The Bioactive Gamma-Oryzanol from Oryza sativa L. Promotes Neuronal Differentiation in Different In Vitro and In Vivo Models
Abstract
1. Introduction
2. Materials and Methods
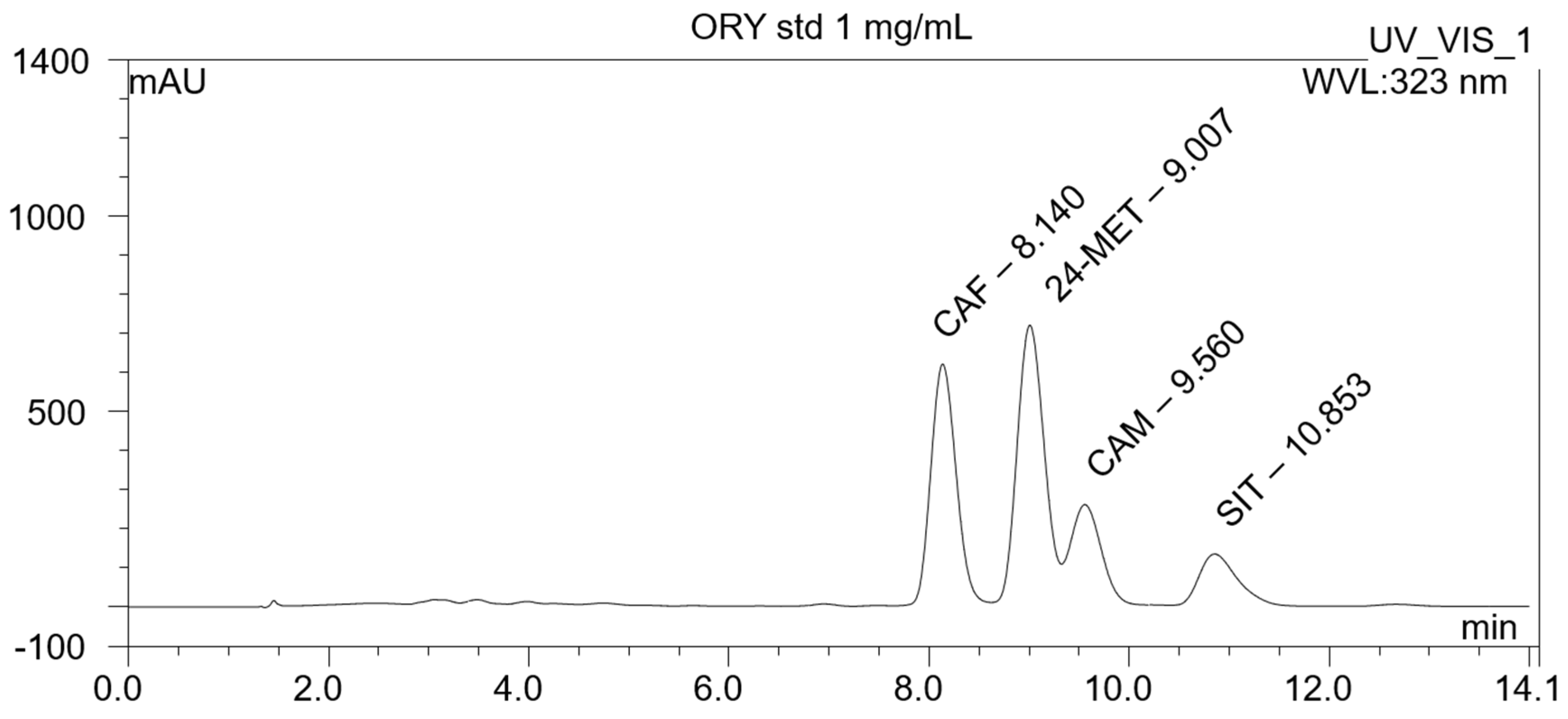
2.1. HPLC Characterization
2.2. Cell Culture and Immunofluorescence Analysis
2.3. Western Blot Analysis
2.4. Subcellular Fractionation for Nrf2 Nuclear Translocation
2.5. Quantitative Real-Time PCR and Gene Expression
2.6. Animals
2.7. Culture of Adult Murine Hippocampal Neural Progenitor Cells
2.8. Neural Progenitor Cell Differentiation
2.9. Zebrafish Maintenance
2.10. Morpholino Microinjection
2.11. Gamma-Oryzanol Microinjection and Fluorescence Evaluation
2.12. Whole Mount In Situ Hybridization (WISH)
2.13. Computational Studies
2.14. Statistical Analysis
3. Results
3.1. Chemical Characterization of the Gamma-Oryzanol Mixture
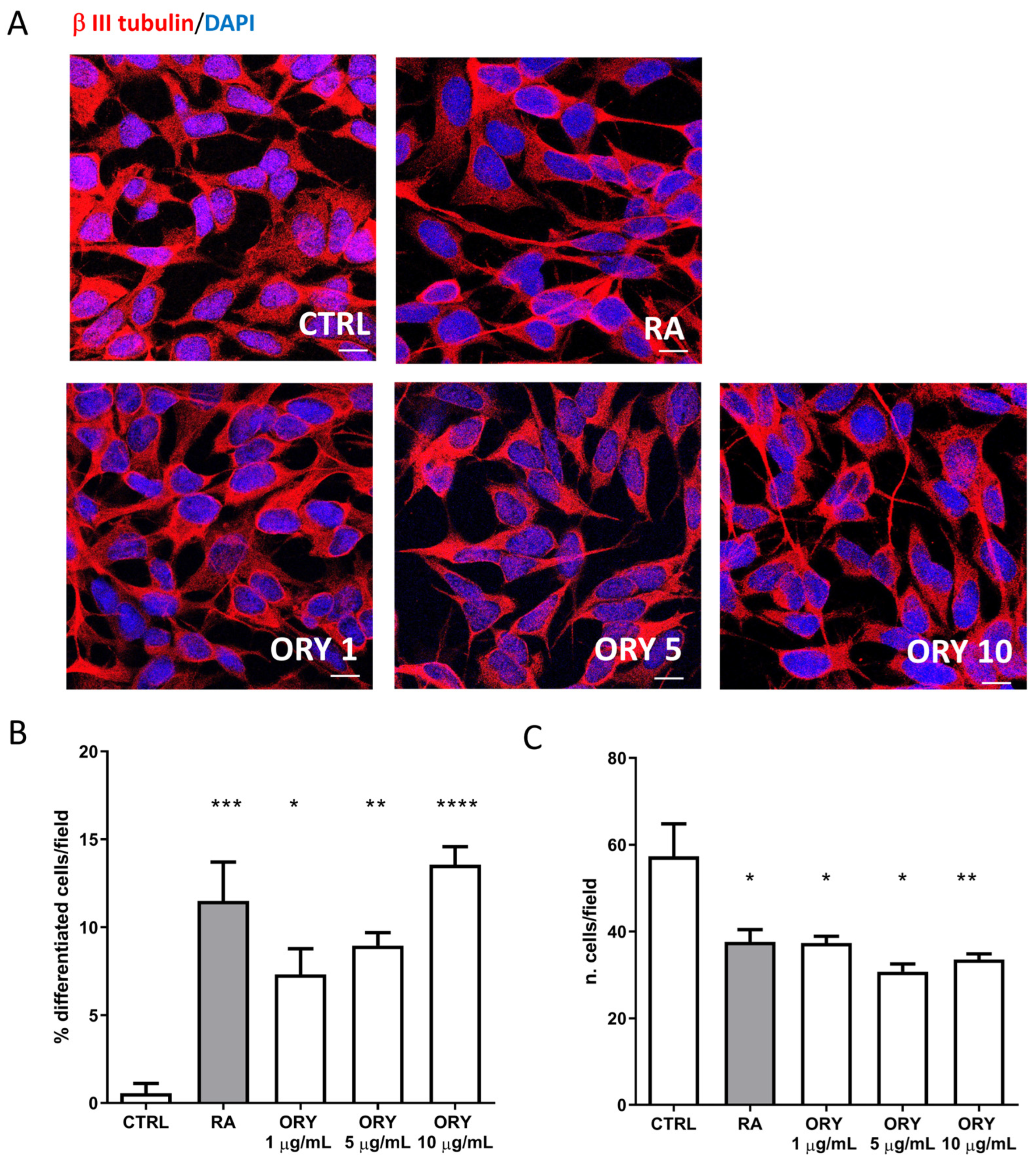
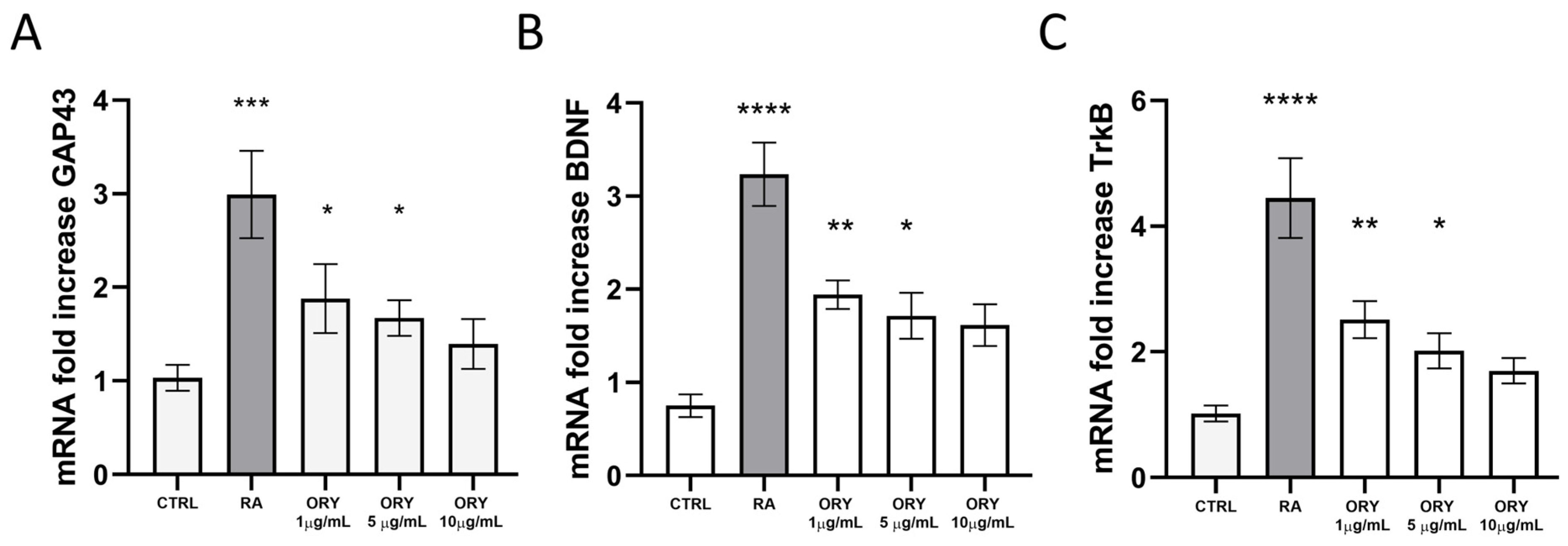
3.2. Gamma-Oryzanol Promotes Neuronal Differentiation in SH-SY5Y Neuroblastoma Cells
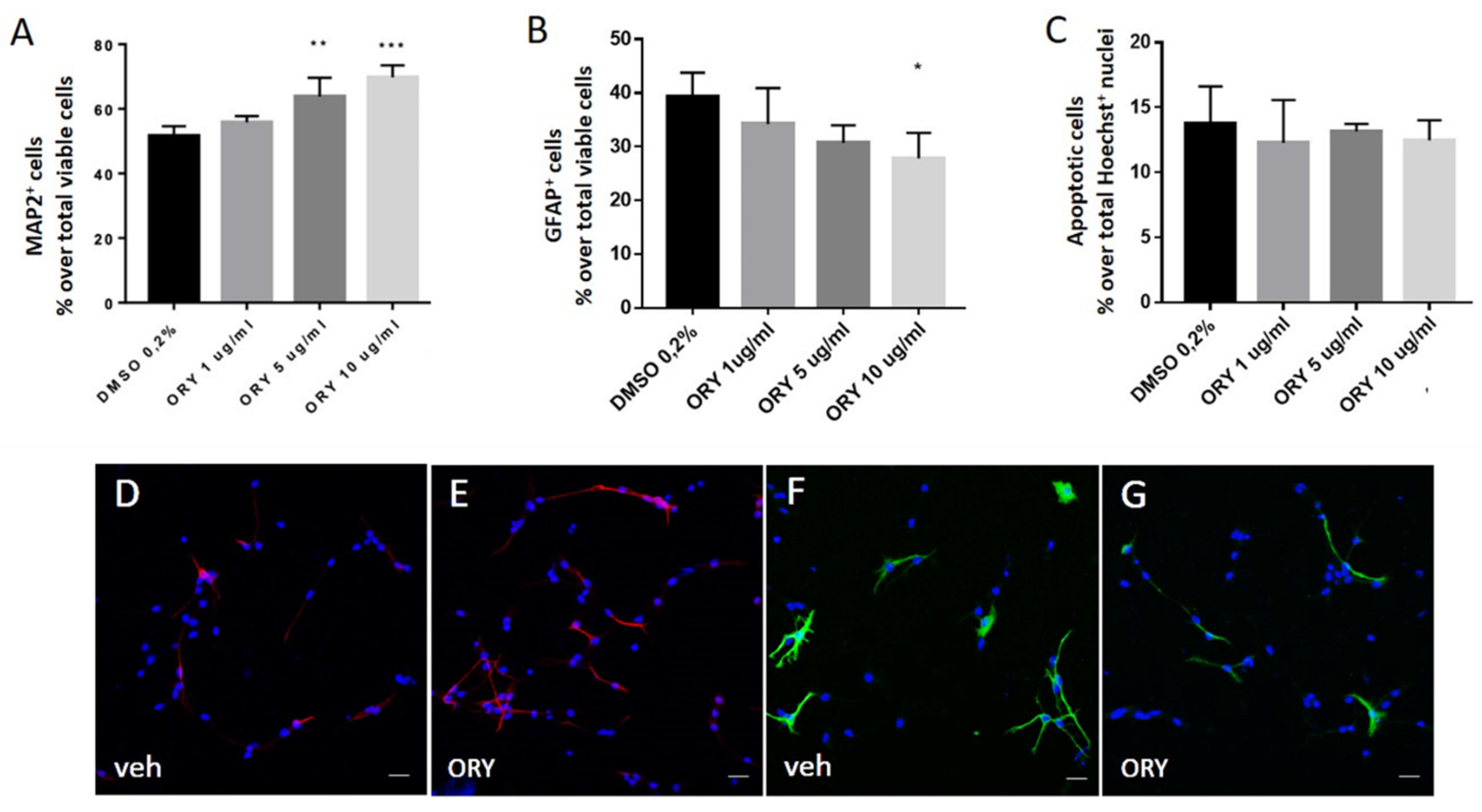
3.3. Gamma-Oryzanol Promotes Neuronal Differentiation in Murine Adult Neural Progenitor Cells
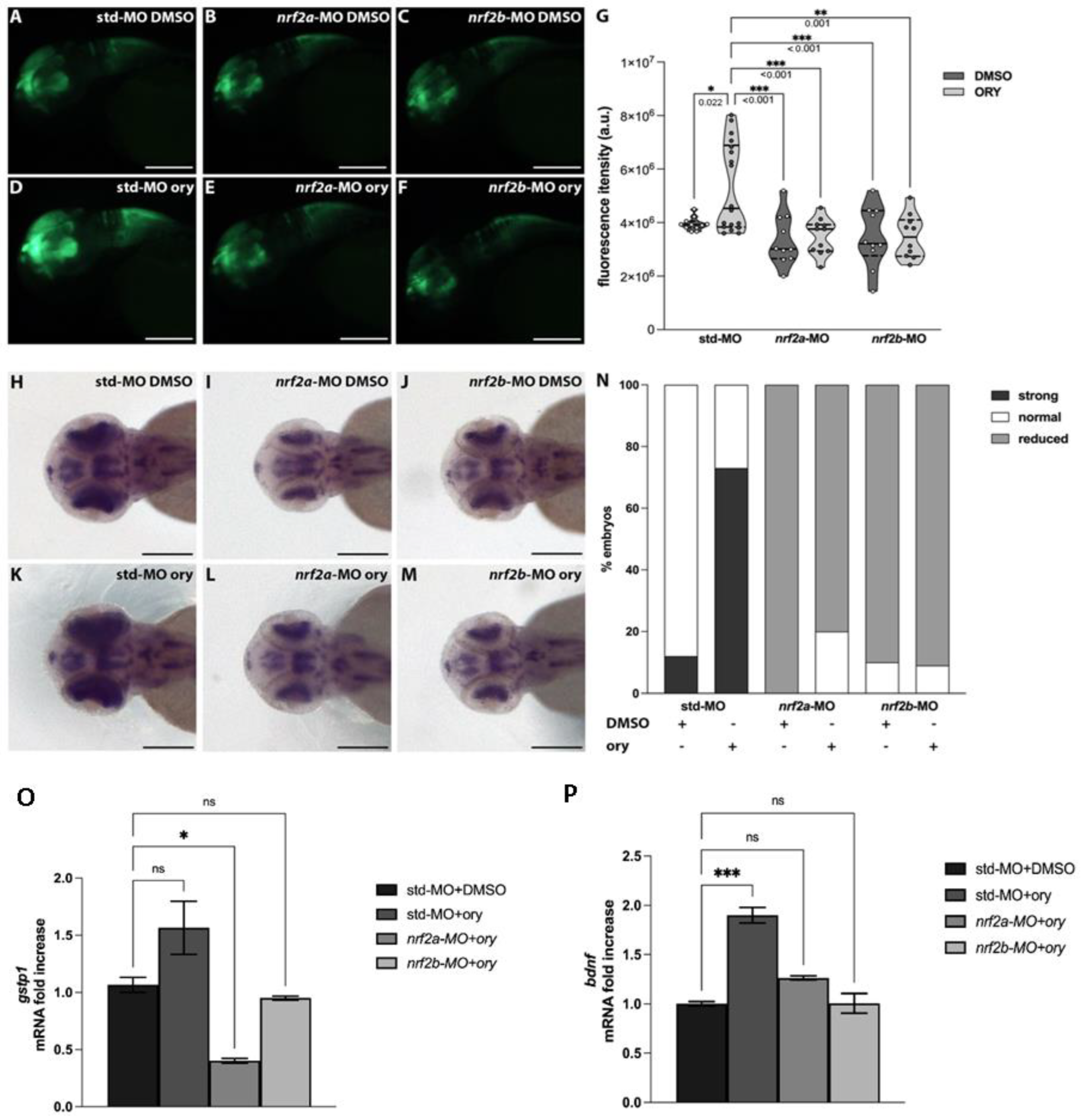
3.4. Gamma-Oryzanol Exerts Pro-Neural Activity in the Presence of Functional Nrf2 Signalling in Zebrafish
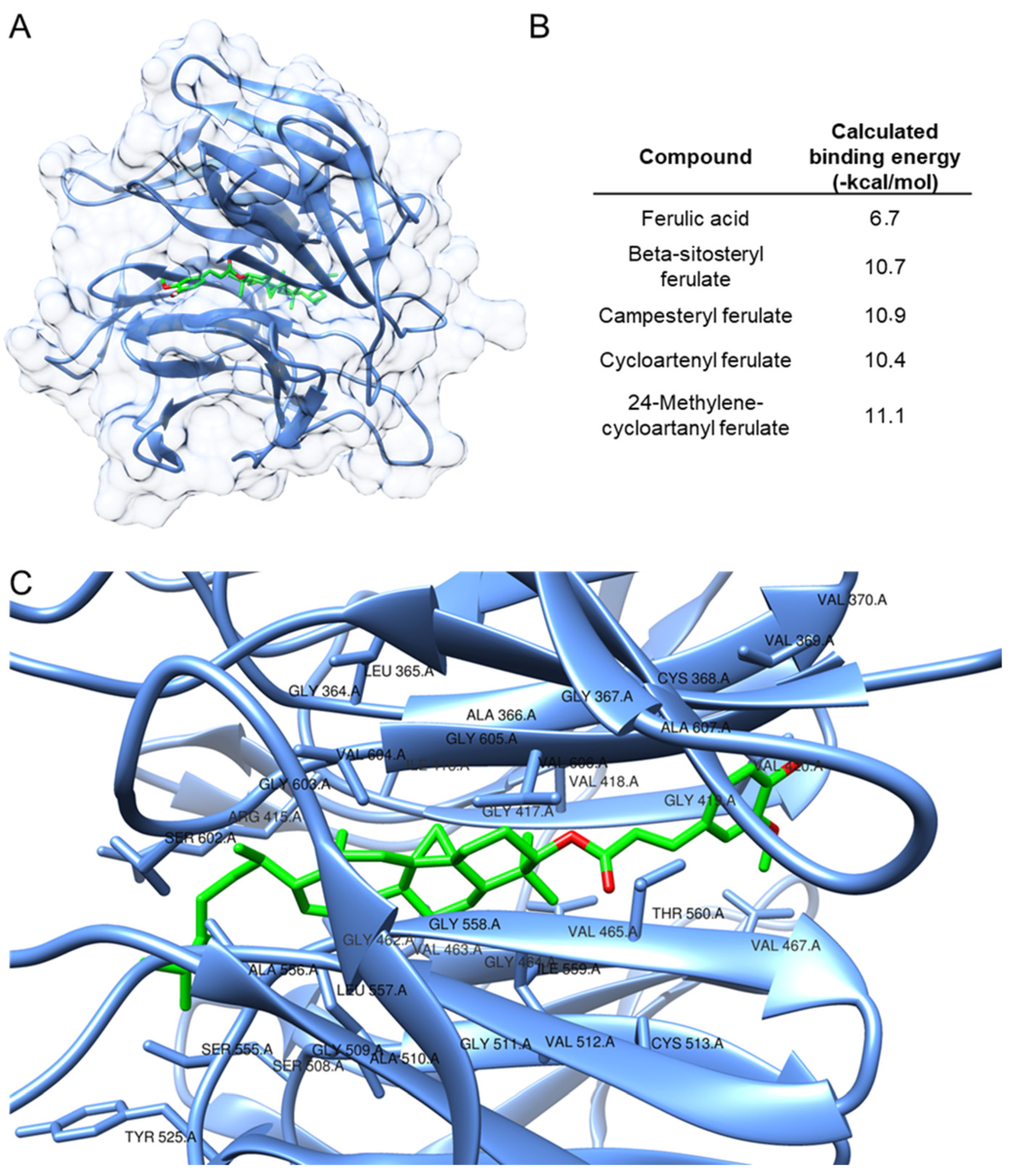
