Direct Molecular Action of Taurine on Hepatic Gene Expression Associated with the Amelioration of Hypercholesterolemia in Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Rat Primary Hepatocyte 3D Culture
2.3. Biochemical Analysis
2.4. Total RNA Extraction and Quantitative Real-Time PCR Analysis (RT-qPCR)
2.5. Microarray Analysis and Data Analysis
2.6. Statistical Analyses
3. Results
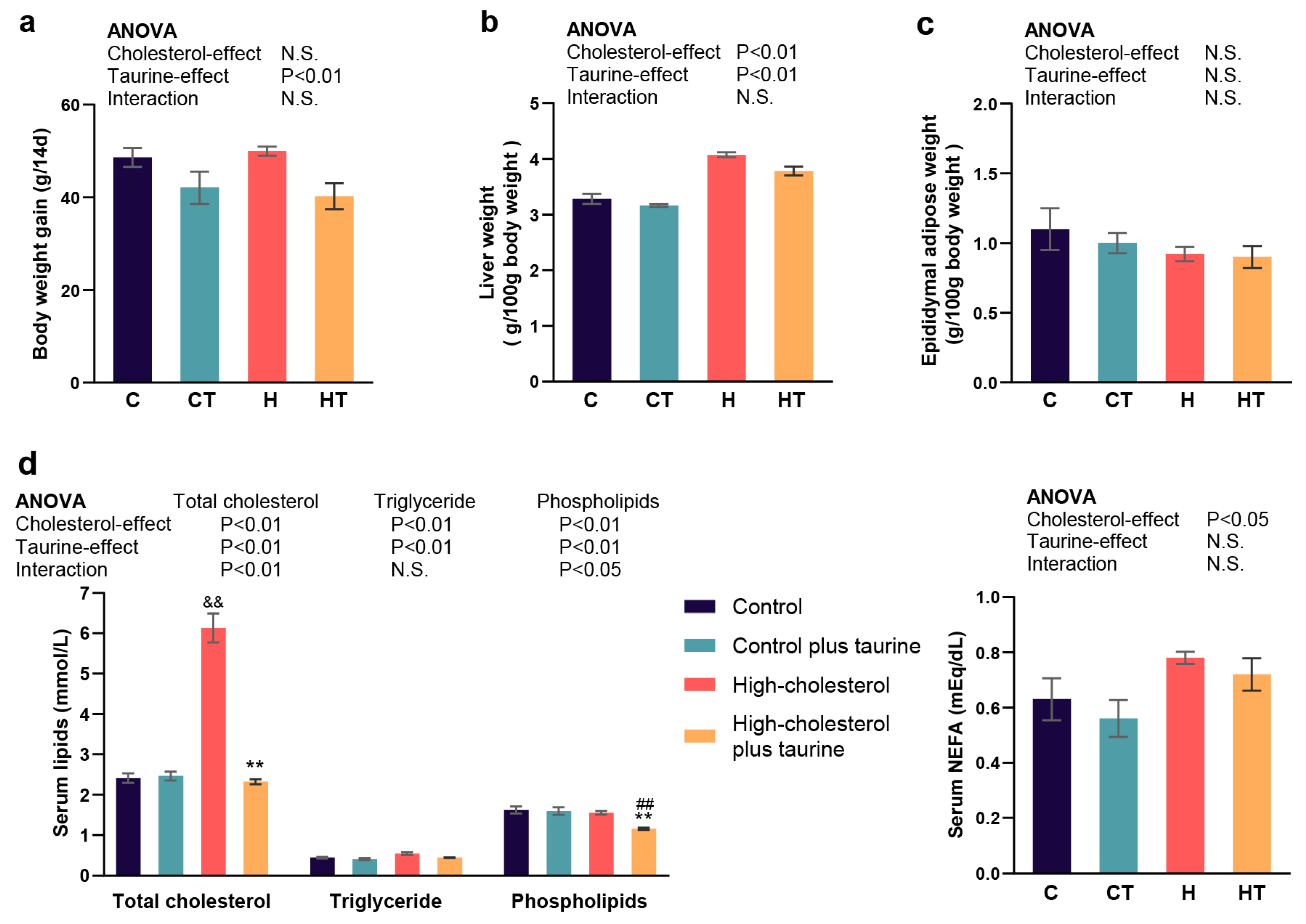
3.1. Effect of Taurine on the Physiological and Biochemical Parameters in Rats Fed a High Cholesterol Diet
3.2. Taurine Increased Hepatic CYP7A1 Gene Expression and Transcription Rate in Rats Fed a High Cholesterol Diet
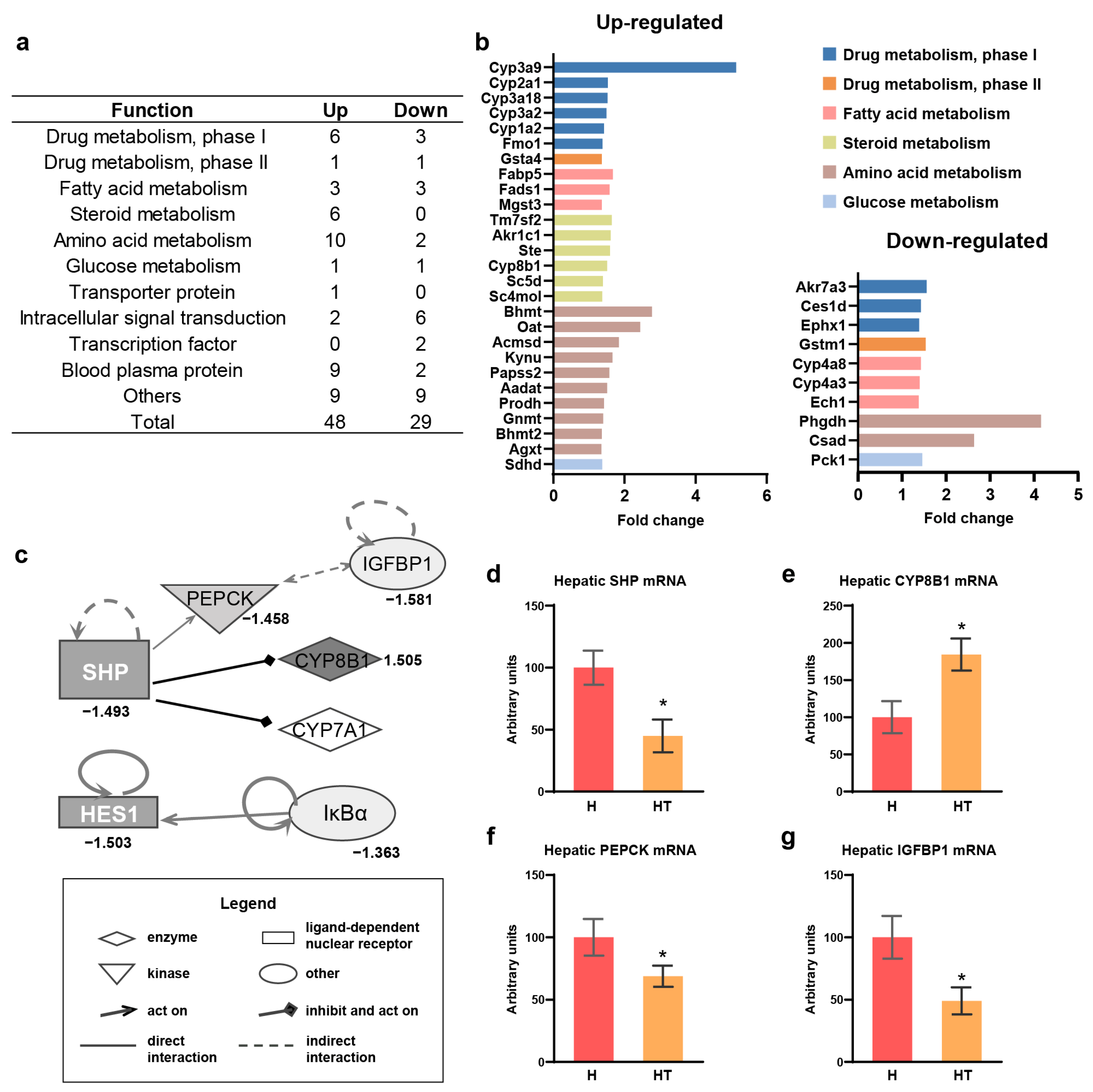
3.3. “Function” and “Pathway” Analyses of Taurine Effect in Transcriptomics of Hepatic RNA in Rats Fed a High Cholesterol Diet
3.4. Taurine Targeted the SHP as an Upstream Factor for Cholesterol Catabolism and Gluconeogenesis in Rats Fed a High Cholesterol Diet
3.5. Effect of Taurine on Gene Expression in Rat 3D-Primary Hepatocytes
3.6. Taurine Direct Target Betaine Homocysteine Methyltransferase (BHMT) and Organic Anion Transporting Polypeptide 2 (OATP2) in Rat 3D-Primary Hepatocytes and Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seidel, U.; Huebbe, P.; Rimbach, G. Taurine: A regulator of cellular redox homeostasis and skeletal muscle function. Mol. Nutr. Food Res. 2019, 63, 1800569. [Google Scholar] [CrossRef] [PubMed]
- McCusker, S.; Buff, P.R.; Yu, Z.; Fascetti, A.J. Amino acid content of selected plant, algae and insect species: A search for alternative protein sources for use in pet foods. J. Nutr. Sci. 2014, 3, e39. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T. Identification of a novel enzyme and the regulation of key enzymes in mammalian taurine synthesis. J. Pharmacol. Sci. 2024, 154, 9–17. [Google Scholar] [CrossRef]
- Schaffer, S.W.; Ito, T.; Azuma, J. Clinical significance of taurine. Amino Acids 2014, 46, 1–5. [Google Scholar] [CrossRef]
- Veeravalli, S.; Phillips, I.R.; Freire, R.T.; Varshavi, D.; Everett, J.R.; Shephard, E.A. Flavin-containing monooxygenase 1 catalyzes the production of taurine from hypotaurine. Drug Metab. Dispos. 2020, 48, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Gollapalli, K.; Mangiola, S.; Schranner, D.; Yusuf, M.A.; Chamoli, M.; Shi, S.L.; Lopes Bastos, B.; Nair, T.; Riermeier, A.; et al. Taurine deficiency as a driver of aging. Science 2023, 380, eabn9257. [Google Scholar] [CrossRef]
