiol in the Lipid Membrane. Antioxidants 2024, 13, 1025. https://doi.org/10.3390/antiox13081025
Villalaín J. Localization and Aggregation of Honokiol in the Lipid Membrane. Antioxidants. 2024; 13(8):1025. https://doi.org/10.3390/antiox13081025
Chicago/Turabian StyleVillalaín, José. 2024. "Localization and Aggregation of Honokiol in the Lipid Membrane" Antioxidants 13, no. 8: 1025. https://doi.org/10.3390/antiox13081025
APA StyleVillalaín, J. (2024). Localization and Aggregation of Honokiol in the Lipid Membrane. Antioxidants, 13(8), 1025. https://doi.org/10.3390/antiox13081025