3.5. Computational Investigation of the Mechanism of Action of Key Compounds in the ORY Mixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rungratanawanich, W.; Abate, G.; Serafini, M.M.; Guarienti, M.; Catanzaro, M.; Marziano, M.; Memo, M.; Lanni, C.; Uberti, D. Characterization of the Antioxidant Effects of γ-Oryzanol: Involvement of the Nrf2 Pathway. Oxid. Med. Cell. Longev. 2018, 2018, 2987249. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Kumar, S.; Sehgal, A.; Singh, S.; Kumari, S.; Brisc, M.C.; Munteanu, M.A.; Brisc, C.; Buhas, C.L.; Judea-Pusta, C.; et al. Rice Bran, an off-Shoot to Newer Therapeutics in Neurological Disorders. Biomed. Pharmacother. 2021, 140, 111796. [Google Scholar] [CrossRef]
- Mastinu, A.; Bonini, S.A.; Rungratanawanich, W.; Aria, F.; Marziano, M.; Maccarinelli, G.; Abate, G.; Premoli, M.; Memo, M.; Uberti, D. Gamma-Oryzanol Prevents LPS-Induced Brain Inflammation and Cognitive Impairment in Adult Mice. Nutrients 2019, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Rungratanawanich, W.; Cenini, G.; Mastinu, A.; Sylvester, M.; Wilkening, A.; Abate, G.; Bonini, S.A.; Aria, F.; Marziano, M.; Maccarinelli, G.; et al. γ-Oryzanol Improves Cognitive Function and Modulates Hippocampal Proteome in Mice. Nutrients 2019, 11, 753. [Google Scholar] [CrossRef]