- Bhat, M.A.; Ahmad, K.; Khan, M.S.A.; Bhat, M.A.; Almatroudi, A.; Rahman, S.; Jan, A.T. Expedition into taurine biology: Structural insights and therapeutic perspective of taurine in neurodegenerative diseases. Biomolecules 2020, 10, 863. [Google Scholar] [CrossRef]
- Santulli, G.; Kansakar, U.; Varzideh, F.; Mone, P.; Jankauskas, S.S.; Lombardi, A. Functional role of taurine in aging and cardiovascular health: An updated overview. Nutrients 2023, 15, 4236. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Chen, W.; Guo, J.; Zhang, Y.; Zhang, J. The beneficial effects of taurine in preventing metabolic syndrome. Food Funct. 2016, 7, 1849–1863. [Google Scholar] [CrossRef]
- Paneni, F.; Diaz Cañestro, C.; Libby, P.; Lüscher, T.F.; Camici, G.G. The aging cardiovascular system. J. Am. Coll. Cardiol. 2017, 69, 1952–1967. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, S.; Kim, H.W. Effects and mechanisms of taurine as a therapeutic agent. Biomol. Ther. 2018, 26, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Toda, Y.; Kitajima, H.; Oda, H.; Nagate, T.; Kameo, K.; Murakami, S. Taurine inhibits development of atherosclerotic lesions in apolipoprotein E-deficient mice. Clin. Exp. Pharmacol. Physiol. 2001, 28, 809–815. [Google Scholar] [CrossRef]
- Vaduganathan, M.; Mensah, G.A.; Turco, J.V.; Fuster, V.; Roth, G.A. The global burden of cardiovascular diseases and risk. J. Am. Coll. Cardiol. 2022, 80, 2361–2371. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Suruga, K.; Nishimura, N.; Gouda, T.; Lam, V.N.; Yokogoshi, H. Comparative regulation of major enzymes in the bile acid biosynthesis pathway by cholesterol, cholate and taurine in mice and rats. Life Sci. 2005, 77, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Yokogoshi, H.; Oda, H. Dietary taurine enhances cholesterol degradation and reduces serum and liver cholesterol concentrations in rats fed a high-cholesterol diet. Amino Acids 2002, 23, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Rais, N.; Parveen, K.; Ahmad, R.; Siddiqui, W.A.; Nadeem, A.; Ved, A. S-Allyl cysteine and taurine revert peripheral metabolic and lipid profile in non-insulin-dependent diabetes mellitus animals: Combination vs monotherapy. Braz. J. Pharm. Sci. 2022, 58, e201183. [Google Scholar] [CrossRef]