- Zhang, H.; Davies, K.J.A.; Forman, H.J. Oxidative Stress Response and Nrf2 Signaling in Aging. Free Radic. Biol. Med. 2015, 88, 314–336. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Lin, S.; Su, J.; Cao, Q.; Chen, Y.; Chen, J.; Zhang, Z.; Hashimoto, K.; Qi, Q.; Zhang, J.-C. Activation of BDNF by Transcription Factor Nrf2 Contributes to Antidepressant-like Actions in Rodents. Transl. Psychiatry 2021, 11, 140. [Google Scholar] [CrossRef]
- Bruna, B.; Lobos, P.; Herrera-Molina, R.; Hidalgo, C.; Paula-Lima, A.; Adasme, T. The Signaling Pathways Underlying BDNF-Induced Nrf2 Hippocampal Nuclear Translocation Involve ROS, RyR-Mediated Ca2+ Signals, ERK and PI3K. Biochem. Biophys. Res. Commun. 2018, 505, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Mann, G.E. When and How Does Brain-Derived Neurotrophic Factor Activate Nrf2 in Astrocytes and Neurons? Neural Regen. Res. 2018, 13, 803–804. [Google Scholar] [CrossRef]
- Ramos-Gomez, M.; Kwak, M.-K.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Talalay, P.; Kensler, T.W. Sensitivity to Carcinogenesis Is Increased and Chemoprotective Efficacy of Enzyme Inducers Is Lost in Nrf2 Transcription Factor-Deficient Mice. Proc. Natl. Acad. Sci. USA 2001, 98, 3410–3415. [Google Scholar] [CrossRef]
- Rojo, A.I.; Pajares, M.; Rada, P.; Nuñez, A.; Nevado-Holgado, A.J.; Killik, R.; Van Leuven, F.; Ribe, E.; Lovestone, S.; Yamamoto, M.; et al. NRF2 Deficiency Replicates Transcriptomic Changes in Alzheimer’s Patients and Worsens APP and TAU Pathology. Redox Biol. 2017, 13, 444–451. [Google Scholar] [CrossRef]