- Venkatesan, N.; Rao, P.V.; Arumugam, V. Inhibitory effect of taurine on puromycin aminonucleoside-induced hyperlipidemia in rats. J. Clin. Biochem. Nutr. 1993, 15, 203–210. [Google Scholar] [CrossRef]
- Dong, Y.; Li, X.; Liu, Y.; Gao, J.; Tao, J. The molecular targets of taurine confer anti-hyperlipidemic effects. Life Sci. 2021, 278, 119579. [Google Scholar] [CrossRef]
- Murakami, S.; Kondo, Y.; Toda, Y.; Kitajima, H.; Kameo, K.; Sakono, M.; Fukuda, N. Effect of taurine on cholesterol metabolism in hamsters: Up-regulation of low density lipoprotein (LDL) receptor by taurine. Life Sci. 2002, 70, 2355–2366. [Google Scholar] [CrossRef]
- Lam, N.V.; Chen, W.; Suruga, K.; Nishimura, N.; Goda, T.; Oda, H.; Yokogoshi, H. Effects of taurine on mRNA levels of nuclear receptors and factors involved in cholesterol and bile acid homeostasis in mice. In Taurine 6; Oja, S.S., Saransaari, P., Eds.; Springer: New York, NY, USA, 2006; Volume 583, pp. 193–202. [Google Scholar]
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Cai, J.; Rimal, B.; Jiang, C.; Chiang, J.Y.L.; Patterson, A.D. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol. Ther. 2022, 237, 108238. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, R.; Kobayashi, M.; Sakurai, M.; Yoshida, M.; Kaneko, H.; Mizunoe, Y.; Nozaki, Y.; Okita, N.; Sudo, Y.; Higami, Y. Long-term dietary taurine lowers plasma levels of cholesterol and bile acids. Int. J. Mol. Sci. 2022, 23, 1793. [Google Scholar] [CrossRef]
- Guo, J.; Gao, Y.; Cao, X.; Zhang, J.; Chen, W. Cholesterol-lowing effect of taurine in HepG2 cell. Lipids Health Dis. 2017, 16, 56. [Google Scholar] [CrossRef]
- Lam, N.V.; Chen, W.; Suruga, K.; Nishimura, N.; Goda, T.; Yokogoshi, H. Enhancing effect of taurine on CYP7A1 mRNA expression in Hep G2 cells. Amino Acids 2006, 30, 43–48. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Up to date on cholesterol 7 alpha-hydroxylase (CYP7A1) in bile acid synthesis. Liver Res. 2020, 4, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Yamajuku, D.; Inagaki, T.; Haruma, T.; Okubo, S.; Kataoka, Y.; Kobayashi, S.; Ikegami, K.; Laurent, T.; Kojima, T.; Noutomi, K.; et al. Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci. Rep. 2012, 2, 439. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; Nozawa, K.; Hitomi, Y.; Kakinuma, A. Laminin-rich extracellular matrix maintains high level of hepatocyte nuclear factor 4 in rat hepatocyte culture. Biochem. Biophys. Res. Commun. 1995, 212, 800–805. [Google Scholar] [CrossRef]
- Nagaoka, S.; Kamuro, H.; Oda, H.; Yoshida, A. Effects of polychlorinated biphenyls on cholesterol and ascorbic acid metabolism in primary cultured rat hepatocytes. Biochem. Pharmacol. 1991, 41, 1259–1261. [Google Scholar] [CrossRef]
- Nakagiri, R.; Oda, H.; Kamiya, T. Small scale rat hepatocyte primary culture with applications for screening hepatoprotective substances. Biosci. Biotechnol. Biochem. 2003, 67, 1629–1635. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Lipson, K.E.; Baserga, R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl. Acad. Sci. USA 1989, 86, 9774–9777. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Armengol, J.; Fajas, L.; Lopez-Mejia, I.C. Inter-organ communication: A gatekeeper for metabolic health. EMBO Rep. 2019, 20, e47903. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.M.; Defina, L.F.; Leonard, D.; Barlow, C.E.; Radford, N.B.; Willis, B.L.; Rohatgi, A.; McGuire, D.K.; De Lemos, J.A.; Grundy, S.M.; et al. Long-term association of low-density lipoprotein cholesterol with cardiovascular mortality in individuals at low 10-year risk of atherosclerotic cardiovascular disease: Results from the cooper center longitudinal study. Circulation 2018, 138, 2315–2325. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Nishimura, N.; Oda, H.; Yokogoshi, H. Effect of taurine on cholesterol degradation and bile acid pool in rats fed a high-cholesterol diet. In Taurine 5; Lombardini, J.B., Schaffer, S.W., Azuma, J., Eds.; Springer: New York, NY, USA, 2003; Volume 526, pp. 261–267. [Google Scholar]
- Nagai, K.; Fukuno, S.; Oda, A.; Konishi, H. Protective effects of taurine on doxorubicin-induced acute hepatotoxicity through suppression of oxidative stress and apoptotic responses. Anti-Cancer Drugs 2016, 27, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Islambulchilar, M.; Asvadi, I.; Sanaat, Z.; Esfahani, A.; Sattari, M. Effect of taurine on attenuating chemotherapy-induced adverse effects in acute lymphoblastic leukemia. J. Cancer Res. Ther. 2015, 11, 426. [Google Scholar] [PubMed]