- Bettinsoli, P.; Ferrari-Toninelli, G.; Bonini, S.A.; Prandelli, C.; Memo, M. Notch Ligand Delta-like 1 as a Novel Molecular Target in Childhood Neuroblastoma. BMC Cancer 2017, 17, 352. [Google Scholar] [CrossRef]
- Pucci, M.; Aria, F.; Premoli, M.; Maccarinelli, G.; Mastinu, A.; Bonini, S.; Memo, M.; Uberti, D.; Abate, G. Methylglyoxal Affects Cognitive Behaviour and Modulates RAGE and Presenilin-1 Expression in Hippocampus of Aged Mice. Food Chem. Toxicol. 2021, 158, 112608. [Google Scholar] [CrossRef]
- Bortolotto, V.; Bondi, H.; Cuccurazzu, B.; Rinaldi, M.; Canonico, P.L.; Grilli, M. Salmeterol, a Β2 Adrenergic Agonist, Promotes Adult Hippocampal Neurogenesis in a Region-Specific Manner. Front. Pharmacol. 2019, 10, 1000. [Google Scholar] [CrossRef]
- Valente, M.M.; Bortolotto, V.; Cuccurazzu, B.; Ubezio, F.; Meneghini, V.; Francese, M.T.; Canonico, P.L.; Grilli, M. A2δ Ligands Act as Positive Modulators of Adult Hippocampal Neurogenesis and Prevent Depression-like Behavior Induced by Chronic Restraint Stress. Mol. Pharmacol. 2012, 82, 271–280. [Google Scholar] [CrossRef]
- Tallafuss, A.; Gibson, D.; Morcos, P.; Li, Y.; Seredick, S.; Eisen, J.; Washbourne, P. Turning Gene Function ON and OFF Using Sense and Antisense Photo-Morpholinos in Zebrafish. Development 2012, 139, 1691–1699. [Google Scholar] [CrossRef]