- Alam, S.S.; Hafiz, N.A.; Abd El-Rahim, A.H. Protective role of taurine against genotoxic damage in mice treated with methotrexate and tamoxfine. Environ. Toxicol. Pharmacol. 2011, 31, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004, 40, 539–551. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.M.; Granner, D.K. PEPCK gene as model of inhibitory effects of insulin on gene transcription. Diabetes Care 1990, 13, 327–339. [Google Scholar] [CrossRef]
- Burgess, S.C.; He, T.; Yan, Z.; Lindner, J.; Sherry, A.D.; Malloy, C.R.; Browning, J.D.; Magnuson, M.A. Cytosolic Phosphoenolpyruvate carboxykinase does not solely control the rate of hepatic gluconeogenesis in the intact mouse liver. Cell Metab. 2007, 5, 313–320. [Google Scholar] [CrossRef]
- Kumar, A.; Schwab, M.; Laborit Labrada, B.; Silveira, M.A.D.; Goudreault, M.; Fournier, É.; Bellmann, K.; Beauchemin, N.; Gingras, A.-C.; Bilodeau, S.; et al. SHP-1 phosphatase acts as a coactivator of PCK1 transcription to control gluconeogenesis. J. Biol. Chem. 2023, 299, 105164. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hsu, D.D.F.; Li, B.; Luo, X.; Alderson, N.; Qiao, L.; Ma, L.; Zhu, H.H.; He, Z.; Suino-Powell, K.; et al. Cytoplasmic tyrosine phosphatase SHP2 coordinates hepatic regulation of bile acid and FGF15/19 signaling to repress bile acid synthesis. Cell Metab. 2014, 20, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Da Costa, K.A.; Lee, S.; Renga, B.; Jaeschke, H.; Yang, Z.; Orena, S.J.; Goedken, M.J.; Zhang, Y.; Kong, B.; et al. Interactions between nuclear receptor SHP and FOXA1 maintain oscillatory homocysteine homeostasis in mice. Gastroenterology 2015, 148, 1012–1023.e14. [Google Scholar] [CrossRef]
- Hagenbuch, B.; Stieger, B. The SLCO (former SLC21) superfamily of transporters. Mol. Asp. Med. 2013, 34, 396–412. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Xiao, Z.; Kanamaluru, D.; Min, G.; Yau, P.M.; Veenstra, T.D.; Ellis, E.; Strom, S.; Suino-Powell, K.; Xu, H.E.; et al. Bile acid signaling pathways increase stability of small heterodimer partner (SHP) by inhibiting ubiquitin–proteasomal degradation. Genes Dev. 2009, 23, 986–996. [Google Scholar] [CrossRef]
- Demetz, E.; Schroll, A.; Auer, K.; Heim, C.; Patsch, J.R.; Eller, P.; Theurl, M.; Theurl, I.; Theurl, M.; Seifert, M.; et al. The arachidonic acid metabolome serves as a conserved regulator of cholesterol metabolism. Cell Metab. 2014, 20, 787–798. [Google Scholar] [CrossRef]
- McCully, K.S. Homocysteine and the pathogenesis of atherosclerosis. Expert Rev. Clin. Pharmacol. 2015, 8, 211–219. [Google Scholar] [CrossRef]