- Sant, K.E.; Hansen, J.M.; Williams, L.M.; Tran, N.L.; Goldstone, J.V.; Stegeman, J.J.; Hahn, M.E.; Timme-Laragy, A. The Role of Nrf1 and Nrf2 in the Regulation of Glutathione and Redox Dynamics in the Developing Zebrafish Embryo. Redox Biol. 2017, 13, 207–218. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-Resolution in Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, L.; Chen, H.; Zhang, W.; Xing, C.; Qu, Z.; Yu, J.; Zhuang, C. Structure-Based Molecular Hybridization Design of Keap1-Nrf2 Inhibitors as Novel Protective Agents of Acute Lung Injury. Eur. J. Med. Chem. 2021, 222, 113599. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Timme-Laragy, A.R.; Karchner, S.I.; Franks, D.G.; Jenny, M.J.; Harbeitner, R.C.; Goldstone, J.V.; McArthur, A.G.; Hahn, M.E. Nrf2b, Novel Zebrafish Paralog of Oxidant-Responsive Transcription Factor NF-E2-Related Factor 2 (NRF2). J. Biol. Chem. 2012, 287, 4609–4627. [Google Scholar] [CrossRef]
- Moreno, R.L.; Williams, K.; Jones, K.L.; Ribera, A.B. Investigation of Islet2a Function in Zebrafish Embryos: Mutants and Morphants Differ in Morphologic Phenotypes and Gene Expression. PLoS ONE 2018, 13, e0199233. [Google Scholar] [CrossRef]