- Ji, C.; Shinohara, M.; Kuhlenkamp, J.; Chan, C.; Kaplowitz, N. Mechanisms of protection by the betaine-homocysteine methyltransferase/betaine system in HepG2 cells and primary mouse hepatocytes. Hepatology 2007, 46, 1586–1596. [Google Scholar] [CrossRef]
- Martínez-Uña, M.; Varela-Rey, M.; Mestre, D.; Fernández-Ares, L.; Fresnedo, O.; Fernandez-Ramos, D.; Juan, V.G.; Martin-Guerrero, I.; García-Orad, A.; Luka, Z.; et al. S-Adenosylmethionine increases circulating very-low density lipoprotein clearance in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 673–681. [Google Scholar] [CrossRef]
- Rome, F.I.; Hughey, C.C. Disrupted liver oxidative metabolism in glycine n-methyltransferase-deficient mice is mitigated by dietary methionine restriction. Mol. Metab. 2022, 58, 101452. [Google Scholar] [CrossRef]
- Zhang, N. Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Anim. Nutr. 2018, 4, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Matye, D.; Nguyen, N.; Zhang, Y.; Li, T. HNF4α regulates CSAD to couple hepatic taurine production to bile acid synthesis in mice. Gene Expr. 2018, 18, 187–196. [Google Scholar] [CrossRef] [PubMed]


| Ingredient (g/kg Diet) | Control (C) | Taurine (CT) | High Cholesterol (H) | High Cholesterol Plus Taurine (HT) |
|---|---|---|---|---|
| Casein 1 | 200 | 200 | 200 | 200 |
| Vitamin mixture 2 | 10 | 10 | 10 | 10 |
| Mineral mixture 3 | 35 | 35 | 35 | 35 |
| Choline chloride | 2 | 2 | 2 | 2 |
| Corn oil 4 | 50 | 50 | 50 | 50 |
| Cellulose | 50 | 50 | 50 | 50 |
| Starch | 435 | 402 | 427 | 394 |
| Sucrose | 218 | 201 | 213.5 | 196.5 |
| Cholesterol | - | - | 10 | 10 |
| Sodium cholate | - | - | 2.5 | 2.5 |
| Taurine | - | 50 | - | 50 |
| Functional Category 2 | p-Value |
|---|---|
| Regulated by taurine | |
| Drug Metabolism | 3.42 × 10−6 |
| Endocrine System Development and Function | 3.42 × 10−6 |
| Lipid Metabolism | 3.42 × 10−6 |
| Small Molecule Biochemistry | 3.42 × 10−6 |
| Molecular Transport | 9.54 × 10−3 |
| Cellular Function and Maintenance | 1.52 × 10−2 |
| Cell Death | 3.02 × 10−2 |
| Carbohydrate Metabolism | 4.50 × 10−2 |
| Hepatic System Disease | 4.50 × 10−2 |
| Upregulated by taurine 3 | |
| Drug Metabolism | 6.59 × 10−7 |
| Endocrine System Development and Function | 6.59 × 10−7 |
| Lipid Metabolism | 6.59 × 10−7 |
| Small Molecule Biochemistry | 6.59 × 10−7 |
| Molecular Transport | 3.32 × 10−3 |