- Jain, A.D.; Potteti, H.; Richardson, B.G.; Kingsley, L.; Luciano, J.P.; Ryuzoji, A.F.; Lee, H.; Krunic, A.; Mesecar, A.D.; Reddy, S.P.; et al. Probing the Structural Requirements of Non-Electrophilic Naphthalene-Based Nrf2 Activators. Eur. J. Med. Chem. 2015, 103, 252–268. [Google Scholar] [CrossRef]
- Georgakopoulos, N.D.; Gatliff, J.; Wells, G. Development of Keap1-Interactive Small Molecules That Regulate Nrf2 Transcriptional Activity. Curr. Opin. Toxicol. 2016, 1, 1–8. [Google Scholar] [CrossRef]
- Shen, G.; Kong, A.-N. Nrf2 Plays an Important Role in Coordinated Regulation of Phase II Drug Metabolism Enzymes and Phase III Drug Transporters. Biopharm. Drug Dispos. 2009, 30, 345–355. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Saha, S.; Buttari, B.; Profumo, E.; Tucci, P.; Saso, L. A Perspective on Nrf2 Signaling Pathway for Neuroinflammation: A Potential Therapeutic Target in Alzheimer’s and Parkinson’s Diseases. Front. Cell. Neurosci. 2022, 15, 787258. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Zhao, F.; Wu, T.; Lau, A.; Jiang, T.; Huang, Z.; Wang, X.-J.; Chen, W.; Wong, P.K.; Zhang, D.D. Nrf2 Promotes Neuronal Cell Differentiation. Free Radic. Biol. Med. 2009, 47, 867–879. [Google Scholar] [CrossRef]
- Williamson, T.P.; Johnson, D.A.; Johnson, J.A. Activation of the Nrf2-ARE Pathway by SiRNA Knockdown of Keap1 Reduces Oxidative Stress and Provides Partial Protection from MPTP-Mediated Neurotoxicity. Neurotoxicology 