| Hepatic System Disease | 2.64 × 10−2 |
| Cancer | 3.50 × 10−2 |
| Gastrointestinal Disease | 3.50 × 10−2 |
| Downregulated by taurine 3 | |
| Cellular Function and Maintenance | 6.36 × 10−3 |
| Cell Death | 1.27 × 10−2 |
| Carbohydrate Metabolism | 1.90 × 10−2 |
| Molecular Transport | 1.90 × 10−2 |
| Category 2 | p-Value | Ratio (%) |
|---|---|---|
| Metabolism of Xenobiotics by Cytochrome P450 | 7.76 × 10−9 | 15.3 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 2.34 × 10−8 | 9.79 |
| Tryptophan Metabolism | 9.55 × 10−7 | 11.0 |
| Arachidonic Acid Metabolism | 2.04 × 10−6 | 11.7 |
| Linoleic Acid Metabolism | 1.15 × 10−5 | 13.5 |
| PXR/RXR Activation | 1.91 × 10−5 | 12.5 |
| Fatty Acid Metabolism | 3.63 × 10−5 | 9.52 |
| Acute Phase Response Signaling | 1.45 × 10−4 | 6.92 |
| Glycine, Serine, and Threonine Metabolism | 3.98 × 10−4 | 11.9 |
| Xenobiotic Metabolism Signaling | 1.29 × 10−3 | 5.14 |
| NRF2-Mediated Oxidative Stress Response | 2.24 × 10−3 | 5.83 |
| Methionine Metabolism | 2.75 × 10−3 | 15.8 |
| Glutathione Metabolism | 3.31 × 10−3 | 9.76 |
| Lysine Biosynthesis | 6.03 × 10−3 | 25.0 |
| Complement System | 6.17 × 10−3 | 12.0 |
| Coagulation System | 8.51 × 10−3 | 10.7 |
| Hepatic Cholestasis | 1.51 × 10−2 | 5.21 |
| Sulfur Metabolism | 1.62 × 10−2 | 15.4 |
| Pantothenate and CoA Biosynthesis | 1.62 × 10−2 | 15.4 |
| Aryl Hydrocarbon Receptor Signaling | 1.86 × 10−2 | 4.95 |
| Inositol Metabolism | 2.69 × 10−2 | 6.98 |
| Biosynthesis of Steroids | 3.02 × 10−2 | 11.1 |
| Insulin Receptor Signaling | 3.31 × 10−2 | 5.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Q.; Kobayashi, S.; Kataoka, Y.; Oda, H. Direct Molecular Action of Taurine on Hepatic Gene Expression Associated with the Amelioration of Hypercholesterolemia in Rats. Antioxidants 2024, 13, 990. https://doi.org/10.3390/antiox13080990
Song Q, Kobayashi S, Kataoka Y, Oda H. Direct Molecular Action of Taurine on Hepatic Gene Expression Associated with the Amelioration of Hypercholesterolemia in Rats. Antioxidants. 2024; 13(8):990. https://doi.org/10.3390/antiox13080990
Chicago/Turabian StyleSong, Qi, Satoru Kobayashi, Yutaro Kataoka, and Hiroaki Oda. 2024. "Direct Molecular Action of Taurine on Hepatic Gene Expression Associated with the Amelioration of Hypercholesterolemia in Rats" Antioxidants 13, no. 8: 990. https://doi.org/10.3390/antiox13080990