2012, 33, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Motohashi, H. Roles of Nrf2 in Cell Proliferation and Differentiation. Free Radic. Biol. Med. 2015, 88, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Yabe, T.; Hirahara, H.; Harada, N.; Ito, N.; Nagai, T.; Sanagi, T.; Yamada, H. Ferulic Acid Induces Neural Progenitor Cell Proliferation in Vitro and in Vivo. Neuroscience 2010, 165, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Holland, R.; Fishbein, J.C. Chemistry of the Cysteine Sensors in Kelch-like ECH-Associated Protein 1. Antioxid. Redox Signal. 2010, 13, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
| Name | Primer Sequence |
|---|---|
| Growth-Associated Protein 43 (GAP-43) [NM_008083] | For: 5′-TTCTTGGTGTTGTTATGGCAA G-3′, Rev: 5′-GAGGAAAGTGGACTCCCACAG-3′ |
| Brain-Derived Neurotrophic Factor (BDNF) [NM_001048141] | For: 5′-CATCCGAGGACAAGGTGGCTTG-3′, Rev: 5′-GCCGAACTTTCTGGTCCTCATC-3′ |
| Tropomyosin receptor kinase B (TrkB) [NM_001025074.3] | For: 5′- GAGCATCATGTACAGGAAAT-3′, Rev: 5′- CTTGATGTTCTTCCTCATGT-3′, |
| Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) [NM_008085] | For: 5′-GAGTCAACGGAT TTGGTCGT-3′, Rev: 5′-TTGATTTTGGAGGGATCTCG-3′. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abate, G.; Pezzotta, A.; Pucci, M.; Bortolotto, V.; Ribaudo, G.; Bonini, S.A.; Mastinu, A.; Maccarinelli, G.; Ongaro, A.; Tirelli, E.; et al. The Bioactive Gamma-Oryzanol from Oryza sativa L. Promotes Neuronal Differentiation in Different In Vitro and In Vivo Models. Antioxidants 2024, 13, 969. https://doi.org/10.3390/antiox13080969
Abate G, Pezzotta A, Pucci M, Bortolotto V, Ribaudo G, Bonini SA, Mastinu A, Maccarinelli G, Ongaro A, Tirelli E, et al. The Bioactive Gamma-Oryzanol from Oryza sativa L. Promotes Neuronal Differentiation in Different In Vitro and In Vivo Models. Antioxidants. 2024; 13(8):969. https://doi.org/10.3390/antiox13080969
Chicago/Turabian StyleAbate, Giulia, Alex Pezzotta, Mariachiara Pucci, Valeria Bortolotto, Giovanni Ribaudo, Sara A. Bonini, Andrea Mastinu, Giuseppina Maccarinelli, Alberto Ongaro, Emanuela Tirelli, and et al. 2024. "The Bioactive Gamma-Oryzanol from Oryza sativa L. Promotes Neuronal Differentiation in Different In Vitro and In Vivo Models" Antioxidants 13, no. 8: 969. https://doi.org/10.3390/antiox13080969
APA StyleAbate, G., Pezzotta, A., Pucci, M., Bortolotto, V., Ribaudo, G., Bonini, S. A., Mastinu, A., Maccarinelli, G., Ongaro, A., Tirelli, E., Zizioli, D., Gianoncelli, A., Memo, M., Grilli, M., & Uberti, D. (2024). The Bioactive Gamma-Oryzanol from Oryza sativa L. Promotes Neuronal Differentiation in Different In Vitro and In Vivo Models. Antioxidants, 13(8), 969. https://doi.org/10.3390/antiox13080969











